Abstract
Following mutations in BRAF and NRAS, the RAC1 c.85C>T single nucleotide variant (SNV) encoding P29S amino acid change represents the next most frequently observed protein-coding hotspot mutation in melanoma. However, the biological and clinical significance of the RAC1 P29S somatic mutation in approximately 4–9% of patients remains unclear. Here, we demonstrate that melanoma cell lines possessing the RAC1 hotspot variant are resistant to RAF inhibitors (vemurafenib and dabrafenib). Enforced expression of RAC1 P29S in sensitive BRAF mutant melanoma cell lines confers resistance manifested by increased viability, decreased apoptosis and enhanced tumor growth in vivo upon treatment with RAF inhibitors. Conversely, RNAi mediated silencing of endogenous RAC1 P29S in a melanoma cell line with a co-occurring BRAF V600 mutation increased sensitivity to vemurafenib and dabrafenib. Our results suggest RAC1 P29S status may offer a predictive biomarker for RAF inhibitor resistance in melanoma patients, where it should be evaluated clinically.
INTRODUCTION
Hotspot mutations in BRAF and NRAS are well-established driver mutations in the MAPK pathway (RAF-MEK-ERK signal transduction cascade), which occurs in over 50% and 20% of melanomas, respectively (1). The identification of oncogenic mutations in BRAF, predominantly at codon 600 (2), was the main driving force in the development of small molecule inhibitors targeting MAPK kinases (MEK) and BRAF in melanoma, which includes vemurafenib and dabrafenib. Patients with BRAF mutant melanomas treated with RAF and MEK inhibitors have significant improvement in progression-free and overall survival as single agents (3–6). Patient survival is further improved with the use of combination treatment of RAF and MEK inhibitors (7). However, most patients treated with vemurafenib and dabrafenib develop disease progression within 6–8 months (reviewed in (8, 9)). In addition, some patients present with intrinsic resistance (often termed de novo) that do not respond at all or have short-lived responses to RAF inhibitors progressing on drug treatment in less than 12 weeks.
Recently, two large-scale whole exome sequencing (WES) studies have revealed that in addition to recurrent mutations in BRAF and NRAS, the next most frequently observed somatic protein-coding hotspot mutation in melanoma of cutaneous origin is the RAC1 c.85C>T single-nucleotide variant (SNV) encoding the P29S amino acid change found in approximately 4% to 9% of melanomas (10, 11). RAC1 is a member of the Rho family of small GTP-binding proteins whose activity has been implicated in tumorigenesis and metastasis (12). RAC1 transduces extracellular signals from growth factor, integrin, and G-protein-coupled receptors, and its best-characterized function is in the regulation of cytoskeleton rearrangement (13). Although the RAC1 P29S mutation has been shown to be oncogenic and biochemically active (10, 11, 14), the biological role and clinical relevance of the RAC1 hotspot mutation in melanoma remains unclear. Recent publications on genomic characterization of matched pre- and post-treated melanoma samples from patients who received RAF and MEK inhibitors (15–19) have confirmed many of the acquired resistance mechanisms identified in preclinical in vitro studies (20–29). In one such study, the authors observed a significant correlation of the RAC1 P29S hotspot mutation and early resistance in patients who received vemurafenib or dabrafenib monotherapy (17). Here, we present functional data supporting a role for the RAC1 P29S mutation in resistance to RAF inhibitors by maintaining activation of the MAPK signaling pathway. Our results suggest that RAC1 P29S status in melanoma may be an important predictor for vemurafenib and dabrafenib resistance in patients, and consequently should be evaluated as a predictive biomarker to such therapies in the clinic.
MATERIALS AND METHODS
Sequenom and Sanger sequencing
gDNA was isolated using DNeasy Blood & Tissue Kit (Qiagen). Mass spectrometric genotyping (Sequenom) was performed as previously described (10, 30). Sequenom was employed to test all cell lines used in this study for RAC1, BRAF, and RAS family member hostpot mutations. Confirmation of the RAC1 c.85C>T SNV encoding for amino acid change P29S was validated by amplifying exon 2 with forward (TGCTAACACCGGGTACCTAAAC) and reverse (TCATCCAGTCTCTGTACCTCAC) primers. PCR products were purified by QIAquick Gel Extraction Kit (Qiagen) followed by bidirectional sequencing with forward (TTTTAACTTAATAGTGAAAGCTA) and reverse (TGGTCAAAGAAATGTGAAAC) primers on ABI 3730 DNA sequencers using Big Dye terminator cycle sequencing chemistry.
Plasmids and shRNA
pDONR RAC1 plasmid was obtained from the hORFeome collection from the Center for Cancer Systems Biology (CCSB) at Dana-Farber Cancer Institute. The c.85C>T RAC1 mutation encoding for amino acid change P29S was generated using Quick-Change Lightning Site-Directed Mutagenesis (Stratagene) according to the manufacturer's protocol. Sub-cloning was performed by Invitrogen Gateway® Technology to a pLENTI6.3-CMV (Invitrogen) and pHAGE-EF1α-IRES GFP expression vector that was kindly provided by Dr. Simona Colla (The University of Texas MD Anderson Cancer Center, Houston, TX). Inducible shRNA RAC1 and control constructs were generated using BLOCK-iT™ Inducible H1 RNAi Entry Vector Kit (Invitrogen). Sub-cloning was performed by Invitrogen Gateway® Technology to a PLKO-Tet-On vector a gift from Dr. Timothy P. Heffernan (The University of Texas MD Anderson Cancer Center, Houston, TX). The hairpin sequences were as follows: shGFP: ACAACAGCCACAACGTCTAT CGAA ATAGACGTTGTGGCTGTTG shRAC1 71: CGCAAACAGATGTGTTCTTAA CGAA TTAAGAACACATCTGTTTGCG; shRAC1 72: CGTGAAGAAGAGGAAGAGAAA CGAA TTTCTCTTCCTCTTCTTCACG. shLuciferease PLKO-Tet-On plasmid was gift from T.P. Heffernan. Lentiviral transduction was essentially performed as previously described (31).
Cell Culture and Cell Viability Assays
A375, MALME-3M, 451Lu, IGR1, CP66, and HMVII melanoma cell lines were maintained in RPMI medium 1640 (Gibco, Life Technologies) and WM3060 cells in Leibovitz's L-15 medium (Gibco, Life Technologies) in 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Life Technologies) at 37°C in a humidified 5% CO2 incubator. Cell lines were authenticated by STR DNA fingerprinting (32) (STR profiles available in Supplementary Table 4). Stably expressing DOX-inducible shRNA cells were cultured in Tet System Approved FBS (Clontech). DOX treated cells were cultured in media at a concentration of 0.4 μg/mL. CellTiter-Glo® Luminescent Cell Viability Assays (Promega) were used to measure viability following cell treatment with dabrafenib (GSK21118436), vemurafenib (PLX4032), trametinib (GSK1120212) and MEK inhibitor (PD325901) treatment (Selleck Chemicals). Briefly, 5000 cells were seeded in 96 well plates in triplicates and 12h later treated with drug with indicated concentrations for 72–96h. % Cell viability calculated by comparison to DMSO no treatment control. Analysis and IC50s calculated by GraphPad Prism 6 software.
Immunoblots and RAC1 Activation Assay
Cells growing in monolayers were lysed using Cell Extraction Buffer (Life Technologies) supplemented with complete protease inhibitors and PhosSTOP phosphatase inhibitor cocktail tablets (Roche). Cell lysates were cleared by centrifugation, protein concentrations were determined by DC Protein Assay (BioRad), and denatured lysates were run on 4–12% Bis-Tris gradient gels (Invitrogen). Gels were transferred to nitrocellulose membranes (BioRad) before being immunoblotted with indicated antibodies. Cleaved-PARP antibody was obtained from Cell Signaling Technology. RAC1 activation assays were performed as previously described according to the manufacturer's protocol (Cell Biolabs) (10).
Xenograft assays
4–6 week old NCR-nude female mice were obtained from Taconic farms. A375 isogenic cell lines overexpressing GFP, RAC1 wild-type and P29S mutant were re-suspended in a solution made up of 2/3 Hank's Balanced Salt Solution (Life Technologies) and 1/3 BD matrigel matrix (BD biosciences). 10 million cells were injected in 100 ul volume in 15 mice per group (GFP, RAC1 WT, and the P29S mutant) and were monitored for tumor formation. An approximate 100–250 mm3 tumor volume was observed post 1 week injection, and all mice were given PLX4720-admixed chow (AIN-76A diet) with a dose of 417 mg / kg diet (Plexxikon and Research Diets Inc.). Body weight and chow was measured to ensure no significant differences in mouse size or intake between groups. Tumor volumes were measured bi-weekly and calculated by the following formula: Volume = 0.5 × (length × (width)2). Survival endpoints were defined by first time point tumor reached 15mm in length or visible evidence of ulceration. Mann-Whitney nonparametric tests were used to determine significance differences in tumor volume. Log-rank (Mantel-Cox) test was used to determine significant differences in survival. Two-tailed Fisher's exact test used for comparison of mouse cohorts that progressed or responded to PLX-4720 treatment. Tumor growth inhibition (TGI) for GFP, WT, and P29S groups were calculated by taking sum of tumor volumes at day 0 / sum of tumor volumes on day 19 on PLX4720 treatment. All mouse experiments were performed with the approval of Institute Institutional Animal Care and Use Committee at MD Anderson.
RESULTS
RAC1 P29S Mutant Melanoma Cell Lines Have Higher Levels of Activated RAC1 and Display Differential Sensitivity to RAF and MEK Inhibitors
To determine the biological role of the RAC1 c.85C>T SNV encoding for the P29S amino acid change, we probed the RAC1 P29S variant status in a panel of melanoma cell lines by mass spectrometry genotyping (sequenom) (10). We identified two melanoma cell lines with SNV encoding for S29 (IGR1 and WM3060), which were confirmed by PCR and Sanger re-sequencing (Supplementary Figure 1). We documented by sequenom genotyping and Sanger re-sequencing that IGR1 cells harbor a co-occurring BRAF c.1798_1799GT>AA dinucleotide variant (DNV) encoding the V600K amino acid change, whereas the WM3060 melanoma possesses a NRAS Q61K mutation (data not shown).
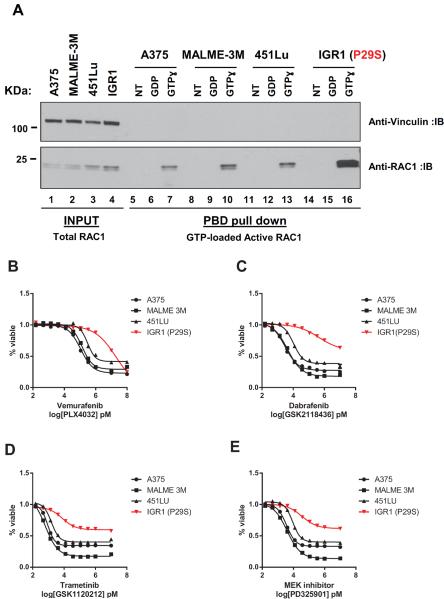
It has been shown that RAC1 P29S is found in a higher active GTP-loaded fraction compared to the wild-type protein (10, 11, 33) as measured by RAC1 interaction with the p21-binding domain (PBD) of p21-activated protein kinase (PAK) (34). We demonstrate that endogenous RAC1 is found in a higher active fraction in PBD-binding assays in cell lines with P29S compared with ones with wild-type (Figure 1A, compare lanes 7, 10, 13, and 16; and Supplementary Figure 2A), confirming that melanoma cell lines with P29S possess higher levels of activated RAC1.
Figure 1. RAC1 P29S mutant melanoma cell lines have higher levels of activated RAC1.
A) Active RAC1 protein levels measured by PAK p21 binding domain (PBD) pull downs in the presence of GDP or a nonhydrolysable GTP analog (GTPɣ) from IGR1 cell lines possessing c.C85T SNV encoding for the P29S amino acid change compared to 3 cells lines lacking this variant (A375, MALME-3M, 451Lu). Log-transformed dose response curves (%viability compared to DMSO treated control) for cell lines treated with RAF inhibitors B) vemurafenib (PLX4032) C) dabrafenib (GSK21118436) and MEK inhibitors D) trametinib (GSK1120212) E) PD325901 for 72h with doses up to 100 000 and 10 000, and serial dilutions of 1 000, 333.33, 111.11, 37.04, 12.35, 4.12, 1.37, 0.46, 0.15 nM. X axis is presented as log transformed drug doses in pM concentrations. Error bars represent standard error means.
To investigate the functional role of RAC1 P29S variant in melanoma, we treated IGR1 and WM3060 cell lines with inhibitors targeting RAF, MEK, PI3K, and mTOR pathways. One observation we noted consistently was that P29S expressing cell lines were less sensitive to both RAF inhibitors (vemurafenib and dabrafenib) and MEK inhibitors (trametinib and PD325901). IGR1 cells had approximately 40 – 185 fold higher IC50s (half maximal inhibitory concentrations) in response to RAF inhibitors and a 4 – 12 fold increase in IC50s in response to MEK inhibitors when compared to RAC1 wild-type BRAF V600E mutant cell lines, A375, MALME-3M and 451Lu (Figure 1B–E and Supplementary Table 1). Furthermore, IGR1 cells did not reach below 50% cell viability even at doses as high as 10μM in response to vemurafenib, dabrafenib, trametinib and PD325901 following 72h treatment (Supplementary Table 1). Similarly, the NRAS Q61K mutant WM3060 cell line with the P29S variant was less sensitive to MEK inhibitor treatment compared to two other NRAS Q61K mutant melanoma cell lines, CP66 and HMVII (Supplementary Figure 2 and Supplementary Table 2). These results suggested that RAC1 P29S modulates sensitivity to inhibitors of the MAPK pathway in melanoma.
RAC1 P29S Mediates Resistance to RAF and MEK Inhibition
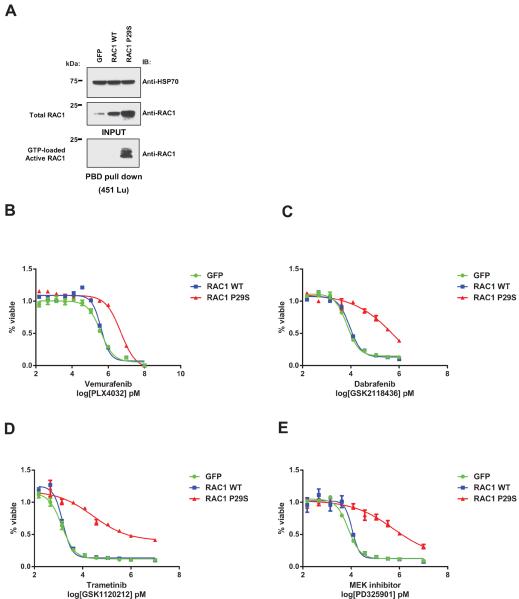
To assess whether RAC1 P29S promotes resistance to RAF and MEK inhibition, we stably infected 451Lu cells with GFP, RAC1 wild-type (WT), and P29S expressing pLENTI6.3-CMV plasmids (Figure 2A). We observed a 13 – 145 fold increase in IC50 of RAC1 P29S mutant expressing cells compared to GFP controls in response to vemurafenib and dabrafenib, and an 19 – 74 fold increase in IC50s following trametinib and PD325901 treatment (Figure 2B–E and Supplementary Table 3). This effect was also observed in isogenic 451Lu cells stably expressing RAC1 P29S and controls from a pHAGE-EF1α plasmid possessing a weaker promoter (Supplementary Figure 3 and Supplementary Table 3). Another independent human melanoma cell line, A375, showed a 3.5 – 7 fold increase in IC50s in response to RAF and MEK inhibitors in presence of P29S mutation (Supplementary Figure 4 and Supplementary Table 3) and similar results were observed in MALME-3M isogenic cell lines (Supplementary Figure 5).
Figure 2. Overexpression of RAC1 P29S increases cell viability to RAF inhibitor treatment.
A) Immunoblot of 451Lu cells stably expressing GFP, RAC1 wild-type (WT) and P29S mutant from pLENTI6.3-CMV plasmids showing active GTP-loaded RAC1 from PBD pull down with corresponding input lysates. Log-transformed dose response curves (%viability compared to DMSO treated control) for 451Lu isogenic cell lines treated with RAF inhibitors B) vemurafenib (PLX4032) C) dabrafenib (GSK21118436) and MEK inhibitors D) trametinib (GSK1120212) E) PD325901 for 96h with doses up to 100 000 and 10 000 nM, and serial dilutions of 1 000, 333.33, 111.11, 37.04, 12.35, 4.12, 1.37, 0.46, 0.15 nM. X axis is presented as log transformed drug doses in pM concentrations. Error bars represent standard error means.
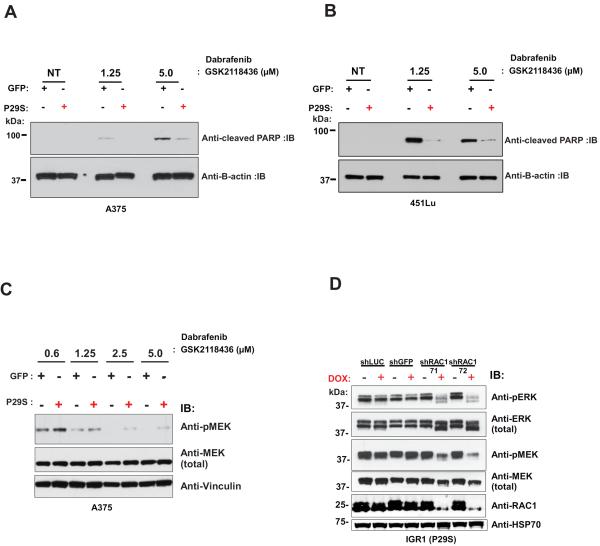
We next examined the apoptotic response to RAF inhibition in our isogenic cell lines by immunoblotting for cleaved-PARP in lysates from GFP and RAC1 P29S overexpressing A375 and 451Lu cells treated with dabrafenib. Consistent with our viability assays, we observed reduced apoptosis in response to RAF inhibition in cell lines overexpressing P29S (Figure 3A and B). Similar results were observed in MALME-3M cell lines overexpressing P29S (Supplementary Figure 5D). In addition, P29S expressing cells maintained elevated MAPK signaling activity as measured by phospho-MAP2K1/MAP2K2 (MEK1/2) S217/S221 levels in response to RAF inhibitor treatment compared to GFP controls (Figure 3C).
Figure 3. RAC1 P29S expression decreases apoptosis and maintains higher levels of MAPK signaling to RAF inhibitor treatment.
A) Immunoblots of cleaved PARP and β-Actin from lysates of A375 and B) 451Lu cells stably expressing GFP and RAC1 P29S treated with 1.25 and 5.0 μM of dabrafenib (GSK2118436). C) Phospho-MEK1/2 S217/S221 and total MEK immunoblots from lysates of A375 cells following dabrafenib (GSK21118436) treatment with indicated doses for 8h are shown. D) Immmunoblot of phospho-ERK1/2 T202/Y204, phospho-MEK1/2 S217/S221 and RAC1 from lysates of stably expressing doxycycline (DOX)-inducible RAC1 shRNA IGR1 cells following 72h of DOX treatment.
Complementing the gain-of-function studies above, we generated IGR1 cells stably expressing doxycycline (DOX)-inducible RAC1 shRNAs. Upon administration of DOX for 72h we observed a decrease in RAC1 protein levels with multiple independent shRNAs compared to two controls (shGFP and shLuciferase) (Fig. 3D and Supplementary Fig. S6A). Along with knockdown of RAC1 expression, we observed concomitant decrease in MAPK signaling as measured by decreased levels of phospho-MEK1/2 S217/S221 and phospho-MAPK3/MAPK1 (ERK1/2) T202/Y204 (Figure 3D). Accordingly, a consistent trend of increased sensitivity to RAF and MEK inhibitor treatment was also observed in IGR1 cells upon RAC1 knockdown (Supplementary Figure 6B–E). Taken together, these in vitro biochemical and drug sensitivity studies suggest that the P29S mutant of RAC1 can confer resistance to RAF and MEK inhibitors by maintaining an elevated level of MAPK activity.
RAC1 P29S Decreases Effect of RAF Inhibitor on Tumor Growth in vivo
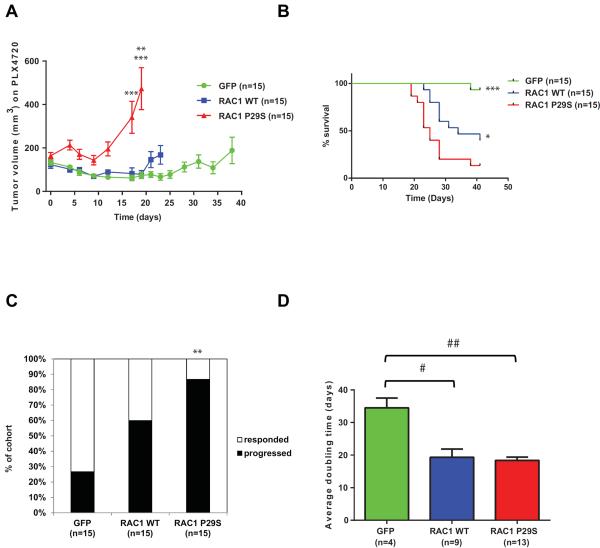
To evaluate how the RAC1 P29S mutant modulates the response to RAF inhibitors in vivo, we implanted the A375 isogenic cells described above in NCR-nude mice. When an approximate 100–250 mm3 tumor volume was observed, mice were switched to chow admixed with PLX4720, a preclinical RAF inhibitor (35). This formulation has previously been shown to deliver high and stable serum concentration through course of treatment in mice (36). As shown in Figure 4A, wild-type and GFP controls showed initial regression followed by stable disease over a 19 day course of PLX4720 treatment (tumor growth inhibition (TGI) of GFP group=55%; WT group=64%; p=not significant). In contrast, RAC1 P29S mutant expressing tumors continued to grow with accelerated rate on treatment (TGI=280%), achieving significantly higher tumor volumes at day 19 (Mann-Whitney test: p<0.005 (GFP vs P29S), p<0.0005 (WT vs P29S)) (Figure 4A and Supplementary Figure 7) when the first mouse in P29S cohort reached experimental endpoint (determined by reaching maximal tumor burden). Mice implanted with A375 cells expressing the RAC1 P29S mutants had a worse overall survival compared to GFP controls on RAF inhibitor treatment (Log-rank (Mantel-Cox) test: p<0.0005 (GFP vs P29S), p<0.05 (WT vs P29S)) (Figure 4B). Four GFP expressing tumors eventually progressed on treatment (determined by criteria of reaching tumor volume twice the size of original volume prior to drug administration); however, only 1 mouse was sacrificed for reaching maximal tumor burden at the end of the experiment on day 41 compared to 13/15 from the P29S mutant cohort (Figure 4B and C, Fisher's exact test: p<0.005 (GFP vs P29S)). An intermediate response was observed for overexpressing RAC1 wild-type tumors, as 9/15 tumors reached maximum tumor burden by day 41 (Log-rank (Mantel-Cox) test: p<0.005 (GFP vs. WT)) (Figure 4B). Furthermore, of the tumors that did progress, GFP controls tumors had an almost twice as long doubling time on PLX4720 treatment (34.5+/− 6.0 days) compared to wild-type (19.33+/−7.6 days, p<0.01) and to P29S mutant overexpressing tumors (18.38+/−3.6 days, p<0.001) (Figure 4D).
Figure 4. RAC1 P29S decreases effect of RAF inhibitor treatment on tumor growth in vivo.
A375 cells expressing GFP, RAC1 wild-type (WT), and RAC1 P29S were injected subcutaneously in flanks of female NCR-Nude mice. An approximate 100–250 mm3 tumor volume was observed post 1 week injection for all three mouse cohorts, and all mice were given PLX4720-admixed chow with a dose of 417 mg / kg diet. A) Tumor volumes measurements in the presence of PLX4720 drug are shown (15 mice in each cohort) up until time point first mouse within cohort reached survival endpoint (GFP: day 38, RAC1 WT: day 23, RAC1 P29S: day 19. Day 17, Mann-Whitney test: *** p<0.0005 (GFP vs P29S), p<0.0005 (WT vs P29S). Day 19, Mann-Whitney test: **p<0.005 (GFP vs P29S), ***p<0.0005 (WT vs P29S)). B) Kaplan–Meier survival curves for the GFP, RAC1 WT and P29S cohorts are shown (Log-rank (Mantel-Cox) test: ***p<0.0005 (GFP vs P29S), *p<0.05 (WT vs P29S), not indicated on graph, p<0.005 (GFP vs WT)). Survival endpoints were maximal one-sided tumor measurement of 15mm or ulceration. C) % of cohort which progressed on PLX4720 treatment as determined by reaching twice the original tumor volume prior to drug administration (Fisher's exact test: **p<0.005 (GFP vs P29S)). D) Doubling times for mice that progressed on treatment in each cohort are shown (Mann-Whitney test: #p<0.01 (GFP vs WT), ##p<0.001 (GFP vs P29S), not significant (WT vs P29S)). Error bars represent standard error means. *p<0.05, **p<0.005, ***p<0.0005. #p<0.01, ##p<0.001.
DISCUSSION
In this study, we identified melanoma cell lines possessing the RAC1 P29S variant and demonstrated that RAC1 is present in a higher GTP-active fraction with cell lines of similar genotype. We should note, although some melanoma oncogenes and tumor suppressors are known to be involved in familial melanoma, the RAC1 P29S somatic mutation is caused by presumptive UVB-induced DNA damage and misrepair (C>T transition) (10, 11), and has not been detected as a germline mutation in the 1000 Genomes databases or among over 2,500 germline exomes sequenced at Yale (11). Interestingly, cell lines expressing RAC1 P29S are more resistant to RAF and MEK inhibitor treatment. Overexpression of the P29S mutant in BRAF mutant melanoma cell lines conferred resistance to MAPK inhibitors in vitro. Knockdown of endogenous P29S RAC1 mutant had an additive effect in decreasing cell viability in response to RAF inhibitors. Finally, RAC1 P29S overexpression in the RAF inhibitor sensitive cell line, A375, resulted in a decreased effect of the drug on tumor growth in vivo. Taken together, these functional and biochemical studies support RAC1 P29S as a RAF inhibitor resistance mutation in melanoma.
Our conclusion is supported by clinical data from a recent study of 45 patients that received vemurafenib and dabrafenib monotherapy where pre- and post-treatment BRAF V600-mutant metastatic samples underwent WES analysis (17). Of the 45 patient data examined, 14 acquired resistance early in the course of treatment. Interestingly, the three patients in this cohort of 45 with RAC1 P29S mutation were all found to belong to this early resistance subgroup. The P29S mutation was not found in any melanoma sample from patients that had a sustained response to therapy in this study (the enrichment in the early resistant group was found to be statistically significant). Together, this clinical correlation in context of our functional results would suggest that RAC1 P29S status in melanoma may not only confer vemurafenib and dabrafenib resistance, but may predict those patients with intrinsic resistance to this class of targeted therapy. However, we acknowledge that an adequately powered study in the clinic will be necessary to definitively demonstrate the utility of RAC1 hotspot mutation in predicting intrinsic resistance to RAF inhibitors.
Mechanistically, our studies suggest that RAC1 P29S may sustain MAPK signaling in the presence of RAF inhibitors. Phospho-MEK1/2 levels were elevated in A375 isogenic cell lines overexpressing RAC1 P29S in response to dabrafenib, and we observed levels of phospho-MEK1/2 S217/S221 and phospho-ERK1/2 T202/Y204 decreased following RAC1 knockdown of endogenous RAC1 P29S in IGR1 cells. This is consistent with previous studies implicating RAC1 in the regulation of MAPK signaling (reviewed in (12)), and a recent study demonstrating RAC1 P29S can induce ERK phosphorylation in melanocytes (11). A number of studies have identified RAF inhibitor resistance mechanisms, which can be broadly characterized into groups that include reactivation of the MAPK pathway, upregulation of the PI3K-PTEN-AKT pathway, and dysregulation of melanocytic signaling (15–29). We cannot exclude the possibility that there may be alternative mechanisms that RAC1 P29S can drive RAF inhibitor resistance, including upstream or parallel to the MAPK pathway. Investigation of the aberrant oncogenic signaling caused by the RAC1 hotspot mutation in melanoma, and potential combination therapies to target this mutation, remains an important area of investigation for future study.
Supplementary Material
Acknowledgements
This work was supported by Cancer Prevention Research Institute of Texas, National Institutes of Health (grant number 5 U01 CA168394 02, PI: Mills) and National Cancer Institute (NCI) (grant Number U24CA143845, PI: Chin), and in part by the Cancer Center Support Grant (CCSG) at The University of Texas MD Anderson Cancer Center. Lynda Chin is a CPRIT Scholar in Cancer Research and is supported by CPRIT funding (R1204). Ian R. Watson is a recipient of the Canadian Institutes of Health Research Fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NCI or the US National Institutes of Health. We would like to thank Eran Hodis, Robert Onofrio, and Levi Garraway from the Broad Institute for assistance with the sequenom assay. We thank Simona Colla, Mary K. Geck Do, Tim P. Heffernan, Trang N. Tieu, Ningping Feng, Jason Gay, Jennifer Greer and Carlo Toniatti from the Institute for Applied Cancer Science and Department of Genomic Medicine at The University of Texas MD Anderson Cancer Center for cloning assistance and help with cell viability, drug and mouse studies.
Footnotes
Conflict of Interest Disclosure Statement: The authors declare no conflict of interest directly related to this study.
REFERENCES
- 1.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes & development. 2012;26:1131–55. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–14. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 5.Hauschild A, Grob JJ, Demidov LV, Jouary T, Gutzmer R, Millward M, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–65. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 6.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–14. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med. 2012;367:1694–703. doi: 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. European journal of cancer. 2013;49:1297–304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Bucheit AD, Davies MA. Emerging insights into resistance to BRAF inhibitors in melanoma. Biochemical pharmacology. 2014;87:381–9. doi: 10.1016/j.bcp.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 10.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–14. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wertheimer E, Gutierrez-Uzquiza A, Rosemblit C, Lopez-Haber C, Sosa MS, Kazanietz MG. Rac signaling in breast cancer: a tale of GEFs and GAPs. Cellular signalling. 2012;24:353–62. doi: 10.1016/j.cellsig.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annual review of cell and developmental biology. 2005;21:247–69. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 14.Kawazu M, Ueno T, Kontani K, Ogita Y, Ando M, Fukumura K, et al. Transforming mutations of RAC guanosine triphosphatases in human cancers. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3029–34. doi: 10.1073/pnas.1216141110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, Hong A, Kong X, Koya RC, Song C, Moriceau G, et al. A Novel AKT1 Mutant Amplifies an Adaptive Melanoma Response to BRAF Inhibition. Cancer discovery. 2014;4:69–79. doi: 10.1158/2159-8290.CD-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer discovery. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, et al. The Genetic Landscape of Clinical Resistance to RAF Inhibition in Metastatic Melanoma. Cancer discovery. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP Kinase Pathway Alterations in BRAF-Mutant Melanoma Patients with Acquired Resistance to Combined RAF/MEK Inhibition. Cancer discovery. 2014;4:61–8. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer discovery. 2012;2:414–24. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–72. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johannessen CM, Johnson LA, Piccioni F, Townes A, Frederick DT, Donahue MK, et al. A melanocyte lineage program confers resistance to MAP kinase pathway inhibition. Nature. 2013;504:138–42. doi: 10.1038/nature12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–4. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montagut C, Sharma SV, Shioda T, McDermott U, Ulman M, Ulkus LE, et al. Elevated CRAF as a potential mechanism of acquired resistance to BRAF inhibition in melanoma. Cancer research. 2008;68:4853–61. doi: 10.1158/0008-5472.CAN-07-6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–7. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–90. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker SR, Theurillat JP, Van Allen E, Wagle N, Hsiao J, Cowley GS, et al. A genome-scale RNA interference screen implicates NF1 loss in resistance to RAF inhibition. Cancer discovery. 2013;3:350–62. doi: 10.1158/2159-8290.CD-12-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antony R, Emery CM, Sawyer AM, Garraway LA. C-RAF mutations confer resistance to RAF inhibitors. Cancer research. 2013;73:4840–51. doi: 10.1158/0008-5472.CAN-12-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paraiso KH, Xiang Y, Rebecca VW, Abel EV, Chen YA, Munko AC, et al. PTEN loss confers BRAF inhibitor resistance to melanoma cells through the suppression of BIM expression. Cancer research. 2011;71:2750–60. doi: 10.1158/0008-5472.CAN-10-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quayle SN, Chheda MG, Shukla SA, Wiedemeyer R, Tamayo P, Dewan RW, et al. Integrative functional genomics identifies RINT1 as a novel GBM oncogene. Neuro-oncology. 2012;14:1325–31. doi: 10.1093/neuonc/nos246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drexler HG, Dirks WG, Matsuo Y, MacLeod RA. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–26. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- 33.Davis MJ, Ha BH, Holman EC, Halaban R, Schlessinger J, Boggon TJ. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:912–7. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. The Journal of biological chemistry. 1999;274:13198–204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 35.Tsai J, Lee JT, Wang W, Zhang J, Cho H, Mamo S, et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3041–6. doi: 10.1073/pnas.0711741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakravarty D, Santos E, Ryder M, Knauf JA, Liao XH, West BL, et al. Small-molecule MAPK inhibitors restore radioiodine incorporation in mouse thyroid cancers with conditional BRAF activation. The Journal of clinical investigation. 2011;121:4700–11. doi: 10.1172/JCI46382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.