Abstract
Selenoprotein K (SelK) is a membrane protein involved in antioxidant defense, calcium regulation and the ER-associated protein degradation pathway. We found that SelK exhibits a peroxidase activity with a rate that is low but within the range of other peroxidases. Notably, SelK reduced hydrophobic substrates, such as phospholipid hydroperoxides, which damage membranes. Thus, SelK might be involved in membrane repair or related pathways. SelK was also found to contain a diselenide bond — the first intramolecular bond of that kind reported for a selenoprotein. The redox potential of SelK was −257 mV, significantly higher than that of diselenide bonds in small molecules or proteins. Consequently, SelK can be reduced by thioredoxin reductase. These finding are essential for understanding SelK activity and function.
1. Introduction
Selenoprotein K (SelK) is a membrane protein linked to development, immunity [1], longevity [2,3], and susceptibility for prostate cancer [4], but its precise function is unknown [1,5]. It is a member of a eukaryotic membrane protein family that contributes to the management of oxidative stress [5], that is the detoxification of reactive oxygen and nitrogen species that can harm cellular components [6]. Members of the family are single-span transmembrane proteins with intrinsically disordered C-terminal domains. SelK is a selenoprotein, a class of enzymes that employ the rare amino acid selenocysteine (Sec, U) [7]. It is localized to the endoplasmic reticulum (ER) membrane, and the Sec (at position 92 of 94 amino acids, scheme 1) is exposed to the cytoplasm [8]. Selenoproteins are often found either to combat reactive oxygen species (ROS) directly or to regulate the cellular response to ROS-related stress [9]. SelK itself has an antioxidant function since it can mitigate the toxicity of ROS both in vitro [10] and in live cells [11,12].
In addition to its antioxidant function, or perhaps as part of this function, SelK is likely to engage in a signaling pathway. Many bitopic proteins, membrane proteins that transverse the bilayer once, are involved in signaling [13], and SelK is particularly rich in Pro (15%) and positively charged residues, a composition common in signaling proteins [14]. Furthermore, it contains at least one, possibly two, SH3 recognition sequences that identify it as a probable signaling protein. Recent reports tied SelK to several binding partners, including palmitoyl acyl transferase ZDHHC6 [15] and components of the ER-associated degradation (ERAD) pathway, such as Derlin-1, p97 ATPase, and selenoprotein S (SelS, VIMP) [5,16,17]. Finally, SelK mediates Ca+2 flux in immune cells and is the target of calpain proteases in myeloid cells, a class of proteases involved in regulation of inflammation and immune response [1,18].
In the search for SelK’s biological function, it is necessary to reconcile two aspects. The majority of selenoproteins are enzymes that operate independently [19]; hence, the presence of Sec suggests an enzymatic function. However, SelK is an intrinsically disordered protein. Such proteins typically function in regulation and signaling but rarely as enzymes [20,21]. Enzymatic catalysis is typically associated with a well-organized active site in which the degrees of motion are tightly controlled. Nevertheless, folding intermediates of several enzymes were shown to still catalyze their reactions though at lower efficiency, and a few intrinsically disordered proteins were also shown to function in the absence of a protein partner [21,22]. Indeed, SelK’s protein partner SelS, which is also disordered, is an efficient reductase even in isolation [23]. In contrast to SelS, however, SelK’s Sec is not in a recognizable redox motif. In most selenoproteins, a vicinal cysteine typically forms a selenenylsulfide bond with the Sec. In SelK the sole Sec is not in a recognizable pattern.
Here we examine whether SelK has similar oxidoreductase activity as that displayed by other selenoproteins: an oxidase, a reductase, an isomerase, or a peroxidase function. Since the in vivo system responsible for acting as an electron donor to SelK is unknown, we tested the two most abundant cytoplasmic systems: thioredoxin reductase/thioredoxin (TrxR/Trx) and glutathione reductase/glutaredoxin (GR/Grx) [24] for their ability to reduce SelK. In addition SelK redox potential was characterized. Finally, we measured the rate in which it can interact with hydroperoxides in isolation.
2. Materials and Methods
2.1 Bacterial Strains, Plasmids, and Chemical Reagents
The Homo sapiens SelK (UniProtKB Q9Y6D0) construct used in this study was modified from the SelK-E expression vector described in our previous publication [25]. SelK-E contains a maltose binding protein (MBP) fused to SelK with a Tabaco Etch Virus (TEV) protease cleavage site (ENLYFQG) between the two proteins to release SelK. Here, to assist in purification, an eight amino acid StrepII tag (WSHPQFEK) was inserted between the TEV protease cleavage site and SelK. The first Met of SelK was deleted to reflect the fact that it is proteolytically cleaved in vivo. Following cleavage of the fusion protein by TEV protease, SelK retains in its N-terminal a GWSHPQFEK peptide. Site-directed mutagenesis was used to prepare mutants of this construct. All other sources of bacterial strains, plasmids, chemical reagents and enzymes were as previously described [25,26], with the addition of 5,5-dimethyl-1,3-cyclohexanedione (dimedone), 5-methyl-2-oxo-4-imidazolidinehexanoic acid (desthiobiotin), sodium deoxycholate, N-ethylmaleimide (NEM), Glycine max lipoxidase, yeast glutathione reductase, 2-linoleoyl-1-palmitoyl-sn-glycero-3-phosphocholine (PLPC) supplied by Sigma-Aldrich and fluorescein-5-maleimide supplied by Life Technologies. Homo sapiens thioredoxin 1, thioredoxin reductase and glutaredoxin were prepared in house [26]. The expression construct for human glutaredoxin 1 (hGrx) was a gift from J. J. Mieyal [27] and that for human thioredoxin 1 (hTrx) was provided by M. A. Marletta [28].
2.2. Expression and purification of SelK and its mutants
SelK U92 was prepared by incorporating Sec into position 92 of a SelK U92C mutant using the cysteine auxotrophic E. coli host cell BL21(DE3) selB::kan cys51E [29]. The protocol was similar to that developed by the Böck group with minor modifications [30]. Cells were grown in modified LB media (10 g tryptone, 5 g NaCI, 5 g yeast extract, 2 g glucose per liter), at 37 °C, with good aeration and the relevant antibiotic selection (100 μg/ml ampicillin + 25 μg/ml kanamycin), and 50 μg/ml L-cysteine. When the Optical Density (OD) at 600 nm reached 1.0, the cells were harvested by centrifugation at 4 °C, 4000g for 10 min. The pellet was washed in sterile buffer (20 mM phosphate, 50 mM NaCI (pH 7.5)) that was chilled on ice, and centrifuged once again. The pellet was resuspended in Studier’s MDAG defined media [31] supplemented with 100 μg/ml ampicillin and 25 μg/ml kanamycin. The volume for the media was the same as the volume of the modified LB media used at the beginning. The cells were allowed to shake at 37 °C for an additional 30 min to consume residual cysteine. The temperature was lowered to 18 °C, and the cells were allowed to shake at the lower temperature for an additional 30 min. Protein expression was induced with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG), and 200 μM L-selenocystine was added to the growth media. Accumulation of SelK in the cells stalled after 10 h at 18 °C; hence, the cells were harvested after 14–16 h. The pellet was resuspended in 50 mM sodium phosphate (pH 7.5), 200 mM NaCI, 1 mM ethylenediaminetetraacetic acid (EDTA), 0.5% Triton X-100, 1 mM benziamidine, and 1 mM phenylmethylsulfonyl fluoride (PMSF) (amylose Buffer); frozen in liquid nitrogen and stored at −80 °C. Protein expression of SelK U92C and SelK U92S mutants was carried out with LB media as previously described [25].
Protein purification of SelK U92, SelK U92C, and SelK U92S was as formerly detailed with the additional step of Strep-Tactin affinity chromatography [25]. In short, cells were defrosted and sonicated in amylose buffer. Cell debris was removed by centrifugation at 20000g for 1 h. The supernatant was loaded on an amylose column, which was washed with the amylose buffer for 5 column volumes (CV) followed by a wash with 50 mM sodium phosphate (pH 7.5), 200 mM NaCI, 1 mM EDTA, and 0.067% n-dodecyl-b-D-maltopyranoside (DDM) (exchange buffer) for 5 CV. The fusion cMBP-SelK was eluted using 5 CV exchange buffer supplemented with 20 mM maltose. Cleavage of the fusion partner, MBP, was carried out by adding a hexahistidine-tagged TEV protease to the dialysis bag overnight at 4 °C. The TEV protease was added at a molar ratio of 1:10 relative to the fusion MBP-SelK protein. Following cleavage, TEV and MBP were removed using immobilized metal ion affinity chromatography. The protein was then loaded on a 5 ml Strep Trap HP (GE Healthcare) equilibrated with exchange buffer. SelK was eluted in the exchange buffer, with a linear gradient of 0–2.5 mM desthiobiotin. The fractions containing SelK were collected, concentrated to 2 mg/mL, loaded onto a sephacryl 200 (GE Healthcare), and eluted with exchange buffer. Protein purity was analyzed using sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and coomassie blue staining. The protein concentrations of SelK U92, SelK U92C, and SelK U92S were determined using an extinction coefficient of 20970 M−1 cm−1 or by bicinchoninic acid (BCA) protein assay. The free thiol/selenol count of SelK U92 and SelK U92C was determined using Ellman’s reagent 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) [32]. The extent of selenium incorporation in a given protein sample was measured by electrospray mass spectrometry as described below. When values were compared to measurements by Inductively Coupled Plasma (ICP) spectroscopy, the agreement was within a factor of 3. We believe that in this specific case mass spectrometry more accurately reflects the actual percent incorporation because ICP also reported sulfur from residual TEV protease. TEV protease contains several Met residues and, hence, can bias the ratio of sulfur to selenium. The incorporation ratio as judged from mass spectrometry varied from 80% to 90% between batches. We refer to this form as SelK U92 (see section 3.1). The concentrations given in the paper are those of the selenium containing SelK in a given preparation.
As a side note, SelK was prone to proteolytic cleavage. Inclusion of protease inhibitors, metals, or EDTA did not reduce the extent of cleavage. The origin of the proteolytic cleavage is being studied. In this paper we employed only protein from fractions of size exclusion chromatography that contained minimal amounts or no truncated forms.
2.3. Electrospray Ionization Mass Spectrometry
Mass spectra were obtained using a quadrupole time-of-flight mass spectrometer QTOF Ultima (Waters), operating under positive electrospray ionization (+ESI) mode and connected to an LC-20AD (Shimadzu). Protein samples were separated from small molecules by reverse phase chromatography on a C4 column (Waters XBridge BEH300), using an acetonitrile gradient from 30–71.4%, with 0.1% formic acid as the mobile phase, in 25 min, at a flow rate of 0.2 ml/min at room temperature. Data were acquired from m/z 350 to 2500 at a rate of 1 sec/scan. The obtained spectra were deconvoluted using maximum entropy in MassLynx (Waters).
2.4. Determination of Redox Potentials
The redox potential of SelK U92 and SelK U92C were determined by redox titrations with the reduced and oxidized forms of DTT. 25 μL SelK U92 (34 μM) was added to aliquots of 25 μL redox buffer containing 100 mM potassium phosphate (pH 7.0), 1 mM EDTA, 0.067% DDM, and various amounts of reduced and oxidized DTT (to a total DTT concentration of 2 mM). SelK U92 runs as a dimer on SDS-PAGE when oxidized and as a monomer when reduced. The reaction of SelK with DTTred/DTTox redox pair (Reaction I) and the corresponding equilibrium constant (Keq), as well as the redox potential (E) of the various DTT buffers are given as follows:
| Reaction 1 |
| Eq. 1 |
| Eq. 2 |
where E0 (DTTred/DTTox)= −327 mV at pH 7.0 and 298 K [33], R is the gas constant (8.315 J K−1 mol−1), T is the absolute temperature (298 K), and n=2. The reaction mixture was equilibrated at 25 °C for 2 h after which the reaction was quenched by adding ice-cold 100% W/V trichloroacetic acid to a final concentration of 20%. The quenched reaction mixture was then spun for 10 min at 16110g. The supernatant was decanted, and the pellet was washed with 0.25 ml ice-cold acetone twice and spun at 16110 g for 10 min at 4 °C after each wash. After the final acetone wash, the pellet was dried by exposing it to air for 10 min and re-suspended in sample loading buffer (50 mM Tris-HCl (pH 6.8), 2% SDS, 12.5 mM EDTA, 10% glycerol) containing 10 mM NEM and analyzed by 16% Tris-glycine SDS-PAGE. Densitometry of coomassie-stained non-reducing gels was performed using ImageJ. The fraction of reduced SelK to oxidized SelK was calculated based on the band intensity (where the dimer’s band intensity was halved to obtain the fraction of the dimer). The midpoint redox potential of SelK U92 was calculated by fitting the known redox potential of the buffer and the measured ratio of reduced SelK U92 to oxidized SelK U92. The successful alkylating of SelK U92 was also validated using fluorescein-5-maleimide.
The redox potential measurement of SelK U92C was similar to that described above but with reduced and oxidized glutathione with a total concentration of 2 mM where E0 (GSH/GSSG) = −240 mV [34].
2.5. Preparation of 1-palmitoyl-2-(13-hydroperoxy-cis-9,trans-11-octadecadienoyl) phosphatidylcholine (PLPCOOH)
PLPCOOH was prepared from PLPC by oxidation with lipoxidase following a published procedure [35,36]. A 500 mL incubation mixture containing 3 mM sodium deoxycholate, 0.13 mM PLPC, 0.1 mg/mL soybean lipoxidase and 0.2 M Tris-HCl buffer (pH 8.8) was continuously stirred aerobically at ambient temperature for 4 h. PLPCOOH was extracted from the reaction mixture using a separatory funnel with ethyl acetate. The PLPCOOH in ethyl acetate was dried by rotary evaporation, redissolved in 5 mL methanol and then purified using reverse phase chromatography on a C18 column (Haisil 100, 5 μm, 250 × 4.6 mm) with a mobile phase of acetonitrile/methanol/water (75:21:4). The purified PLPCOOH was dried, redissolved in methanol, and stored at −20 °C. The concentrations of hydroperoxides were determined using an extinction coefficient of 25000 M−1 cm−1 at 232 nm. The identity of PLPCOOH was confirmed by mass spectrometry.
2.6. Peroxidase Activity Assays
Peroxidase activity was measured via a coupled reaction with human thioredoxin reductase/human thioredoxin (TrxR/Trx) or yeast glutathione reductase/human glutaredoxin 1 (GR/Grx) as previously described [37]. The reaction mixture contained 100 mM potassium phosphate (pH 7.0), 0.067% DDM, 2 mM EDTA, 150 μM NADPH, and 5 μM SelK. For the TrxR/Trx assays, the reaction also included 8 nM hTrxR and 5 μM hTrx. For the GR/Grx assays, the reaction also included 1 mU/μL yGR, 1 mM GSH, and 5 μM hGrx. The temperature was kept constant at 25 °C. The reaction mixture was incubated for three min, and the reaction was initiated by adding 200 μM H2O2 to the cuvette. The enzymatic oxidation of NADPH was monitored spectroscopically by recording its consumption at 340 nm.
To test the effect of detergents on SelK peroxidase activity, the DDM detergent was exchanged by superdex 200 (GE Healthcare) to the following detergents (all at 10 the critical micelle concentration (CMC)): 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) (5%), Triton X-100 (0.1%), and 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine (LMPC) (0.017%). The peroxidase activity was measured using the GR/Grx assays: 100 mM potassium phosphate (pH 7.0), 2 mM EDTA, 150 μM NADPH, 1 mU/μL yGR, 1 mM GSH, 5 μM hGrx, and 5 μM SelK in either 0.067% DDM, 0.017% LMPC, 0.1% Triton X-100, or 5% CHAPS. The reaction was triggered by adding 200 μM H2O2 or 60 μM 15(S)-hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic acid (HpETE) to the cuvette. The initial NADPH oxidation rate was calculated by monitoring the absorbance at 340 nm.
To determine the substrate specificity of SelK’s peroxidase activity, a variety of potential peroxide substrates were tested in GR/Grx reaction mixture with 0.067% DDM as described above. The reaction was triggered by addition of 60 μM H2O2, cumene hydroperoxide (COOH), tert-Butyl hydroperoxide (tBuOOH), HpETE, or PLPCOOH respectively. HpETE and PLPCOOH were dissolved in the buffer used for the activity assays. All experiments were repeated three times, and the size of the error bar is one standard deviation. The P value in Figs. 4 and 5 was calculated using a one-way analysis of variance (ANOVA) where the four proteins were compared. The p-value calculated this way gives the probability that the individual samples taken from the different conditions tested against each other do actually not show any difference and belong instead to the same population, i.e. the observed difference is a pure statistical artifact.
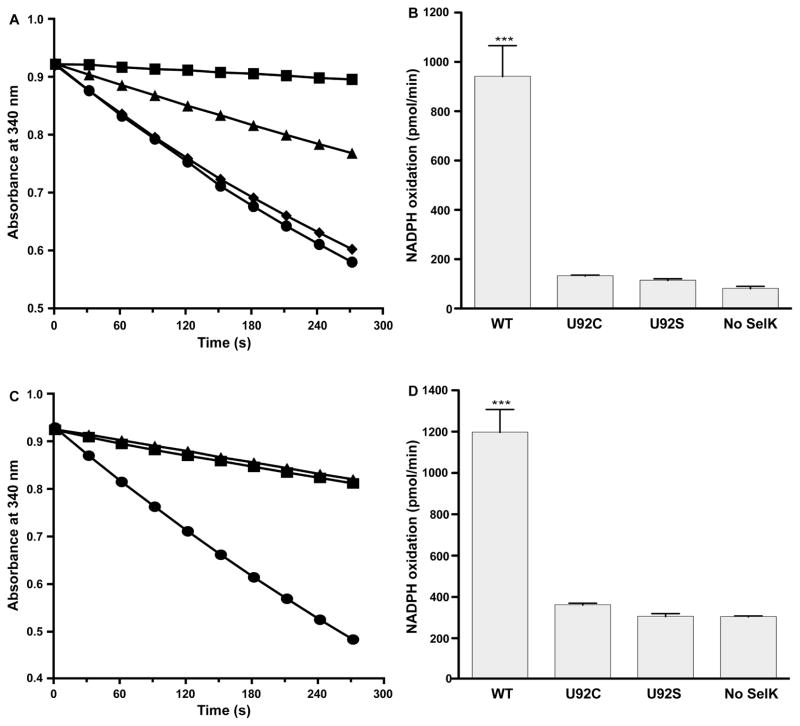
Fig. 4.
SelK peroxidase activity assays. (A) NADPH consumption is monitored in the presence of 8 or 64 nM hTrxR, with and without 5 μM hTrx, 200 μM H2O2, and 5 μM of SelK U92. The reaction with 5 μM SelK U92, 5 μM hTrx and 8 nM hTrxR is shown in circles, with 5 μM SelK U92 and 64 nM hTrxR is shown in diamonds, with 5 μM SelK U92 and 8 nM hTrxR in triangles, and with 5 μM hTrx and 8 nM hTrxR but in the absence of SelK in squares. (B) The activity of different mutants was compared using the TrxR/Trx system: 5 μM SelK U92 (U92), 5 μM SelK U92C (U92C), 5 μM SelK U92S (U92S) and in the absence of SelK (No SelK). (C) NADPH consumption is monitored in the presence of 1 mU/μL yGR, 5 μM hGrx, 1 mM GSH, with or without 5 μM of SelK U92. The reaction with both 5 μM SelK and 5 μM hGrx is shown in circles, in the absence of SelK in squares, and in the absence of hGrx in triangles. (D) Same as B, but activity was tested using the GR/Grx system. In either system, SelK peroxidase activity depends on the presence of Sec (***, P<0.001, n=3).
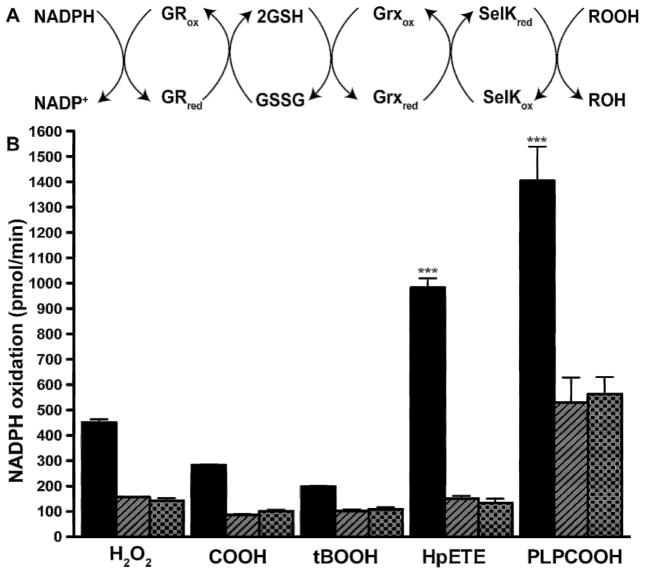
Fig. 5.
Substrate specificity of SelK’s peroxidase activity. (A) Reaction scheme. (B) NADPH for the reaction with SelK and hGrx (black bars), the reaction excluding hGrx (striped bars), and the reaction excluding SelK (dotted bars). Of the five substrates tested, HpETE and PLPCOOH have the highest reaction rates (***, p<0.001, n=3).
2.7. Steady State Kinetics
Steady-state kinetic analysis was performed using the GR/Grx peroxidase assay described above. Briefly, the assays were carried out with varying concentrations of H2O2 (0 to 600 μM), PLPCOOH (0 to 60 μM), and hGrx (0 to 5 μM). The reaction was initiated by the addition of H2O2 or PLPC-OOH. The rate of the initial NADPH oxidation was measured by monitoring the absorbance at 340 nm. The reaction mixture without SelK served as a blank. Double reciprocal plots were prepared by plotting [E0]/V0 versus 1/[S] and fitted to the transformed Lineweaver-Burk equation (Eq. 3). The plots were used to determine apparent kinetic parameters Km, kcat. Each condition was repeated three times, and the size of the error bar is one standard deviation.
| Eq. 3 |
2.8. Dimedone Inhibition Assay and Trapping of Selenenic Acid
GR/Grx peroxidase activity assays were carried out in 100 mM potassium phosphate (pH 7.0), 0.067% DDM, 2 mM EDTA, 150 μM NADPH, 1 mU/μL yGR, 1 mM GSH, 5 μM hGrx with or without 5 μM SelK, and with 0, 0.5, 1, 2, 5, 10 mM dimedone. The reaction was initiated with 200 μM H2O2. The rate of the initial NADPH oxidation was measured by monitoring the absorbance at 340 nm. After 10 min, the reaction mixture was subjected to analysis by mass spectrometry. Experiments were repeated three times, and the size of the error bar is one standard deviation.
3. Results
3.1. Sec incorporation into SelK
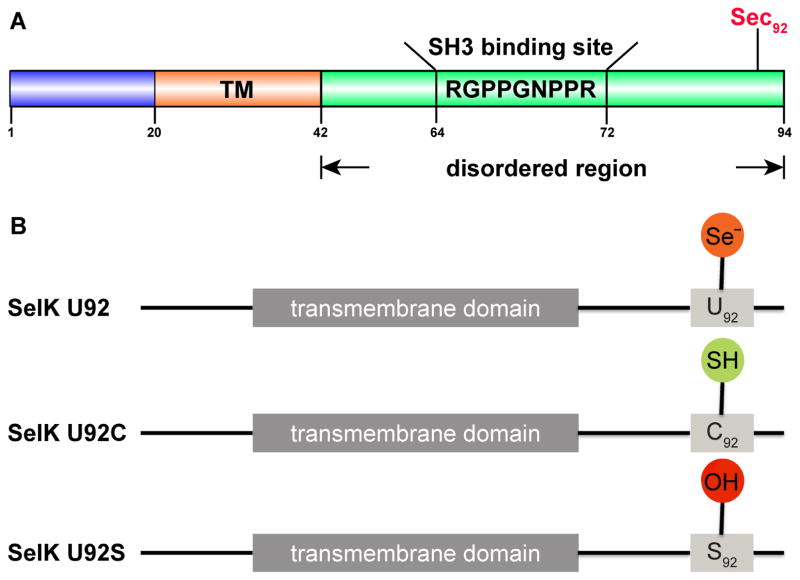
Sec is the catalytic residue in selenoproteins, and its presence is essential for studies of their enzymatic activity. Incorporation of Sec into proteins can be carried out by several methods [38], but the most frequently employed approach is heterologous expression in E. coli that harnesses its dedicated selenium incorporation machinery [39]. However, as Sec is encoded by a UGA codon [40], which is also a stop codon, this approach leads not only to the desired selenoprotein but also to its truncated form [41]. The truncated form in our preparations still dimerized with the full-length form and cannot be separated. Since that may lead to structural and functional changes, preparation methods based on genetic incorporation of Sec are not a viable procedure for SelK. In its place Sec was incorporated by an alternative method in which Cys in the protein of interest are substituted for Sec when an E. coli cysteine auxotroph strain is grown on media supplemented with Sec [29,30]. Since Cys and Sec share similar physiochemical properties, Sec will be misloaded onto the cysteyl-tRNA and, thus, will get incorporated into proteins as Sec moiety. SelK has no native Cys residues, which makes it possible to use a SelK U92C mutant to incorporate Sec at position 92 (Fig. 1A) and obtain the Sec-containing SelK. Since it is impossible to prevent Cys recycling in the cell the sample does contain a small percent of the Cys-containing SelK. However, it is possible to characterize the Cys-containing protein separately to isolate its potential contribution to enzymatic activity. The dominant form in our samples is the homodimer wild-type SelK (Fig. 2). We label this selenium-rich preparation of the enzyme SelK U92 (Fig. 1B).
Fig. 1.
Schematic representation of SelK as well as the mutants used in this study. (A) SelK’s domain organization. In blue, the N terminal domain is located in the ER lumen; In purple, the hydrophobic segment formed by residues 20–42 is predicted to contain a single transmembrane helix (TM); In green, the intrinsically disordered domain facing the cytoplasm. This domain contains an SH3 binding site as well as the catalytic selenocysteine (Sec). The location of the Sec is shown in red. SelK has no additional cysteines or selenocysteines besides the one outlined. (B) Proteins used in this study. SelK U92 was prepared by misloading of cysteyl-tRNA with Sec by a cysteine auxotrophic strain and a media with Sec. About 10% of this sample is SelK U92C generated from remaining pools of Cys in the cell. SelK U92C and SelK U92S were prepared by conventional heterologous expression and are purely the mutant form.
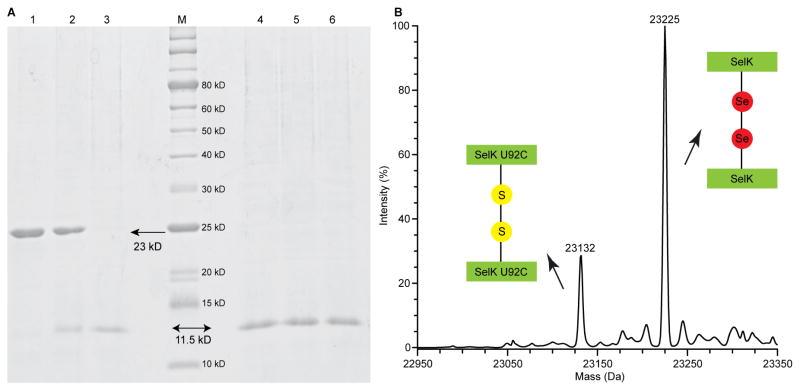
Fig. 2.
SelK contains an intermolecular diselenide bond. (A) SDS-PAGE analysis of SelK, SelK U92C, and SelK U92S run under reducing and non-reducing conditions. Lanes 1–3 were run under non-reducing conditions: lane 1, SelK; lane 2, SelK U92C; lane 3, SelK U92S; lane M, protein molecular mass standards (the molecular mass is noted on the right of the corresponding band). Lanes 4–6 were run under reducing conditions: lane 4, SelK; lane 5, SelK U92C; lane 6, SelK U92S. The arrows point to the monomer and dimer forms of SelK. (B) SelK ionizes as a dimer in electrospray ionization mass spectrum. Molecular masses: 23132 Da for homodimer SelK U92C, and 23225 Da for homodimer SelK.
3.2 SelK forms an intermolecular diselenide bond
In DDM micelles, SelK U92 was at minimum a homodimer containing an intermolecular diselenide bond. As can be observed from a SDS-PAGE run under non-reducing conditions (Fig. 2A), SelK U92 migrated as a dimer of 23 kDa, SelK U92C migrated mostly as a dimer, and SelK U92S migrated mostly as a monomer. All three proteins migrated as a monomer of 11.5 kDa under reducing conditions. A thiol count using Ellman’s reagent confirmed that SelK U92 contained no exposed thiols/selenols. Furthermore, when examined by electrospray mass spectrometry, SelK U92 ionized as a dimer under non-reducing conditions (Fig. 2B). The dimerization is not exclusively dependent on the presence of a diselenide bond since SelK U92S, which has no intermolecular bond, can run as both a monomer and dimer on denaturing SDS-PAGE when its concentration is sufficiently high (Supplementary data, Fig. S1). Bitopic proteins’ oligomerization is frequently driven by the transmembrane segment, and the majority are non-covalent oligomers [42].
3.3. SelK’s redox potential
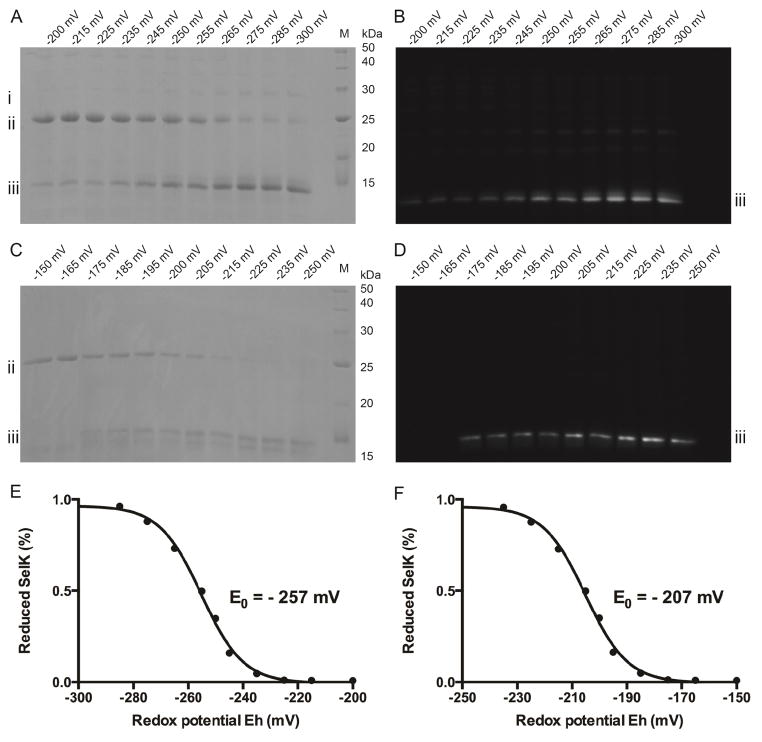
Numerous bitopic membrane proteins change oligomerization states depending on the protein functional state. Therefore, SelK is likely to engage in self-interactions as well as interactions with protein partners to fulfill its role. This suggests that the intermolecular diselenide bond is physiologically relevant and that a cytoplasmic protein is capable of reducing it. To assess which proteins can potentially reduce SelK, we measured the redox potential of SelK by gel shift assays using maleimide alkylation of U92. A fluorescein tagged maleimide was employed to validate that the protein is indeed alkylated.
SelK U92 was equilibrated in buffers with known redox potentials, the reaction was quenched with acid and SelK U92 was then alkylated with fluorescein-5-maleimide and analyzed by SDS-PAGE. It was feasible to measure the ratio of oxidized and reduced (and subsequently alkylated) proteins by SDS-PAGE analysis since the alkylated protein run as a monomer while the oxidized protein run as a dimer (Fig. 3A). Furthermore, only the alkylated protein will be fluorescent (Fig. 3B). The ratios were subsequently used to determine the redox potentials of the Sec- and Cys-containing SelK using the Nernst equation (see material and methods). Using this assay the redox potential of SelK U92 was determined to be −257 mV. We have also validated the results with alkylation with both N-ethylmaleimide (NEM) and iodoacetamide, IAA. The results were independent of the alkylating reagent. Note that a small amount of TEV forms an oligomer with SelK that dissociated as the redox potential is lowered. Also, due to the method employed for Sec incorporation, a small percent of the protein in these samples is the Cys-containing protein (in Fig. 3A and 3B the percent of the Cys-containing SelK is less than 5%). However, because SelK U92C redox potential is −207 mV (Fig. 3C and 3D), all of it is reduced at redox buffers used to determine the redox potential of the SelK U92. Hence, it does not bias the measurements.
Fig. 3.
The redox potential of SelK’s intermolecular diselenide bond is −257 mV. Gel shift assays based on alkylation with fluorescein-5-maleimide. The proteins were separated by SDS-PAGE. The protein bands are labeled to the right of each image to simplify identification. M denotes molecular weight markers (given in kDa); i TEV protease (bands with higher molecular weights are complexes of TEV protease with SelK); ii oxidized SelK present as a dimer; iii alkylated SelK present as a monomer (bands with lower molecular weights are truncated SelK generated by an E. coli protease). (A) Percent of reduced and oxidized wild-type SelK. Visualized by coomassie brilliant blue stain using white light imaging. (B) Alkylation pattern of wild-type SelK. Visualized by fluorescence imaging. Only alkylated protein is detected. (C) Same as A for SelK U92C. (D) Same as B for SelK U92C. (E) Wild-type SelK redox potential titration curve (reduction potential of −257 mV) and (F) SelK U92C redox potential titration curve (reduction potential of −207 ± 1 mV). The fraction of reduced protein is plotted against the buffer redox potential.
3.4. SelK can reduce phospholipids hydroperoxides
Selenoproteins possess enzymatic activity in practically all characterized cases, with the majority being oxidoreductases. Thus, we tested SelK for reductase and peroxidase activities. SelK doesn’t have disulfide reductase activity in an insulin reduction assay (Supplementary data, Fig. S2) [43]. This assay is based on the aggregation of insulin chain B following the reduction of insulin’s intermolecular disulfide bond. It is a general assay for reductase activity as DTT is included as the reducing reagent to generate SelK. SelK also does not have an isomerase or oxidase activity as expected from its redox potential. SelK did exhibit peroxidase activity as assayed by the consumption of NADPH in the presence of human thioredoxin reductase (hTrxR) and human thioredoxin 1 (hTrx) or yeast glutathione reductase (yGR) and human glutaredoxin 1 (hGrx) (Fig. 4) [37]. We have elected to test the activity using both the Trx and Grx systems as both are highly abundant in the cytoplasm and can theoretically couple with SelK. Indeed, both systems efficiently reduced SelK’s diselenide bond, allowing SelK to act as a peroxidase. hTrxR, which is known to exhibit a broad substrate selectivity [44], reduced SelK directly and does not require Trx (Fig 4A). As will be discussed later, hTrxR is the most efficient electron donor. In the GR/Grx system, the NADPH oxidation rate in the presence of SelK and yGR but without hGrx is the same as that of the control, i.e. NADPH oxidation in buffer (Fig. 4C). This implies that in the time scale of this experiment, SelK U92 is reduced more efficiently by hGrx than by GSH. Since the Grx system and hTrxR alone efficiently reduce SelK U92 (section 3.6), we use both systems interchangeably in the remaining characterization of the enzymatic activity.
In both systems, the peroxidase activity was dependent on the presence of Sec, and SelK U92C and SelK U92S mutants didn’t exhibit significant activity at pH 7.0 (Fig. 4B and 4D and Supplementary data, Fig. S3). All assays were carried out in DDM since in our previous publication we concluded that SelK behaved best in the presence of this detergent [25]. However, since the activity of membrane proteins depends on the choice of detergents, we also included studies of the effect of detergent on the activity in our assays (Supplementary data, Table S1). Among the detergents tested, SelK had a higher activity in the presence of the mild detergents DDM, Triton X-100, and the zwitterionic detergent LMPC. All experiments were carried at 10 times the critical micelle concentration (CMC) because SelK is not stable at detergents concentration close to the CMC. Activity was lower in the presence of CHAPS, which is a harsh anionic detergent. However, in all detergents tested, SelK showed only a weak peroxidase activity as will be detailed below.
3.5. Substrate specificity
Because SelK is a membrane protein, we tested whether it is able to reduce hydrophobic substrates. Among the selenium-containing peroxidases, only GPx4, GPx3, and SelP can catalyze the reduction of phospholipid hydroperoxides [45,46]. We used similar substrates as those employed for the characterization of SelP’s substrate specificity [45,46]. Those include the hydrophilic substrates hydrogen peroxide, tertiary butyl hydroperoxide (tBOOH), cumene hydroperoxide (COOH), and the hydrophobic substrates 15(S)-hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic acid (HpETE), and 2-linoleoyl-1-palmitoyl-sn-glycero-3-phosphocholine hydroperoxide (PLPCOOH). As Fig. 5 shows the hydrophobic HpETE and PLPCOOH are the preferred substrates.
3.6. Steady state kinetics
To evaluate the catalytic efficiency of SelK we have measured its reaction rate with lipid hydroperoxides. We utilized a steady-state kinetic analysis, as described by Dalziel [47], because peroxidases often exhibit a bi-substrate mechanism without an apparent saturation (see Table 1 of [48]). In this case there is no enzyme-substrate complex and Vmax and Km are infinite.
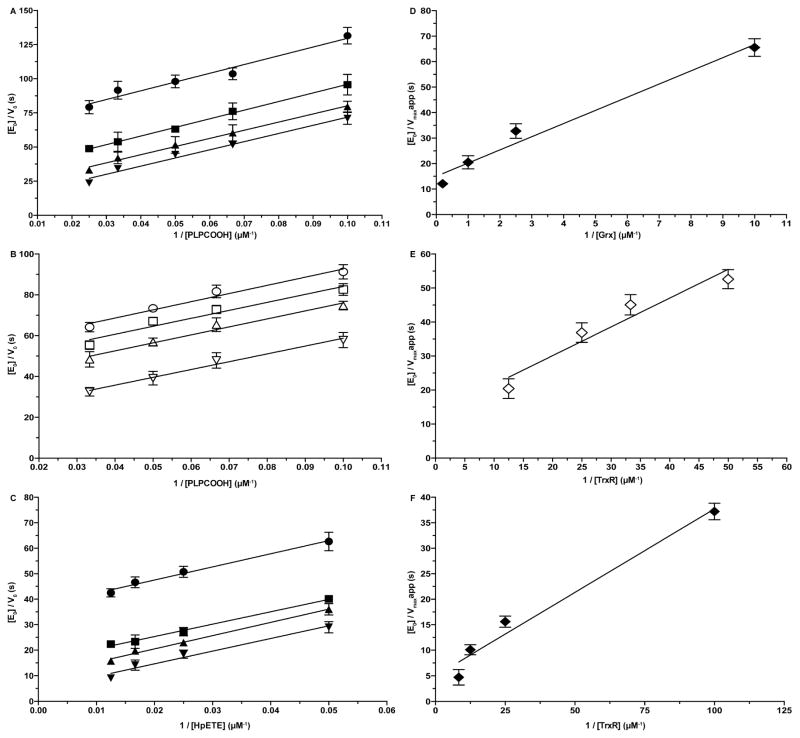
To evaluate SelK’s catalytic efficiency we have recorded the initial rate for the reduction of HpETE or PLPCOOH as a function of either the concentration of the substrate or the concentration of the reducing substrate (either Grx or TrxR). The primary Dalziel plots, which are the double reciprocal plots of the initial rate versus substrate concentration, displayed characteristic parallel lines of a ping-pong mechanism (Fig. 6A, C, and E). The reciprocal slope corresponds to the rate constant k1 for the reduction of hydroperoxides. The rate of the reaction between reduced SelK U92 and the hydrophobic substrates is on the order of 103 M−1 s−1, which is on the low range reported for lipid hydroperoxides’ peroxidases (see discussion). The kinetic parameters are listed in Table 1.
Fig. 6.
Dalziel plots for reduction of PLPCOOH catalyzed by SelK by coupling Grx/GR or TrxR reduction system. (A) [E0]/V0 vs. 1/[PLPCOOH] at various concentrations of Grx: 0.1 μM (●), 0.4 μM (■), 1 μM (▲), and 5 μM (▼). (B) [E0]/V0 vs. 1/[PLPCOOH] at various concentrations of TrxR: 20 nM (○), 30 nM (□), 40 nM (△), and 80 nM (▽). (C) [E0]/V0 vs. 1/[HpETE] at various concentrations of TrxR: 10 nM (●), 40 nM (■), 80 nM (▲), and 120 nM (▼). (D) Secondary plot of the intercepts of the primary plot A versus the reciprocal Grx concentrations. (E) Secondary plot of the intercepts of the primary plot B versus the reciprocal TrxR concentrations. (F) Secondary plot of the intercepts of the primary plot C versus the reciprocal TrxR concentrations.
Table 1.
Kinetic parameters of SelK activity with PLPCOOH as substrate
| Reducing substrate | ROOH | k1 (ROOH) (M−1 s−1) × 103 | k2 (RS) (M−1 s−1) × 106 | kcat (ROOH) (s−1) | Km ROOH (μM) | kcat (RS) (s−1) | Km (RS) (μM) |
|---|---|---|---|---|---|---|---|
| Grx | PLPCOOH | 1.624± 0.060 | 0.193± 0.021 | 0.066± 0.013 | 41.0± 8.3 | 0.066± 0.009 | 0.343± 0.028 |
| TrxR | PLPCCOH | 2.556± 0.047 | 1.18± 0.23 | 0.076± 0.031 | 30± 12 | 0.076± 0.016 | 0.0640± 0.0046 |
| HpETE | 1.984± 0.063 | 3.06± 0.36 | 0.201± 0.080 | 101± 41 | 0.201± 0.027 | 0.0660± 0.0040 |
To evaluate whether the reaction follows saturation or non-saturation kinetics, secondary Dalziel plots were employed. That is the concentration of the enzyme divided by the apparent maximal velocity is plotted against the second substrate, which was either hGrx or hTrxR. Secondary Dalziel plots of unstarurated kinetics have an intercept with the Y axis of zero. SelK displayed saturation kinetics in both Grx/GR and TrxR systems, as shown in Fig. 6B, D, and F. When the kinetics is saturated, the reciprocal intercept gives the value for the maximum velocity (kcat) and the reciprocal slope corresponds to the rate constant k2 for the reduction of SelK by Grx or TrxR. The rate constant k2 for hGrx and hTrxR is 105 M−1 s−1 and 106 M−1 s−1, respectively. This shows that both the Grx/GR system and hTrxR alone are proficient at reducing SelK, with the later being most efficient.
3.7. Trapping of SelK’s selenenic acid intermediate
The reaction mechanism of selenoperoxidases contains the reactive species selenenic acid (SeOH) [48]. We have previously shown that a selenenic acid intermediate can be trapped by the alkylating reagent 5,5-dimethyl-1,3-cyclohexanedione (dimedone) in a mutant of human SelS lacking cysteines but not in the wild-type enzyme [26]. We have also examined here, for SelK, whether a selenenic acid intermediate can be trapped in the wild type. When 10 mM dimedone was added to the peroxidase reaction mixture with 200 μM H2O2 all of SelK U92 was alkylated by dimedone as evident by a mass gain of 138 Da (Fig. 7A). Dimedone also reacts with sulfenic acids, which are expected to evolve in the SelK U92C mutant. However, due to its lower reactivity, the SelK U92C mutant was unmodified in similar incubations with 200 μM H2O2. Trapping of a sulfenic acid intermediate by dimedone in SelK U92C was detected only when incubating with 1 mM H2O2 (Supplementary data, Fig. S4). Overall, the data provides unambiguous proof that a selenenic acid intermediate is formed in SelK’s peroxidase reaction (Supplementary data, Scheme S1).
Fig. 7.
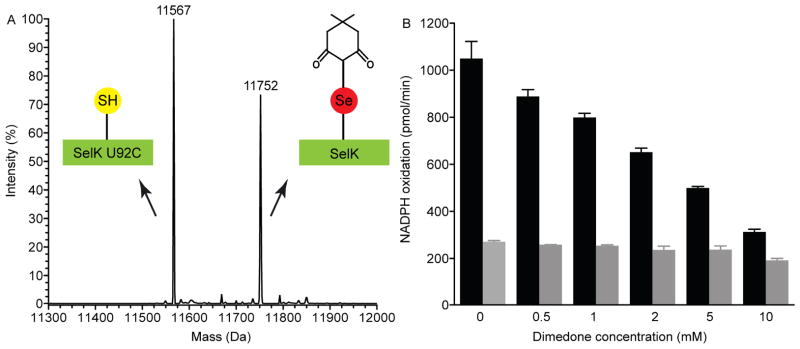
Selenenic acid contributes to the reaction mechanism. (A) Alkylation of selenenic acid by dimedone. Incubation of SelK with hydrogen peroxide and dimedone leads to a covalent adduct with dimedone. Mass spectrum shows that while SelK U92C (11567 Da) did not react with dimedone, SelK fully reacted with it (11752 Da). (B) Inhibition of SelK’s peroxidase activity by dimedone. The initial rate of NADPH oxidation was measured in the presence of H2O2, various concentrations of dimedone with SelK (black bar) and without SelK (gray bar).
In order to probe the rate of diselenide bond formation from the reactive selenenic acid species, the inhibition of SelK by dimedone was characterized. Since SelK will be inactivated by alkylation with dimedone, the activity will diminish in direct proportion to the rate in which dimedone reacted and inactivated SelK. In contrast, if the diselenide bond can reform faster than alkylation of the selenenic acid intermediate, then the enzyme remains active. As expected we found that the peroxidase activity of SelK was inhibited in a dimedone concentration dependent fashion (Fig. 7B). The data suggests that the reformation of the diselenide bond following the formation of the selenenic acid is facile, as evidenced by the concentration of dimedone necessary to fully abolish activity.
4. Discussion
Here we report that, in isolation, SelK’s oligomerization state is at least a homodimer containing an intermolecular diselenide bond. SelK has a short N-terminal single-pass transmembrane helix that resides in the ER membrane, and its transmembrane helix is unusually rich in polar residues (including a Glu and an Asp). This composition is a strong indication that SelK oligomerizes and is likely to associate in the membrane with a protein partner [49]. It is also quite possible that, like other bitopic proteins, it is capable of forming both homo- and hetero-oligomers. Many bitopic proteins dynamically exchange between different oligomerization states [50]. The close proximity of Sec from opposing monomers will facilitate the formation of diselenide bonds across the dimer interface. However, it is also possible that SelK pairs with a protein partner forming a selenylsulfide (Se-S) bond during function.
If such homo-oligomerization takes place under physiological conditions, then SelK represents the first example of a human selenoprotein possessing a diselenide bond. An intramolecular diselenide linkage (UXXU) was shown to occur in selenoprotein L (SelL) [51], which does not occur in humans. It is also possible, though not yet corroborated, that internal diselenide bonds can form in selenoproteins that have more than one Sec: the selenium storage and delivery enzyme selenoprotein P [52] and Metridium senile’s methionine sulfoxide reductase [53]. However, these examples of multiple Sec residues are rare as the majority of selenoproteins utilize only one Sec residue. Thus, SelK’s intermolecular diselenide bond stands out as an exception.
The redox potential of SelK’s intermolecular diselenide bond, −257 mV, is considerably higher than that reported for selenoglutathione (−407 mV [54]); selenocystine (−383 mV [55]); a diselenide bond in a peptide (−381 mV [56]); or selenoproteins with intramolecular disulfide bond. For example, Danio rerio SelL’s native UXXU redox motif was not reduced by DTT (E0′ = −327 mV) [51]. Similarly, the redox potential of non-native E. coli Grx3 and Trx, into which the diselenide bond was introduced, had a redox potential of −309 mV (Grx3) [57] and between −280 mV and −327 mV (Trx) [29,57]. Thus, the redox potential of SelK’s diselenide bond is higher than values reported so far for a diselenide bond in biological molecules. We propose that the protein molecular environment tailors it to be in the range accessible to the cytoplasmic proteins TrxR, Trx, and Grx. SelK was most efficiently reduced by hTrxR. The observation that hTrxR is capable of reducing a diselenide bond is in agreement with previous reports that it reduces diselenide bond in ebselen, a GPx mimic [58].
In regard to SelK’s enzymatic activity, our data demonstrates that SelK in micelles has no obvious oxidoreductase function other than a weak peroxidase activity. The peroxidase activity relies on the formation of an intermolecular diselenide bond. In addition, SelK most efficiently reduces lipid hydroperoxides. This is notable, as polyunsaturated fatty acids are a major target of ROS, which leads to disruption of membrane organization [59] and related diseases [59]. However, when the peroxidase activity of SelK is contrasted with other Sec-containing peroxidases, its efficiency is comparatively low. Human selenoproteins with peroxidase activity range from the highly efficient glutathione peroxidases (GPxs) with the Sec reacting with hydroperoxides at a rate of 105–107 M−1 s−1 [45,46], to moderate peroxidases such as selenoprotein P (SelP) [43], SelS [24], and TrxR [60,61], whose bimolecular rate constant for the reaction of the reduced protein with hydroperoxides are lower than 105 M−1 s−1. Accordingly, at least for SelS and TrxR, the peroxidase activity is considered secondary to their primary functions as reductases. Non-selenium peroxidases, like peroxiredoxin 6 [62,63], were also reported to react with lipid hydroperoxides at a rate of 106 M−1 s−1. The reaction resolution (that is regeneration of the enzyme after the reaction with the hydroperoxide) typically takes place at a bimolecular rate of 104 to 105 M−1 s−1 [45]. The rate of SelK’s oxidation by PLPCOOH is 103 M−1 s−1. While this value is still within the range exhibited by peroxidases, it calls into question whether SelK’s peroxidase activity is its primary enzymatic function. It is possible that since this moderate catalytic efficiency was measured using the isolated SelK in detergent micelles, the activity might increase if SelK interacts with a protein partner or in the presence of its native membrane environment. Similarly, a phosphorylation on Y60 was previously reported [58], but it is unclear how this posttranslational modification governs SelK conformation, activity, and interactions with physiological partners and membranes. It is another possibility that SelK acts as a lipid hydroperoxide sensor. The mechanisms of lipid hydroperoxides sensing in ROS signaling are not well understood [64] but are clearly critical for health. However, the link between SelK’s ability to reduce lipid hydroperoxides and its physiological function remains an open question.
We demonstrated that when the Sec was exposed it reacted with hydroperoxides to form a selenenic acid since this specie was captured by alkylation with dimedone. That is contrasted with the case of SelS, a closely related protein partner with an intramolecular selenylsulfide bond, whose selenenic acid could only be trapped in a mutant, but not in the wild-type enzyme [24]. It raises the intriguing prospect that SelK might be able to form a selenylsulfide bond with a protein partner.
Overall, our work established the redox properties of SelK, the bonding interactions of the sole Sec and that dimerization does not require the presence of the intermolecular diselenide bond. It demonstrated that it is premature to conclusively assign an enzymatic activity in the absence of protein partners and/or the membrane environment. However, SelK can interact with lipid hydroperoxides. Future experiments will examine the role of known protein partners on SelK’s oligomerization and activity.
Supplementary Material
Highlights.
SelK contains an intermolecular diselenide bond.
SelK’s redox potential is −257 mV, which is unusually high for a diselenide bond.
SelK is a weak peroxidase but can reduce phospholipid hydroperoxides.
Acknowledgments
We gratefully acknowledge assistance from Ms. Fei Li with mass spectrometry and service from the University of Delaware’s protein characterization core facility. This work was supported by the National Institute for General Medical Sciences grant P30 GM103519 and by the National Science Foundation under Grant No. MCB-1054447 “CAREER: Reactivity of Selenoproteins”.
Abbreviations
- NEM
N-ethylmaleimide
- HpETE
15(S)-hydroperoxy-6(E),8(Z),11(Z),14(Z)-eicosatetraenoic acid
- tBOOH
tert-butyl hydroperoxide
- yGR
yeast glutathione reductase
- hTrxR
human thioredoxin reductase 1
- DDM
n-dodecyl-β-D-maltopyranoside
- Dimedone
5,5-dimethyl-1,3-cyclohexanedione
- COOH
cumene hydroperoxide
- PLPC
2-linoleoyl-1-palmitoyl-sn-glycero-3-phosphocholine
- PLPCOOH
1-palmitoyl-2-(13-hydroperoxy-cis-9,trans-11-octadecadienoyl) phosphatidylcholine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Verma S, Hoffmann FW, Kumar M, Huang Z, Roe K, Nguyen-Wu E, Hashimoto AS, Hoffmann PR. Selenoprotein K knockout mice exhibit deficient calcium flux in immune cells and impaired immune responses. J Immunol. 2011;186:2127–2137. doi: 10.4049/jimmunol.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morozova N, Forry EP, Shahid E, Zavacki AM, Harney JW, Kraytsberg Y, Berry MJ. Antioxidant function of a novel selenoprotein in Drosophila melanogaster. Genes Cells. 2003;8:963–971. doi: 10.1046/j.1365-2443.2003.00687.x. [DOI] [PubMed] [Google Scholar]
- 3.Lai CQ, Parnell LD, Lyman RF, Ordovas JA, Mackay TFC. Candidate genes affecting Drosophila life span identified by integrating microarray gene expression analysis and QTL mapping. Mech Ageing Dev. 2007;128:237–249. doi: 10.1016/j.mad.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Meplan C, Rohrmann S, Steinbrecher A, Schomburg L, Jansen E, Linseisen J, Hesketh J. Polymorphisms in thioredoxin reductase and selenoprotein k genes and selenium status modulate risk of prostate cancer. Plos One. 2012;7 doi: 10.1371/journal.pone.0048709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shchedrina VA, Everley RA, Zhang Y, Gygi SP, Hatfield DL, Gladyshev VN. Selenoprotein K binds multiprotein complexes and is involved in the regulation of endoplasmic reticulum homeostasis. J Biol Chem. 2011;286:42937–48. doi: 10.1074/jbc.M111.310920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finkel T, Serrano M, Blasco MA. The common biology of cancer and ageing. Nature. 2007;448:767–774. doi: 10.1038/nature05985. [DOI] [PubMed] [Google Scholar]
- 7.Lu J, Holmgren A. Selenoproteins. J Biol Chem. 2009;284:723–727. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- 8.Chen CL, et al. G-rich, a Drosophila selenoprotein, is a Golgi-resident type III membrane protein. Biochem Biophys Res Commun. 2006;348:1296–1301. doi: 10.1016/j.bbrc.2006.07.203. [DOI] [PubMed] [Google Scholar]
- 9.Hatfield DL, Tsuji PA, Carlson BA, Gladyshev VN. Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem Sci. 2014;39:112–120. doi: 10.1016/j.tibs.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roseler A, et al. Insight into the selenoproteome of the malaria parasite plasmodium falciparum. Antioxid Redox Signaling. 2012;17:534–543. doi: 10.1089/ars.2011.4276. [DOI] [PubMed] [Google Scholar]
- 11.Lu CL, et al. Identification and characterization of selenoprotein K: An antioxidant in cardiomyocytes. FEBS Lett. 2006;580:5189–5197. doi: 10.1016/j.febslet.2006.08.065. [DOI] [PubMed] [Google Scholar]
- 12.Du SQ, Zhou J, Jia Y, Huang KX. SelK is a novel ER stress-regulated protein and protects HepG2 cells from ER stress agent-induced apoptosis. Arch Biochem Biophys. 2010;502:137–143. doi: 10.1016/j.abb.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Hubert P, Sawma P, Duneau JP, Khao J, Henin J, Bagnard D, Sturgis J. Single-spanning transmembrane domains in cell growth and cell-cell interactions More than meets the eye? Cell Adh Migr. 2010;4:313–324. doi: 10.4161/cam.4.2.12430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shchedrina VA, Zhang Y, Labunskyy VM, Hatfield DL, Gladyshev VN. Structure-function relations, physiological roles, and evolution of mammalian ER-resident selenoproteins. Antioxid Redox Signal. 2010;12:839–849. doi: 10.1089/ars.2009.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meiler S, et al. Selenoprotein K is required for palmitoylation of CD36 in macrophages: implications in foam cell formation and atherogenesis. J Leukocyte Biol. 2013;93:771–780. doi: 10.1189/jlb.1212647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christianson JC, et al. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turanov AA, et al. Selenoprotein S is involved in maintenance and transport of multiprotein complexes. Biochem J. 2014 doi: 10.1042/BJ20140076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Z, Hoffmann FW, Norton RL, Hashimoto AC, Hoffmann PR. Selenoprotein K Is a novel target of m-calpain, and cleavage is regulated by toll-like receptor-induced calpastatin in macrophages. J Biol Chem. 2011;286:34830–34838. doi: 10.1074/jbc.M111.265520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: Molecular Pathways and Physiological Roles. Physiol Rev. 2014;94:739–777. doi: 10.1152/physrev.00039.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uversky VN. A decade and a half of protein intrinsic disorder: Biology still waits for physics. Protein Sci. 2013;22:693–724. doi: 10.1002/pro.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vendruscolo M. Enzymatic activity in disordered states of proteins. Curr Opin Chem Biol. 2010;14:671–675. doi: 10.1016/j.cbpa.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 22.Zambelli B, Cremades N, Neyroz P, Turano P, Uversky VN, Ciurli S. Insights in the (un)structural organization of Bacillus pasteurii UreG, an intrinsically disordered GTPase enzyme. Molecular Biosystems. 2012;8:220–228. doi: 10.1039/c1mb05227f. [DOI] [PubMed] [Google Scholar]
- 23.Liu J, Li F, Rozovsky S. The intrinsically disordered membrane protein selenoprotein S is a reductase in vitro. Biochemistry. 2013;52:3051–3061. doi: 10.1021/bi4001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–7. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Srinivasan P, Pham DN, Rozovsky S. Expression and purification of the membrane enzyme selenoprotein K. Protein Expr Purif. 2012;86:27–34. doi: 10.1016/j.pep.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Rozovsky S. Contribution of selenocysteine to the peroxidase activity of selenoprotein S. Biochemistry. 2013;52:5514–5516. doi: 10.1021/bi400741c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jao SC, Ospina SME, Berdis AJ, Starke DW, Post CB, Mieyal JJ. Computational and mutational analysis of human glutaredoxin (thioltransferase): Probing the molecular basis of the low pK(a) of cysteine 22 and its role in catalysis. Biochemistry. 2006;45:4785–4796. doi: 10.1021/bi0516327. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell DA, Morton SU, Fernhoff NB, Marletta MA. Thioredoxin is required for S-nitrosation of procaspase-3 and the inhibition of apoptosis in Jurkat cells. Proc Natl Acad Sci USA. 2007;104:11609–11614. doi: 10.1073/pnas.0704898104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muller S, Senn H, Gsell B, Vetter W, Baron C, Bock A. The formation of diselenide bridges in proteins by incorporation of selenocysteine residues - biosynthesis and characterization of (Se)(2)-thioredoxin. Biochemistry. 1994;33:3404–3412. doi: 10.1021/bi00177a034. [DOI] [PubMed] [Google Scholar]
- 30.Strub MP, Hoh F, Sanchez JF, Strub JM, Bock A, Aumelas A, Dumas C. Selenomethionine and selenocysteine double labeling strategy for crystallographic phasing. Structure. 2003;11:1359–1367. doi: 10.1016/j.str.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Ellman GL. Tissue sulfhydryl groups. Arch Biochem Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 33.Lees WJ, Whitesides GM. Equilibrium-constants for thiol disulfide interchange reactions - a coherent, corrected set. J Org Chem. 1993;58:642–647. [Google Scholar]
- 34.Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 35.Roveri A, Maiorino M, Ursini F. Enzymatic and immunological measurements of soluble and membrane-bound phospholipid-hydroperoxide glutathione-peroxidase. Oxygen Radicals in Biological Systems, Pt C. 1994;233:202–212. doi: 10.1016/s0076-6879(94)33023-9. [DOI] [PubMed] [Google Scholar]
- 36.Ursini F, Bonaldo L, Maiorino M, Gregolin C. High-performance liquid-chromatography of hydroperoxy derivatives of stearoyllinoleoylphosphatidylcholine and of their enzymatic reduction products. J Chromatogr. 1983;270:301–308. [Google Scholar]
- 37.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Cloning and sequencing of thiol-specific antioxidant from mammalian brain - alkyl hydroperoxide reductase and thiol-specific antioxidant define a large family of antioxidant enzymes. Proc Natl Acad Sci USA. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rozovsky S. 77Se NMR spectroscopy of selenoproteins. In: Bayse CA, Brumaghim JL, editors. Biochalcogen Chemistry: The Biological Chemistry of Sulfur, Selenium, and Tellurium. ACS Press; 2013. pp. 127–142. [Google Scholar]
- 39.Johansson L, Gafvelin G, Arner ESJ. Selenocysteine in proteins - properties and biotechnological use. Biochim Biophys Acta. 2005;1726:1–13. doi: 10.1016/j.bbagen.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 40.Yoshizawa S, Bock A. The many levels of control on bacterial selenoprotein synthesis. Biochim Biophys Acta. 2009;1790:1404–1414. doi: 10.1016/j.bbagen.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Arner ESJ, Sarioglu H, Lottspeich F, Holmgren A, Bock A. High-level expression in Escherichia coli of selenocysteine-containing rat thioredoxin reductase utilizing gene fusions with engineered bacterial-type SECIS elements and co-expression with the selA, selB and selC genes. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 42.Langosch D, Arkin IT. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 2009;18:1343–1358. doi: 10.1002/pro.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem. 1979;254:9627–9632. [PubMed] [Google Scholar]
- 44.Gromer S, Urig S, Becker K. The thioredoxin system - From science to clinic. Med Res Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 45.Saito Y, Hayashi T, Tanaka A, Watanabe Y, Suzuki M, Saito E, Takahashi K. Selenoprotein P in human plasma as an extracellular phospholipid hydroperoxide glutathione peroxidase - Isolation and enzymatic characterization of human selenoprotein P. J Biol Chem. 1999;274:2866–2871. doi: 10.1074/jbc.274.5.2866. [DOI] [PubMed] [Google Scholar]
- 46.Takebe G, Yarimizu J, Saito Y, Hayashi T, Nakamura H, Yodoi J, Nagasawa S, Takahashi K. A comparative study on the hydroperoxide and thiol specificity of the glutathione peroxidase family and selenoprotein P. J Biol Chem. 2002;277:41254–41258. doi: 10.1074/jbc.M202773200. [DOI] [PubMed] [Google Scholar]
- 47.Dalziel K. Initial steady state velocities in the evaluation of enzyme-coenzyme-substrate reaction mechanisms. Acta Chem Scand. 1957;11:1706–1723. [Google Scholar]
- 48.Toppo S, Flohe L, Ursini F, Vanin S, Maiorino M. Catalytic mechanisms and specificities of glutathione peroxidases: Variations of a basic scheme. Biochim Biophys Acta. 2009;1790:1486–1500. doi: 10.1016/j.bbagen.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 49.Gayen S, Li QX, Kang CB. Solution NMR study of the transmembrane domain of single-span membrane proteins: opportunities and strategies. Curr Protein Peptide Sci. 2012;13:585–600. doi: 10.2174/138920312803582979. [DOI] [PubMed] [Google Scholar]
- 50.Vostrikov VV, Mote KR, Verardi R, Veglia G. Structural dynamics and topology of phosphorylated phospholamban homopentamer reveal its role in the regulation of calcium transport. Structure. 2013;21:2119–2130. doi: 10.1016/j.str.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shchedrina VA, Novoselov SV, Malinouski MY, Gladyshev VN. Identification and characterization of a selenoprotein family containing a diselenide bond in a redox motif. Proc Natl Acad Sci USA. 2007;104:13919–13924. doi: 10.1073/pnas.0703448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma SG, Hill KE, Burk RF, Caprioli RM. Mass spectrometric determination of selenenylsulfide linkages in rat selenoprotein P. J Mass Spectrom. 2005;40:400–404. doi: 10.1002/jms.801. [DOI] [PubMed] [Google Scholar]
- 53.Lee BC, Lobanov AV, Marino SM, Kaya A, Seravalli J, Hatfield DL, Gladyshev VN. A 4-Selenocysteine, 2-Selenocysteine Insertion Sequence (SECIS) element methionine sulfoxide reductase from metridium senile reveals a non-catalytic function of selenocysteines. J Biol Chem. 2011;286:18747–18755. doi: 10.1074/jbc.M111.229807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Beld J, Woycechowsky KJ, Hilvert D. Selenoglutathione: Efficient oxidative protein folding by a diselenide. Biochemistry. 2007;46:5382–5390. doi: 10.1021/bi700124p. [DOI] [PubMed] [Google Scholar]
- 55.Nauser T, Dockheer S, Kissner R, Koppenol WH. Catalysis of electron transfer by selenocysteine. Biochemistry. 2006;45:6038–6043. doi: 10.1021/bi0602260. [DOI] [PubMed] [Google Scholar]
- 56.Besse D, Siedler F, Diercks T, Kessler H, Moroder L. The redox potential of selenocystine in unconstrained cyclic peptides. Angew Chem. 1997;36:883–885. [Google Scholar]
- 57.Metanis N, Keinan E, Dawson PE. Synthetic seleno-glutaredoxin 3 analogues are highly reducing oxidoreductases with enhanced catalytic efficiency. J Am Chem Soc. 2006;128:16684–16691. doi: 10.1021/ja0661414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao R, Holmgren A. A novel antioxidant mechanism of ebselen involving ebselen diselenide, a substrate of mammalian thioredoxin and thioredoxin reductase. J Biol Chem. 2002;277:39456–62. doi: 10.1074/jbc.M206452200. [DOI] [PubMed] [Google Scholar]
- 59.Niki E. Lipid peroxidation: Physiological levels and dual biological effects. Free Radical Biol Med. 2009;47:469–484. doi: 10.1016/j.freeradbiomed.2009.05.032. [DOI] [PubMed] [Google Scholar]
- 60.Zhong LW, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 61.Bjornstedt M, Hamberg M, Kumar S, Xue J, Holmgren A. Human thioredoxin reductase directly reduces lipid hydroperoxides by NADPH and selenocystine strongly stimulates the reaction via catalytically generated selenols. J Biol Chem. 1995;270:11761–11764. doi: 10.1074/jbc.270.20.11761. [DOI] [PubMed] [Google Scholar]
- 62.Fisher AB, Dodia C, Manevich Y, Chen JW, Feinstein SI. Phospholipid hydroperoxides are substrates for non-selenium glutathione peroxidase. J Biol Chem. 1999;274:21326–34. doi: 10.1074/jbc.274.30.21326. [DOI] [PubMed] [Google Scholar]
- 63.Manevich Y, Shuvaeva T, Dodia C, Kazi A, Feinstein SI, Fisher AB. Binding of peroxiredoxin 6 to substrate determines differential phospholipid hydroperoxide peroxidase and phospholipase A(2) activities. Arch Biochem Biophys. 2009;485:139–149. doi: 10.1016/j.abb.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Girotti AW. Translocation as a means of disseminating lipid hydroperoxide-induced oxidative damage and effector action. Free Radic Biol Med. 2008;44:956–68. doi: 10.1016/j.freeradbiomed.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.