Abstract
SIRT6 is a histone deacetylase that has been proposed as a potential therapeutic target for metabolic disorders and the prevention of age-associated diseases. Thus the identification of compounds that modulate SIRT6 activity could be of great therapeutic importance. We have previously reported on the identification of quercetin and vitexin as SIRT6 inhibitors, using SIRT6-coated magnetic beads. In this study, we have immobilized SIRT6 onto the surface of an open tubular capillary and characterized the quercetin binding site using frontal displacement chromatography. Structurally related flavonoids were tested for their activity on SIRT6, including apigenin, naringenin, luteolin and kaempferol. In addition to obtaining their binding activity using frontal affinity chromatographic techniques, we also ranked the compounds based on their ability to displace quercetin. The data suggest that a single displacement curve is representative of the enzymatic activity of the tested ligand. In addition, using the inhibition data obtained in this study, we developed a preliminary pharmacophore model that confirmed the experimental data.
Keywords: frontal displacement chromatography, pharmacophore model, luteolin, Histone deacetylation assay
Introduction
Epigenetic modifications control gene expression by altering the chromatin structure to control the access of transcriptional activators or repressors to a gene locus. The acetylation of histone tails is one example of such epigenetic marks that is associated with activation of gene expression [1]. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are the enzymes that add or remove this posttranslational modification, and they typically exert their activity on large sets of genes to which they are recruited [2]. Alterations in the levels of such histone modifying enzymes or mutations that alter their function have been associated with a large number of diseases including developmental and neurological disorders, cancers, and aging [3]. Importantly the etiology of these diseases is likely caused by the deregulation of a large number of target genes throughout the genome.
Sirtuins are a family of NAD+-dependent HDACs that have been implicated to be important regulators in the aging process, cancers and metabolic diseases [4]. SIRT1 is the most-studied member of this protein family and the overexpression of its homolog in yeast, fly, worms has been associated with lifespan extension and its absence with a reduction in lifespan [5]. The biological roles of the other sirtuins are less well understood, and we decided to focus on SIRT6. In mice, deletion of the SIRT6 gene leads to a striking premature aging phenotype including decreased serum glucose and insulin-like growth factor IGF-1 levels [6]. The broad range of phenotypes is likely caused by the deregulation of a large set of genes in the absence of SIRT6 as well as alterations in the telomere structure which affect chromosomal stability [7,8]. Importantly, it has been proposed that SIRT6 might be a good therapeutic target in the context of age-associated diseases as well as metabolic disorders [9,10]. Thus the identification of compounds that modulate SIRT6 activity could be of therapeutic relevance. We have previously reported that quercetin and vitexin inhibit H3K9 deacetylase activity [11]. In addition to these flavonoids and nicotinamide, a recent study [12] has also identified a series of inhibitors for the SIRT6, however, these inhibitors are carrying a charged nitrogen as opposed to the polyphenols considered in this study.
The aim of this study was to generate a screening method for the identification of modulators of the SIRT6 protein that is more rapid than the current deacetylation assay on the SIRT6-Magnetic Beads (SIRT6-MB) and to characterize the quercetin binding site of SIRT6. To this end, we have immobilized the SIRT6 protein onto the surface of an open tubular (OT) capillary generating the SIRT6-OT column. The immobilized SIRT6 remained catalytically active and was characterized using frontal chromatographic techniques. In addition to quercetin and vitexin, the flavanols apigenin, kaempferol and luteolin and the flavanone naringenin were studied as they are structurally related to quercetin and vitexin.
Materials and methods
Materials
2-(N-morpholino)ethanesulfonic acid (MES) was purchased from EMD-Calbiochem (Gibbstown, NJ). Adenosine, adenosine 5′-diphosphoribose (ADP ribose), adenosine 5′-monophosphate (AMP), p-aminohippuric acid, 1-ethyl-3-(3-methylaminopropyl)carbodiimide (EDC), gluteraldehyde, hydroxylamine hydrochloride, nicotinamide, nicotinamide adenine dinucleotide (NAD+), Dithiothreitol (DTT) potassium phosphate dibasic, pyridine (99.8%), sodium azide, sodium cyanoborohydride, and sodium phosphate monobasic, quercetin, vitexin, naringenin, kaempferol, apigenin were obtained from Sigma-Aldrich Chemical Co. (Milwaukee, WI). N-Hydroxysulfosuccinimide (Sulfo-NHS) was from Pierce (Rockford, IL). Acetylated histone H3 (K9) peptide (H3K9Ac) were from Upstate/Millipore (Temecula, CA). Solutions were prepared using purified water from a Millipore MilliQ system (Millipore Corporation, Bedford, MA). BcMag amine-terminated magnetic beads (50 mg/mL, 1 μm) were purchased from Bioclone, Inc. (San Diego, CA). The manual magnetic separator Dynal MPC-S was from Invitrogen Corporation (Carlsbad, CA).
Cloning of expression construct and Protein Purification
The full-length cDNA of chicken SIRT6 was excised from pMH6-SIRT6 [11] using BamHI and XhoI, and ligated into the pGEX4T1 plasmid (GE Healthcare) that was linearized with the same restriction enzymes. The resulting pGEX SIRT6 plasmid was verified by DNA sequencing.
Expression and Purification of GST-Tagged SIRT6 Proteins in E. coli
Recombinant SIRT6 protein was expressed in E. coli (BL21, Rosetta strain, Novagen) and purified. An overnight 5 mL culture of a single bacterial colony harboring the pGEX SIRT6 plasmid was used to inoculate 1 L of Luria Broth medium (10 g/L tryptone, 5 g/L yeast extract, 10 g/L NaCl, pH=7.0) containing 2 g/L glucose. The cultures were grown at 37°C and 200 rpm, and protein production was induced at an OD600=0.6 by adding IPTG (final concentration 50μM). After 3 hours the bacteria were pelleted and flash frozen. Bacterial pellets were resuspended in 12.5 mL ice-cold lysis buffer (20mM Tris pH=8.0, 200mM NaCl, 1mM EDTA, 5mM β-mercaptoethanol, 10% glycerol) including protease inhibitors (10 μg/mL leupeptin, 100 μg /mL aprotinin, 10 μg/mL pepstatin A, 1mM PMSF) and lysed by sonication (3× 15 seconds bursts with 30 seconds intervals on ice using a Branson digital sonifier). The GST-SIRT6 fusion proteins were bound to 400 μL glutathione sepharose resin (GE Healthcare) for 2 hours at 4°C on a rotator. The resin was washed twice with ice-cold lysis buffer, twice with ice-cold cleavage buffer (20mM Tris pH 8.0, 150mM NaCl, 1mM CaCl2, 5 mM β-mercaptoethanol, 10% glycerol), and finally resuspended in 600 μL of cleavage buffer. The SIRT6 protein was released from the resin be adding 4μL of thrombin (1 U/μL, Novagen) and incubation at 4° overnight. The resin was pelleted, and the supernatant was added to 50μL benzamidine-agarose (Sigma) to remove the thrombin. After 30 minutes at room temperature, the agarose was pelleted, and the supernatant was further purified by size-exclusion chromatography using a Superose 6 column in an AKTA FPLC system (GE Healthcare). Fractions containing monomeric SIRT6 protein were pooled and dialyzed over-night against 1× PBS + 20% glycerol, and finally stored as frozen aliquots at -80°C.
Preparation of SIRT6 (CT)-Open tubular capillary
The SIRT6 C-terminus coupled (CT) open tubular (OT) capillary was prepared by a previously published protocol with slight modification [12,13]. Briefly, the open tubular capillary (30 cm × 100 μm i.d.) was washed with MES [100 mM, pH 5.5] for 20 min using a Rabbit peristaltic pump (Rainin, France) with a setting of 85. A solution 1 mL of MES [100 mM, pH 5.5] containing of 700 μL of SIRT6 (44 μg/mL) with 100 μl of EDC (500 mg/mL) and 50 μl of Sulpho-NHS (340 mg/mL) was passed through the column. Both tips of the capillary were submerged into the solution for 18 h at 4°C. After which MES buffer [100mM, pH 5.5] was passed through for 10 min.
Frontal Chromatography
The SIRT6(CT)-OT column was attached to the chromatographic system Series 1100 Liquid Chromatography/Mass Selective Detector (Agilent Technologies, Palo Alto, CA, USA) equipped with a vacuum de-gasser (G 1322 A), a binary pump (1312 A), an autosampler (G1313 A) with a 20 μL injection loop, a mass selective detector (G1946 B) supplied with atmospheric pressure ionization electrospray and an on-line nitrogen generation system (Whatman, Haverhill, MA, USA). The chromatographic system was interfaced to a 250 MHz Kayak XA computer (Hewlett-Packard, Palo Alto, CA, USA) running ChemStation software (Rev B.10.00, Hewlett-Packard). In the chromatographic studies, the mobile phase consisted of ammonium acetate [10 mM, pH 7.4]: methanol (90:10v/v) containing 0.2 mM NAD+ delivered at 0.05 mL min-1 at room temperature. Pumps A, C and D were used to apply a series of ligands: quercetin (2 μM, 5 μM, 11 μM, 15 μM,100 μM), naringenin (1.25 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM), vitexin (0.125 μM, 0.625 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM, 80 μM), apigenin (1.25 μM, 2.5 μM, 5 μM, 10 μM, 20 μM, 40 μM), kaempferol (1.25 μM, 2.5 μM, 10 μM, 20 μM, 40 μM) and luteolin (1.25 μM, 2.5 μM, 10 μM, 20 μM, 50 μM, 100 μM). Quercetin was monitored in the negative ion mode using single ion monitoring at m/z = 301.00 [MW - H]- ion for quercetin, with the capillary voltage at 3000 V, the nebulizer pressure at 35 psi, and the drying gas flow at 11 L/min at a temperature of 350°C. Naringenin was monitored in the negative ion mode using single ion monitoring at m/z = 271.00 [MW - H]- ion for naringenin, with the capillary voltage at 3000 V, the nebulizer pressure at 35 psi, and the drying gas flow at 11 L/min at a temperature of 350°C. Vitexin was monitored in the negative ion mode using single ion monitoring at m/z = 431.00 [MW - H]- ion for vitexin, with the capillary voltage at 3000 V, the nebulizer pressure at 35 psi, and the drying gas flow at 11 L/min at a temperature of 350°C. Apigenin was monitored in the negative ion mode using single ion monitoring at m/z = 269.00 [MW - H]- ion for apigenin, with the capillary voltage at 3000 V, the nebulizer pressure at 35 psi, and the drying gas flow at 11 L/min at a temperature of 350°C. Kaempferol and Luteolin were monitored in the negative ion mode using single ion monitoring at m/z = 285.00 [MW - H]- ion for kaempferol and luteolin, with the capillary voltage at 3000 V, the nebulizer pressure at 35 psi, and the drying gas flow at 11 L/min at a temperature of 350°C.
Data Analysis
The dissociation constants, Kd, for the displacer ligands were determined using a previously reported approach [14]. The experimental paradigm is based upon the effect of escalating approach of a competitive binding ligand on the retention volume. For example, the displacer ligands (D) dissociation constant, Kd, as well as the number of the active binding sites of the immobilized SIRT6, Bmax, can be calculated using equation (1):
| (1) |
where: V is the retention volume of ligand, Vmin is the retention volume of ligand when the specific interaction is completely suppressed and P=Bmax (Kd/KdM), when Displacer and marker are the same compound P=Bmax. The KD for D is obtained from the plot of [D] (V-Vmin) versus [D]. The data were analysed by nonlinear regression with a one-site binding (hyperbola) equation using Prism 4 software (Graph pad Software, Inc., San Diego, CA, USA) running on a personal computer.
Histone deacetylation assay by mass spectrometry
SIRT6 C-terminus coupled magnetic beads were incubated with 50 μL of HDAC assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT and 0.2 mM NAD+) containing 5 μg of acetylated histone H3(K9) peptide (amino acid residues 1–21; H3K9Ac) for 4 h at 37°C. After incubation, 10 μL of 120 μM p-aminohippuric acid was added as an internal standard. After magnetic separation, the supernatant was collected and analyzed by mass spectrometry using a system composed of an Agilent Technologies 1100 LC/MSD equipped with a G1322A degasser, G1312A binary pump, G1367A autosampler, G1316A column thermostat, G1315A diode array detector and G1946D mass spectrometer equipped with an electrospray ionization (ESI) interface. Selected ion monitoring (SIM) chromatograms were acquired using Chemstation software, Rev. A.10.02. For the separation of compounds, a reversed-phase Zorbax XDB-C18 analytical column (50 × 4.6 mm i.d., 1.8 μm particles) was used. The column was operated at 25°C. The mobile phase was a mixture of 0.05% trifluoroacetic acid (TFA) in water (A) and 0.02% TFA in acetonitrile (B), in linear gradient mode, as followed: 2-min hold at 100% A, followed by a linear gradient to 20% B from 2–22 min and then up to 80% B within 0.1 min, held for 2 min, and then re-equilibrated at initial conditions for 4 min. The flow rate was 1.2 mL/min and the injection volume was 50 μL. The substrates and metabolites were monitored in the positive-ion mode for SIM at m/z 922.4 ([M+3H]3+ of H3K9Ac), m/z 908.4 ([M+3H]3+ of deacetylated peptide; H3K9).
Molecular Modeling
All our training set compound structures were downloaded from pubchem database (http://www.ncbi.nlm.nih.gov/pubmed). Downloaded structures were energy minimized using Minimize Ligands module in Discovery Studio (ver. 3.1; Accelrys Inc, San Diego, CA). CHARMm forcefield as implemented in Discovery Studio was used for this step (see Suppl. information for more information). Common Feature Pharmacophore Generation module in Discovery Studio (ver. 3.1; Accelrys Inc, San Diego, CA) was used to create the pharmacophore models. Details of pharmacophore modeling had been explained in many reviews and articles [15,16]. For details in Pharmacophore generation in Discovery Studio please see [ref 16], as the pharmacophore model is only a minor component of this manuscript, it will not be discussed in further detail.
Common Feature pharmacophore modeling produced 10 models (Supplemental Table 1). In this step, the test set compounds, vitexin, daidzein and genistein, were mapped to all 10 pharmacophore models generated earlier to determine how they fit the models. This was carried out using Ligand Profiler Module in Discovery Studio. The results are shown in the form of heat map (suppl. Fig S1). Most of the pharmacophore models including the top-ranked model identified the experimental trend correctly. Currently, three crystal structures for the deacetylase sirtuin-type domain of human SIRT6 are available19 (PDB IDs: 3k35, 3pki and 3pkj; www.rcsb.org). However, as Pan et al [17] had pointed out SIRT6 function is markedly different from other known SIRT types especially in how the cofactor (NAD+) binding lacks the necessity for acetyl lysine substrate and possibly a different role for the key residue, H131, along with the possibility of a conformational modification during ligand binding. Keeping these issues in mind we have not considered the crystal structures for this study but rather focused on generating a preliminary pharmacophore model.
Results and Discussion
In our previous studies, we developed a novel SIRT6 deacetylase assay and identified quercetin and vitexin from fenugreek seed extract as inhibitors of the SIRT6 protein [11]. Although, this method was useful, it is time consuming and requires extensive resources in order to determine a compounds activity on the SIRT6 protein. We have previously demonstrated that bioaffinity columns can be used to fully characterize the immobilized protein using frontal affinity chromatographic techniques [18], and that the kinetics can be determined on-line. In the current study, the SIRT6 protein was immobilized onto the surface of an open tubular capillary generating the SIRT6-OT column, for the first time. Although, in general the protein could have been immobilized using any other surface exposed acidic residues, the choice of C-terminal end was based on our structural analysis (PDB ID: 3pki, SIRT6 crystal structure in complex with ADP ribose). No attempt was made to immobilize the protein through the N-terminus [11], as we had previously shown that this method resulted in an inactive enzyme.
After immobilization, the SIRT6 catalytic deacetylase activity was tested by monitoring the production of H3K9 from the deacetylation of the acetylated H3K9ac tail of H3K9 peptide substrate. It was determined that the SIRT6 protein was active after immobilization onto the OT column (data not shown). During the process of optimization, it was determined that the addition of 200 μM NAD+ to the mobile phase was necessary to carry out frontal displacement studies on the immobilized SIRT6-OT column with quercetin. The elution profile of a sub-saturating concentration of quercetin (2 μM) is represented in Figure 1, where an initial flat portion representing specific binding to the column followed by a breakthrough curve and plateau representing saturation of the column. The midpoint of the breakthrough curve occurred at 105 min on the SIRT6-OT column, representing breakthrough volume of 5.25 mL (Fig 1). Using increasing concentrations of quercetin (2 μM to 100 μM) in the mobile phase, a decrease in the retention volume was observed representing specific binding to the SIRT6-OT column. The binding affinity of quercetin for SIRT6 was calculated to be 4.1 μM using a non-linear regression model. The respective binding affinity of vitexin, an apigenin flavone glucoside, was determined to be 15.27 μM (Table 1, Scheme 1), and the binding affinities of three other flavanols (kaempferol, luteolin and apigenin) and the flavanone (naringenin) were also measured, and are shown in Table 1.
Figure 1.
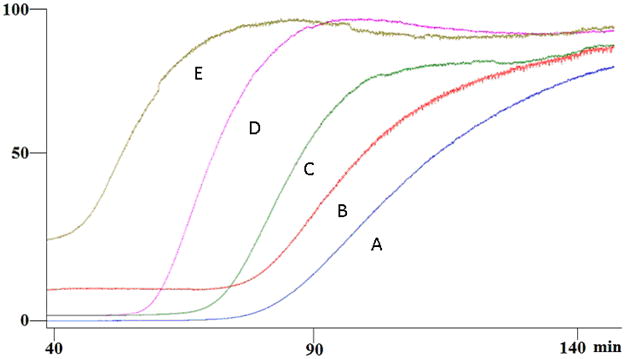
Frontal elution profiles of 2 μM (A), 5 μM (B), 11 μM (C), 15 μM (D) and 100 μM (E) quercetin on the SIRT6-OT column on the agilent LC-MSD. Running buffer consisted of ammonium acetate [10 mM, pH 7.4]: methanol (90:10v/v) containing 0.2 mM NAD+ and the flow rate was 50 μl/min.
Table 1.
The binding affinities for the flavonoids for the SIRT6 protein determined by frontal affinity chromatographic techniques on the SIRT6-OT column (Kd) and the percent inhibition of deacetylation relative to control. SIRT6-MB was incubated with 50 μL of HDAC assay buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM DTT and 0.2 mM NAD+) containing 5 μg of acetyl-histone H3(K9) peptide (amino acid residues 1–21; H3K9Ac) and 10 μM of each compound for 4h at 37°C.
| Kd (μM) | % inhibition | |
|---|---|---|
| Luteolin | 1.78 ± 0.64; r2=0.922 | 53.7 ± 17.7 |
| Apigenin | 1.92 ± 0.50; r2=0.938 | 39.0 ± 5.9 |
| Kaempferol | 3.88 ± 0.57; r2=0.993 | 48.6 ± 6.0 |
| Quercetin | 4.09 ± 2.26; r2=0.895 | 46.6 ± 2.3 |
| Naringenin | 6.62 ± 0.88; r2=0.993 | 31.0 ± 5.3 |
| Vitexin | 15.27 ± 3.87; r2=0.982 | 24.9 ± 0.9 |
Scheme 1.
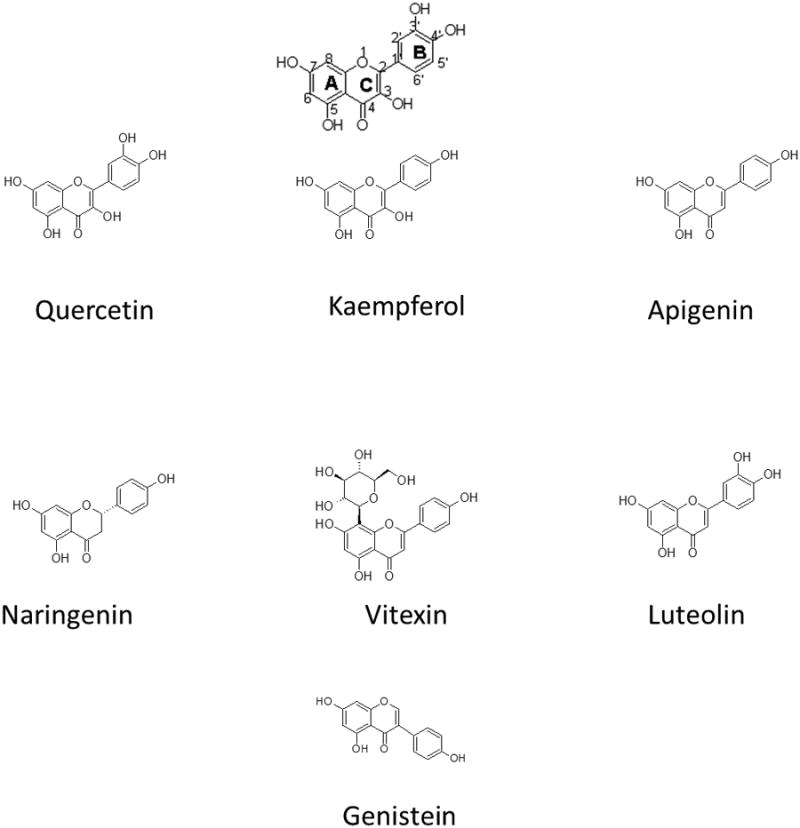
Scaffold and structure of the Quercetin, Vitexin, Naringenin, Apigenin, Luteolin, Genistein and Kaempferol.
The binding affinities ranged from ∼2 μM for luteolin to 15 μM for the apigenin flavone glucoside vitexin. The removal of the 3′ hydroxyl group on the B ring (Figure 1), did not result in any change in affinity for the SIRT6 protein c.f. quercetin vs kaempferol (3.9 vs 4.1 μM) and apigenin vs luteolin (1.9 vs 1.8 μM). A slight increase in affinity was observed with the removal of the C-3 hydroxy on the C ring c.f. kaempferol vs luteolin (3.9 vs 1.8 μM). The addition of the glucoside at C-8 on ring A resulted in a significant decrease in the binding affinity to the SIRT6 protein which could result from steric hindrance c.f. vitexin vs apigenin (15.3 vs 1.9 μM). Saturation of the double bond in the C-3 position of the C ring resulted in a 3-fold decrease in affinity for the SIRT6 protein (naringenin vs apigenin (6.6 vs 1.9 μM, respectively). The binding affinities, Kd, determined by frontal affinity chromatographic techniques, does not necessarily represent the binding of these compounds to the quercetin binding site, but rather to the SIRT6 protein.
To establish whether these polyphenols compete with quercetin for the quercetin binding site of SIRT6, and to determine whether a single displacement concentration could correctly predict the functional activity of the immobilized enzyme, frontal displacement chromatographic experiments were carried out. Namely, 10 μM concentration of each polyphenol was placed in the mobile phase in the presence of 5 μM quercetin (Figure 2). The change in retention volume was obtained to rank the compounds in order of affinity. Based on these observations the compounds with the strongest displacements were luteolin> kaempferol> quercetin> apigenin> naringenin> vitexin (Figure 2), with luteolin having a displacement of 38.1 min and vitexin displacing quercetin by 16.5 min, genistein, a known non-binder had a displacement of only 2.5 min, which is within the margin of error, indicating little or no binding. While, this is not in full agreement with the Kd's determined by frontal affinity chromatography which ranked them as luteolin >apigenin> kaempferol> quercetin> naringenin> vitexin, considering the standard error on the binding affinities obtained by frontal affinity chromatography, there is not a statistically significant difference between the binding affinities of luteolin, apigenin, kaempferol and quercetin and therefore it is not suprising to see apigenins movement. Naringenin and vitexin were both significantly weaker and this was consistent with both methods. In addition, when compared to the order obtained using our previously developed histone tail deacetylation assay [11] using SIRT6 coated magnetic beads (Table 1), the level of inhibition of the deacetylation activity of SIRT6 for each of these polyphenols correlated with the change in retention time relative to quercetin, and a linear correlation was found with an r2=0.9363 (y=-0.7584x + 71.17). Therefore, a single concentration frontal displacement chromatogram can correctly predict the enzymatic activity of SIRT6 inhibitors, allowing for a more rapid screening of compounds for activity at the SIRT6 protein as compared to the Histone deacetylation assay, which is also more costly.
Figure 2.
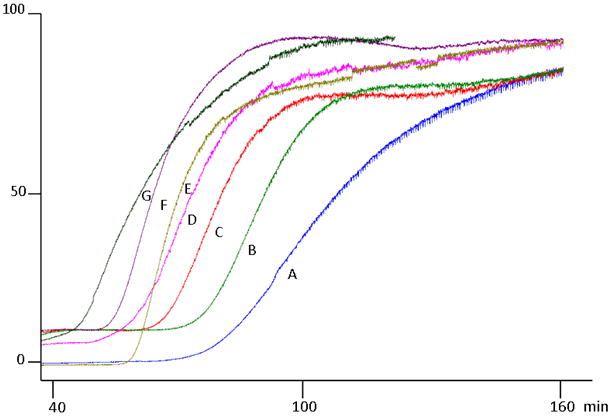
Frontal displacement profiles, demonstrating the effect of the addition of 10 μM vitexin (B), 10 μM naringenin (C), 10 μM apigenin (D), 10 μM quercetin (E), 10 μM kaempferol (F) and 10 μM luteolin (G) to the chromatographic retention of 5 μM quercetin (A). Running buffer consisted of ammonium acetate [10 mM, pH 7.4]: methanol (90:10v/v) containing 0.2 mM NAD+ and 5 μM quercetin the flow rate was 50 μl/min.
As an extension of the experimental data, preliminary pharmacophore models were generated (Suppl Table S1, Fig. S1) with the test compounds in an attempt to identify key functional features of the quercetin binding site of the SIRT6 protein. The top ranked pharmacophore model generated (Figure 3A, suppl Fig. S1) contains three hydrogen bond donor (HBD) features and one hydrogen bond acceptor (HBA). The spatial constraints in this model are detailed in Figure 3A. The conformation of the strong binder (luteolin) as mapped to the model is presented in Figure 3B. Based on our analysis, the features HBD1 and HBD2 were consistently misaligned when the weak binders are mapped to the pharmacophore models (data not shown). Overall, based on our analysis and comparison with the experimental results, the top ranked model (Model-1, sup. Fig. S1), correlates most closely with the experimentally determined Kd's, however, as this is only a preliminary model, further studies are being carried out to construct a more robust pharmacophore model with a larger experimental cohort and will be reported elsewhere.
Figure 3.
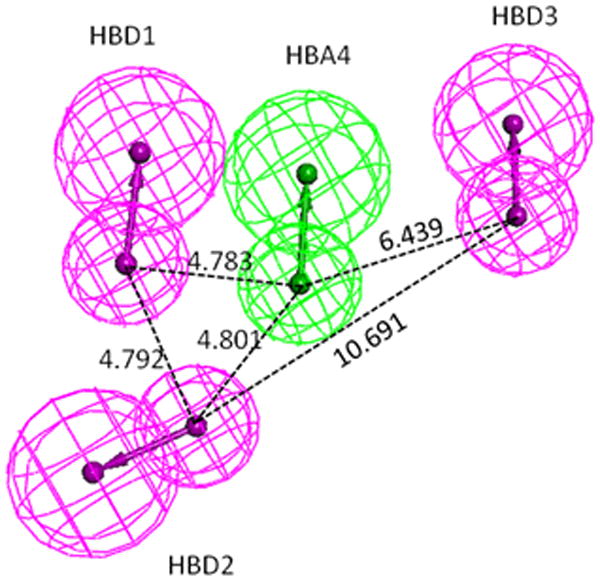
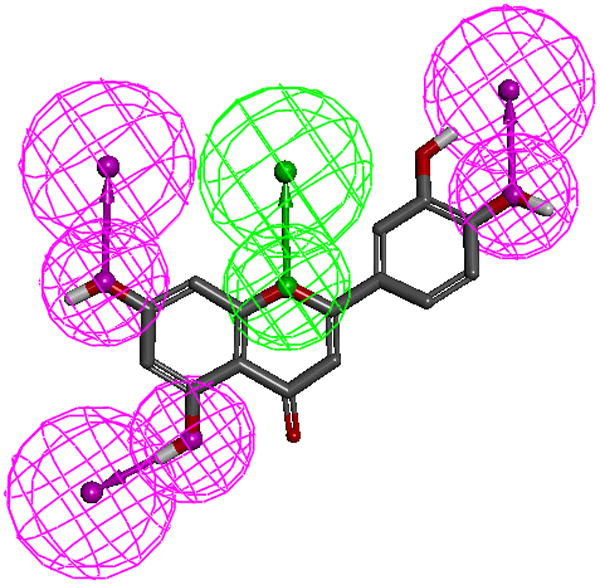
A.The top-ranked pharmacophore model (1, see supplemental Table 1). Pharmacophore model-1 consists of three hydrogen bond donors (magenta spheres) and one hydrogen bond acceptor (green spheres). Inter-spatial distances are shown in Angstroms.
B. Luteolin mapped to pharmacophore model-1. Note only polar hydrogens are displayed on the molecules.
Conclusion
In this report, we describe the development of a SIRT6 OT column that can be used to not only study the pharmacological properties of SIRT6 protein but also as a predictive tool of its deacetylation activity. In an extension from our previous work, we have characterized the quercetin binding site by determining the affinity of five additional polyphenols. In addition, we have demonstrated that a SIRT6-OT column can be used as a rapid screening tool to rank inhibitors by affinity.
Supplementary Material
Acknowledgments
This work was supported in part by funds from the NIA Intramural Research Program (RM), and from the National Cancer Institute/National Institutes of Health contract No. HHSN261200800001E. SR would like to thank Katalin Nadassy, Tina Yeh and Jodi Shaulsky from Accelrys Inc for many discussions during the analysis of pharmacophore modeling results.
Abbreviations Used
- HATS
histone acetyltransferases
- HDACs
histone deacetylases
- OT
open tubular
- MES
2-(N-morpholino)ethanesulfonic acid
- ADP ribose
Adenosine 5′-diphosphoribose
- AMP
Adenosine 5′-monophosphate
- DTT
Dithiothreitol
- EDC
1-ethyl-3-(3-methylaminopropyl)carbodiimide
- Sulfo-NHS
N-Hydroxysulfosuccinimide
- H3K9Ac
Acetylated histone H3 (K9) peptide
- NAD+
Nicotinamide adenine dinucleotide
- CT
C-terminus coupled
- Kd
Dissociation constants
- D
displacer ligands
- V
Retention volume of ligand
- Vmin
Retention volume of ligand when the specific interaction is completely suppressed
- ESI
Electrospray ionization
- TFA
trifluoroacetic acid
- SIM
Selected ion monitoring
- HBA
Hydrogen bond acceptor
- HBD
Hydrogen bond donor
Footnotes
There are no financial conflicts.
References
- 1.Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 3.Butler JS, Koutelou E, Schibler AC, Dent SY. Histone-modifying enzymes: regulators of developmental decisions and drivers of human disease. Epigenomics. 2012;2:163–177. doi: 10.2217/epi.12.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donmez G, Guarente L. Aging and disease: connections to sirtuins. Aging Cell. 2010;2:285–290. doi: 10.1111/j.1474-9726.2010.00548.x. [DOI] [PubMed] [Google Scholar]
- 6.Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 7.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, Chua KF. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodgers JT, Puigserver P. Certainly can't live without this: SIRT6. Cell Metabolism. 2006;3:77–82. doi: 10.1016/j.cmet.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Zhong L, D'Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack D, Guimaraes A, Marinelli B, Widstrom JD, Nir T, Clish CB, Vaitheesvaran B, Illiopoulos O, Kurland I, Dor Y, Weissleder R, Shirihai OS, Ellisen LW, Espinosa JM, Mostoslavsky R. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yasuda M, Wilson DR, Fugmann SD, Moaddel R. The Synthesis and characterization of SIRT6 protein coated magnetic beads: Identification of a novel inhibitor of SIRT6 deacetylase from medicinal plant extracts. Anal Chem. 2011;83:7400–7407. doi: 10.1021/ac201403y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schlicker C, Boanca G, Lakshminarasimhan M, Steegborn C. Structure-based development of novel sirtuin inhibitors. Aging. 2011;3:852–872. doi: 10.18632/aging.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanghvi M, Moaddel R, Frazier C, Wainer IW. Synthesis and Characterization of Liquid Chromatographic Columns Containing the Immobilized Ligand Binding Domain of the Estrogen Related Receptor α and Estrogen Related Receptor γ. J Pharm Biomed Anal. 2010;53:777–780. doi: 10.1016/j.jpba.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura T, Perry J, Anzai N, Pritchard J, Moaddel R. Development and Characterization of Immobilized Human Organic Anion Transporter Based Liquid Chromatographic Stationary Phase: hOAT1 and hOAT2. J Chrom B. 2007;859:267–271. doi: 10.1016/j.jchromb.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutter J, Li J, Maynard AJ, Goupil A, Luu T, Nadassy K. New features that improve the pharmacophore tools from Accelrys. Curr Comput Aided Drug Des. 2011;7:173–180. doi: 10.2174/157340911796504305. [DOI] [PubMed] [Google Scholar]
- 16.Vaidyanathan J, Vaidyanathan TK, Ravichandran S. Computer simulated screening of dentin bonding primer monomers through analysis of their chemical functions and their spatial 3D alignment. J Biomed Mater Res B Appl Biomater. 2009;88:447–457. doi: 10.1002/jbm.b.31134. [DOI] [PubMed] [Google Scholar]
- 17.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. The Journal of biological chemistry. 2011;286:14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moaddel R, Wainer IW. The preparation and development of cellular membrane affinity chromatography columns. Nat Protocol. 2009;4:197–205. doi: 10.1038/nprot.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


