Abstract
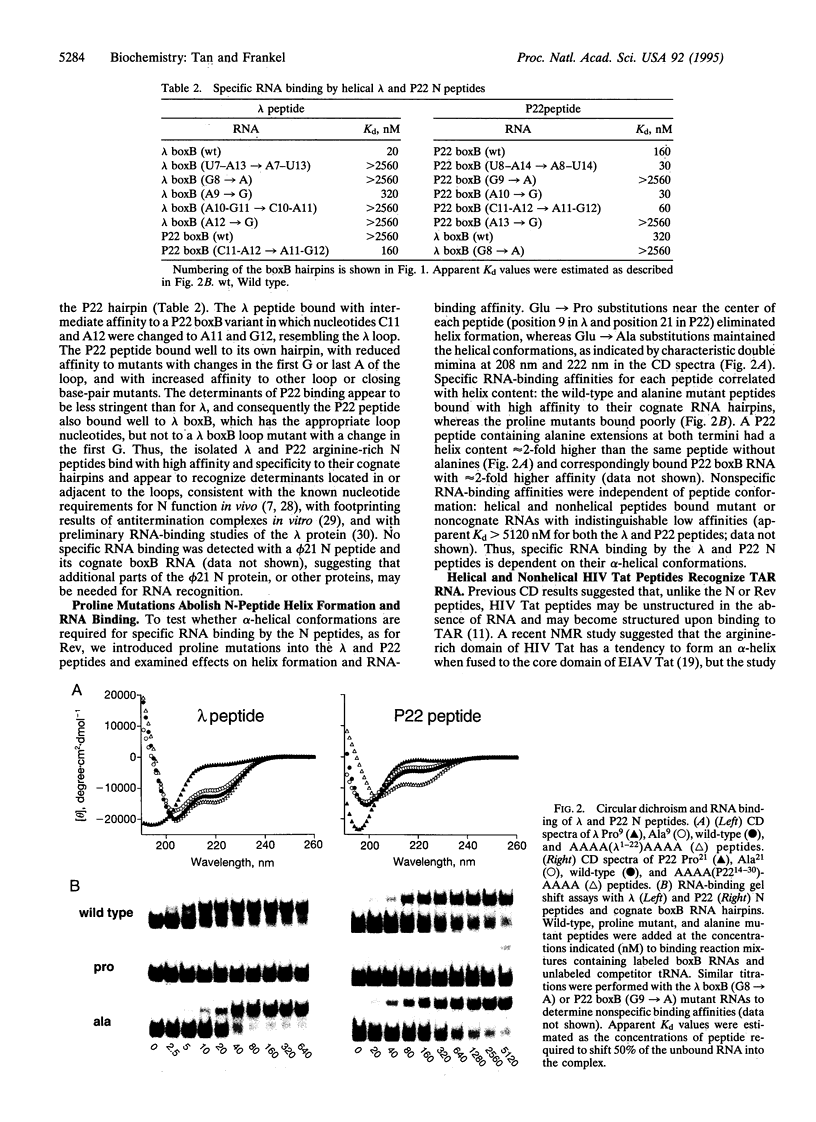
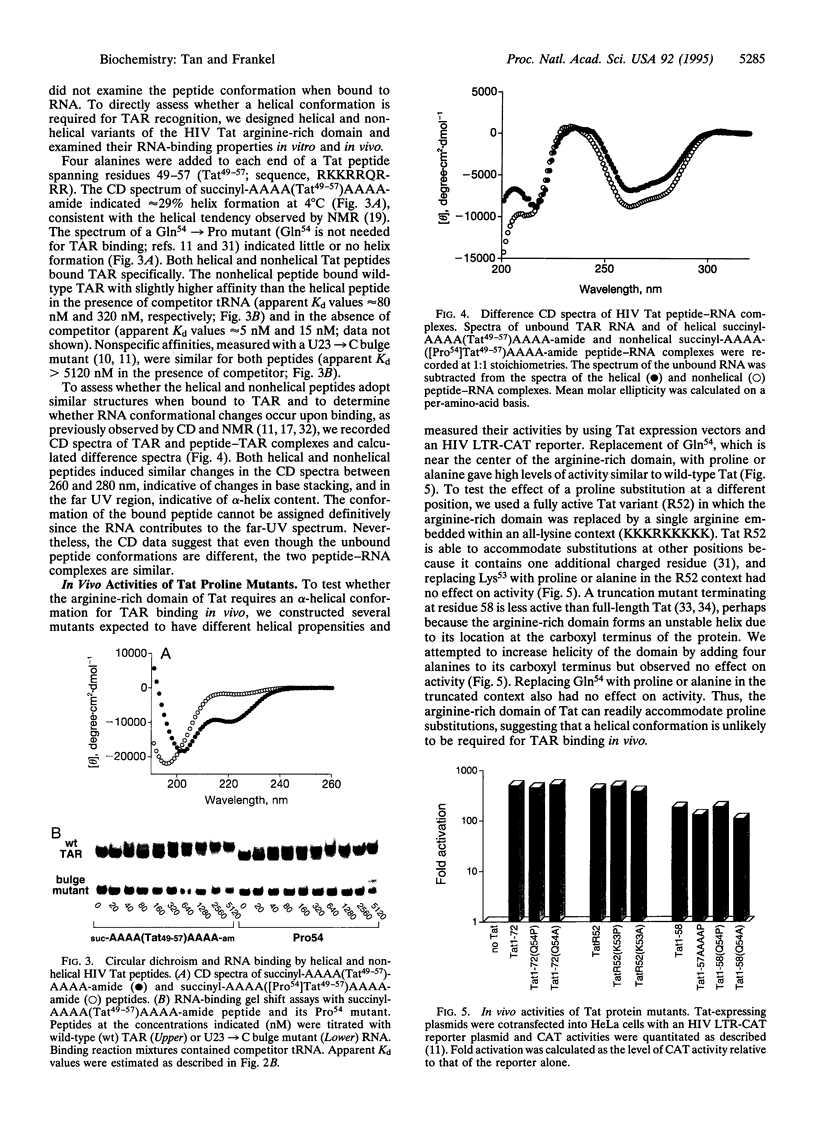
Arginine-rich domains are used by a variety of RNA-binding proteins to recognize specific RNA hairpins. It has been shown previously that a 17-aa arginine-rich peptide from the human immunodeficiency virus Rev protein binds specifically to its RNA site when the peptide is in an alpha-helical conformation. Here we show that related peptides from splicing factors, viral coat proteins, and bacteriophage antiterminators (the N proteins) also have propensities to form alpha-helices and that the N peptides require helical conformations to bind to their cognate RNAs. In contrast, introducing proline mutations into the arginine-rich domain of the human immunodeficiency virus Tat protein abolishes its potential to form an alpha-helix but does not affect RNA-binding affinity in vitro or in vivo. Based on results from several peptide-RNA model systems, we suggest that helical peptides may be used to recognize RNA structures having particularly wide major grooves, such as those found near loops or large bulges, and that nonhelical or extended peptides may be used to recognize less accessible grooves.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Battiste J. L., Tan R., Frankel A. D., Williamson J. R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry. 1994 Mar 15;33(10):2741–2747. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994 Mar 11;263(5152):1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Chen L., Frankel A. D. A peptide interaction in the major groove of RNA resembles protein interactions in the minor groove of DNA. Proc Natl Acad Sci U S A. 1995 May 23;92(11):5077–5081. doi: 10.1073/pnas.92.11.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Frankel A. D. An RNA-binding peptide from bovine immunodeficiency virus Tat protein recognizes an unusual RNA structure. Biochemistry. 1994 Mar 8;33(9):2708–2715. doi: 10.1021/bi00175a046. [DOI] [PubMed] [Google Scholar]
- Chen Y. H., Yang J. T., Chau K. H. Determination of the helix and beta form of proteins in aqueous solution by circular dichroism. Biochemistry. 1974 Jul 30;13(16):3350–3359. doi: 10.1021/bi00713a027. [DOI] [PubMed] [Google Scholar]
- Churcher M. J., Lamont C., Hamy F., Dingwall C., Green S. M., Lowe A. D., Butler J. G., Gait M. J., Karn J. High affinity binding of TAR RNA by the human immunodeficiency virus type-1 tat protein requires base-pairs in the RNA stem and amino acid residues flanking the basic region. J Mol Biol. 1993 Mar 5;230(1):90–110. doi: 10.1006/jmbi.1993.1128. [DOI] [PubMed] [Google Scholar]
- Das A. How the phage lambda N gene product suppresses transcription termination: communication of RNA polymerase with regulatory proteins mediated by signals in nascent RNA. J Bacteriol. 1992 Nov;174(21):6711–6716. doi: 10.1128/jb.174.21.6711-6716.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J. H., Franklin N. C. Effects of all single base substitutions in the loop of boxB on antitermination of transcription by bacteriophage lambda's N protein. Nucleic Acids Res. 1989 Jul 25;17(14):5565–5577. doi: 10.1093/nar/17.14.5565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal R., Hall T. C. Identification of domains in brome mosaic virus RNA-1 and coat protein necessary for specific interaction and encapsidation. J Virol. 1993 Nov;67(11):6406–6412. doi: 10.1128/jvi.67.11.6406-6412.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D., Biancalana S., Hudson D. Activity of synthetic peptides from the Tat protein of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1989 Oct;86(19):7397–7401. doi: 10.1073/pnas.86.19.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin N. C. Clustered arginine residues of bacteriophage lambda N protein are essential to antitermination of transcription, but their locale cannot compensate for boxB loop defects. J Mol Biol. 1993 May 20;231(2):343–360. doi: 10.1006/jmbi.1993.1287. [DOI] [PubMed] [Google Scholar]
- Greenblatt J., Nodwell J. R., Mason S. W. Transcriptional antitermination. Nature. 1993 Jul 29;364(6436):401–406. doi: 10.1038/364401a0. [DOI] [PubMed] [Google Scholar]
- Heus H. A., Pardi A. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science. 1991 Jul 12;253(5016):191–194. doi: 10.1126/science.1712983. [DOI] [PubMed] [Google Scholar]
- Lazinski D., Grzadzielska E., Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989 Oct 6;59(1):207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- Leclerc F., Cedergren R., Ellington A. D. A three-dimensional model of the Rev-binding element of HIV-1 derived from analyses of aptamers. Nat Struct Biol. 1994 May;1(5):293–300. doi: 10.1038/nsb0594-293. [DOI] [PubMed] [Google Scholar]
- Legrain P., Choulika A. The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J. 1990 Sep;9(9):2775–2781. doi: 10.1002/j.1460-2075.1990.tb07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj I. W. RNA recognition: a family matter? Cell. 1993 Jun 4;73(5):837–840. doi: 10.1016/0092-8674(93)90265-r. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Mujeeb A., Bishop K., Peterlin B. M., Turck C., Parslow T. G., James T. L. NMR structure of a biologically active peptide containing the RNA-binding domain of human immunodeficiency virus type 1 Tat. Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8248–8252. doi: 10.1073/pnas.91.17.8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell J. R., Greenblatt J. The nut site of bacteriophage lambda is made of RNA and is bound by transcription antitermination factors on the surface of RNA polymerase. Genes Dev. 1991 Nov;5(11):2141–2151. doi: 10.1101/gad.5.11.2141. [DOI] [PubMed] [Google Scholar]
- Peterson R. D., Bartel D. P., Szostak J. W., Horvath S. J., Feigon J. 1H NMR studies of the high-affinity Rev binding site of the Rev responsive element of HIV-1 mRNA: base pairing in the core binding element. Biochemistry. 1994 May 10;33(18):5357–5366. doi: 10.1021/bi00184a001. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tan R., Calnan B. J., Frankel A. D., Williamson J. R. Conformation of the TAR RNA-arginine complex by NMR spectroscopy. Science. 1992 Jul 3;257(5066):76–80. doi: 10.1126/science.1621097. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sacher R., Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989 Nov;63(11):4545–4552. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigel L. J., Ratner L., Josephs S. F., Derse D., Feinberg M. B., Reyes G. R., O'Brien S. J., Wong-Staal F. Transactivation induced by human T-lymphotropic virus type III (HTLV III) maps to a viral sequence encoding 58 amino acids and lacks tissue specificity. Virology. 1986 Jan 15;148(1):226–231. doi: 10.1016/0042-6822(86)90419-8. [DOI] [PubMed] [Google Scholar]
- Tan R., Chen L., Buettner J. A., Hudson D., Frankel A. D. RNA recognition by an isolated alpha helix. Cell. 1993 Jun 4;73(5):1031–1040. doi: 10.1016/0092-8674(93)90280-4. [DOI] [PubMed] [Google Scholar]
- Tan R., Frankel A. D. Circular dichroism studies suggest that TAR RNA changes conformation upon specific binding of arginine or guanidine. Biochemistry. 1992 Oct 27;31(42):10288–10294. doi: 10.1021/bi00157a016. [DOI] [PubMed] [Google Scholar]
- Tan R., Frankel A. D. Costabilization of peptide and RNA structure in an HIV Rev peptide-RRE complex. Biochemistry. 1994 Dec 6;33(48):14579–14585. doi: 10.1021/bi00252a025. [DOI] [PubMed] [Google Scholar]
- Tao J., Frankel A. D. Electrostatic interactions modulate the RNA-binding and transactivation specificities of the human immunodeficiency virus and simian immunodeficiency virus Tat proteins. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1571–1575. doi: 10.1073/pnas.90.4.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Frankel A. D. Specific binding of arginine to TAR RNA. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valegård K., Murray J. B., Stockley P. G., Stonehouse N. J., Liljas L. Crystal structure of an RNA bacteriophage coat protein-operator complex. Nature. 1994 Oct 13;371(6498):623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. Major groove accessibility of RNA. Science. 1993 Sep 17;261(5128):1574–1577. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]
- Willbold D., Rosin-Arbesfeld R., Sticht H., Frank R., Rösch P. Structure of the equine infectious anemia virus Tat protein. Science. 1994 Jun 10;264(5165):1584–1587. doi: 10.1126/science.7515512. [DOI] [PubMed] [Google Scholar]
- Zamore P. D., Patton J. G., Green M. R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992 Feb 13;355(6361):609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- van der Graaf M., Scheek R. M., van der Linden C. C., Hemminga M. A. Conformation of a pentacosapeptide representing the RNA-binding N-terminus of cowpea chlorotic mottle virus coat protein in the presence of oligophosphates: a two-dimensional proton nuclear magnetic resonance and distance geometry study. Biochemistry. 1992 Sep 29;31(38):9177–9182. doi: 10.1021/bi00153a009. [DOI] [PubMed] [Google Scholar]







