Abstract
Precise control of lineage-specific gene expression in the neural stem/progenitor cells is crucial for generation of the diversity of neuronal and glial cell types in the central nervous system (CNS). The mechanism underlying such gene regulation, however, is not fully elucidated. Here, we report that a 377 bp evolutionarily conserved DNA fragment (CR5), located approximately 32 kbp upstream of Olig2 transcription start site, acts as a cis-regulator for gene expression in the development of the neonatal forebrain. CR5 is active in a time-specific and brain region-restricted manner. CR5 activity is not detected in the embryonic stage, but it is exclusively in a subset of Sox5+ cells in the neonatal ventral forebrain. Furthermore, we show that Sox5 binding motif in CR5 is important for this cell-specific gene regulatory activity; mutation of Sox5 binding motif in CR5 alters reporter gene expression with different cellular composition. Together, our study provides new insights into the regulation of cell-specific gene expression during CNS development.
Keywords: Gene regulation, Cis-element, Trans-acting factor, Sox5, Olig2, Neural stem/progenitor cells
Introduction
During the development of the central nervous system (CNS), the cellular diversity emerges largely from controlled spatiotemporal segregation of cell type-specific molecular regulators (Butt et al., 2005; Fode et al., 2000; Hoshino, 2012; Lo et al., 2002; Molyneaux et al., 2007; Parras et al., 2002). A large number of different neurons and glial cells are derived from a population of self-renewing stem and progenitor cells. Well-orchestrated lineage-specific gene expression in the neural stem/progenitor cells is crucial for the generation of these neuronal and glial cell types (Dessaud et al., 2008; Panman et al., 2011). However, it is not quite clear how the differentiation of neural progenitors and the acquisition of their cell-fate is processed and programmed.
Different types of neurons and glial cells in the brain originate from separate progenitor pools in distinct areas (Marin and Rubenstein, 2001). Many transcription factors are important regulators during this neural differentiation process. One group of such transcription factors are the basic helix-loop-helix (bHLH) transcription factors which involved in the determination of neural cell fates (Akagi et al., 2004; Bertrand et al., 2002). Oligodendrocyte transcription factor (Olig) is a family of bHLH proteins that has received great attention for its essential role in neural cell specification and differentiation (Lu et al., 2001; Lu et al., 2002; Yu et al., 2013; Zhou and Anderson, 2002). The expression of the Olig gene family is predominantly restricted to the CNS (Lu et al., 2000; Zhou et al., 2000). Olig2, a member of Olig gene family, is required for the formation of oligodendrocyte and motoneuron progenitors; Olig2 null mouse embryos do not form oligodendrocytes and die at birth (Lu et al., 2002; Zhou and Anderson, 2002). Although the role of Olig2 in the development of the CNS has been well established, little is known about the molecular mechanism underlies spatiotemporal Olig2 expression during development. Other factors involved in neurogenesis during the CNS development include the Sox family transcription factors (Azim et al., 2009; Wegner and Stolt, 2005). Sox5 is a member of the Sox D group widely expressed in the developing forebrain and involved in the formation of the cephalic neural crest, in the control of cell cycle progression in neural progenitors, and of the sequential generation of distinct corticofugal neuron subtypes (Lai et al., 2008; Martinez-Morales et al., 2010; Perez-Alcala et al., 2004).
Neurogenesis is a developmental process highly conserved across a wide range of species (Finlay and Darlington, 1995; Gomez-Skarmeta et al., 2006). The evolutionarily conserved non-coding component of the genome is known to play an essential role in regulating this developmental process and has been receiving increased attention because of its predicted function in regulation of transcription, DNA replication, chromosome pairing, and chromosome condensation (Blackwood and Kadonaga, 1998; Jeziorska et al., 2009; Long and Miano, 2007). DNA sequences involved in gene regulation through the binding of transcriptional factors, termed as cis-regulatory elements, enhance or suppress gene expression in a spatiotemporal manner. The regulatory function is independent of orientation or position relative to the transcription sites (Blackwood and Kadonaga, 1998; Jeziorska et al., 2009). Several non-coding DNA fragments have been demonstrated to influence the expression of the Olig1 and/or Olig2 genes (Friedli et al., 2010; Sun et al., 2006). However, the trans-acting factors that activate these DNA fragments still need to be identified.
In search for distant cis-elements of Olig2 gene, we identified a highly evolutionarily conserved non-coding DNA element (CR5) upstream of the Olig2 gene transcription start site. CR5 plays essential roles in regulating gene expression in a subpopulation of Sox5+ cells that exclusively located in the ventral forebrain during the neonatal CNS development. We present evidence that the binding motif for Sox5 is important for the regulatory activity of this cis-element. Our findings may provide new insights into molecular mechanism underlying cell-specific gene expression.
Material and methods
Sequence alignment analysis
The sequence and annotation of the mouse Olig2 gene along with its homologs from the human, rat, cow and zebrafish genomes were retrieved using NCSRS (Doh et al., 2007). The sequences were analyzed by VISTA (Frazer et al., 2004) to identify highly conserved regions (CR). The percent identity and the length of the conserved sequence were used to calculate a score for each conserved region (score=percent identity+(length/60)). A limit of 2 kb in sequence length was implemented in order to isolate individual cis-elements for this study. Based on this scoring system, the percent identity was more heavily weighted to ensure that shorter and highly conserved sequences are not ranked below longer sequences with lower levels of conservation (Fig. S1).
Reporter plasmid constructions
Computationally-predicted conserved regions were amplified using the Taq PCR Kit (New England Biolabs, MA) following the routine Taq PCR reaction protocol. Primers used were summarized in Table 1. Mouse genomic DNA (Swiss Webster strain) was extracted from an adult mouse tail and used as the PCR template for all primers. A random extension sequence (CGATATAT) and the SpeI recognition sequence (ACTAGT) was added to the 5′ end of the forward primer, plus a random extension sequence and FseI recognition sequence (GGCCGGCC) was added to the 5′ end of the reverse primer. Then, the sticky end inserts were digested, gel purified, and ligated into the βGP-GFP backbone which was linearized with FseI and SpeI to generate experimental constructs (Fig. S1).
Table 1.
A list of computationally predicted conserved regions.
| cis-element | Chr16 start position | Chr16 end position | Conserved region (bp) | Primer sequence | |
|---|---|---|---|---|---|
| CR1 | 91062084 | 91062946 | 863 | forward | ATGCAAGCCTGTGTCTCTGACGAT |
| reverse | TTGAGCAGTCTGGGTGAGGACAAA | ||||
| CR2 | 91064822 | 91065087 | 266 | forward | AAGCCGCTGTCCACTATCCTTCAT |
| reverse | ACAGCTGTGGTCCAGGTGAATCTT | ||||
| CR3 | 91080246 | 91081050 | 805 | forward | ATGAGAACACGTCATTGGCTTCGG |
| reverse | TGACTTTGGGAAGAGGGAGAGAGT | ||||
| CR4 | 91081310 | 91081883 | 574 | forward | TGTGTAGGCCACAAACAGGAGACT |
| reverse | GCGAGCCGCTTAAGCTTCATCAAT | ||||
| CR5 | 91082197 | 91082573 | 377 | forward | ATTTGCGCTCTAAGGATGGCACAC |
| reverse | ACCTTGACTCCCAAGTAGCCCTAT | ||||
| CR6 | 91095187 | 91095771 | 585 | forward | TGCGGATCACGAGTAGCTTCCATT |
| reverse | TGCCGCACCATTTGAACTCTGAAG | ||||
| CR7 | 91097808 | 91098766 | 959 | forward | CCTTGCTTGCCAGGAGCATGAAAT |
| reverse | TGTTGACAATGTGGTGTTTGCGGG | ||||
| CR8 | 91100392 | 91100662 | 271 | forward | GTGCACCAATGCTCACTGAAATGC |
| reverse | TGGGTATGTTGAGTTGGAGGCACA | ||||
| CR9 | 91111442 | 91111756 | 315 | forward | TCGCTGCCTGAATGCTAGTAGGAA |
| reverse | TGGGAATGGAGAGTTCACTTGCCT | ||||
| CR10 | 91112578 | 91112794 | 217 | forward | TCCTCACATGCCCAAGCTCCTAAT |
| reverse | GCGATTGCTCCTCATTTCGTGCTA | ||||
| CR11 | 91121114 | 91121281 | 168 | forward | ACTGGCTTAGCTCCAACAGGGAAA |
| reverse | TGGTCTCTCTCCGCAAGCATGAAT | ||||
| CR12 | 91135088 | 91135543 | 456 | forward | TGACTCTTGTTCCCAGCCCTTTCA |
| reverse | TTGCCACTCTGGACACTTACAGCA | ||||
Animals and ethics statement
For in vivo and in utero electroporation experiments, Swiss Webster mice were purchased from Charles River Laboratories (Wilmington, MA) and maintained on a 12 h/12 h (7:00 a.m. to 7:00 p.m.) light/dark schedule from the time of arrival until the time of the experiment. Pregnancies were timed from the day at which a vaginal plug was detected, which was designated as embryonic day 0 (E0). By this convention, birth would normally occur on E19. This strain was also used as recipient to implant 0.5 dpc (days post coitum) embryos for transgenic animal studies. Mice were randomly assigned to distinct experimental groups. All studies were conducted in accordance with the NIH guidelines for the care and use of animals with approved animal protocol from the Institutional Animal Care and Use Committees at the Rutgers University.
In vivo electroporation
Individual experimental plasmid DNA constructs (2–3 μg/μl) were mixed with the control plasmid (2–3 μg/μl) to make the working DNA mixture. 1 μl DNA mixture was delivered into the mouse brain at postnatal day 0 (P0) targeting the SVZ progenitors (Fig. S1) with a Hamilton syringe. Five square pulses (80 V) of 50 ms duration with 950 ms intervals were then applied using a pulse generator ECM 830 (BTX Harvard Apparatus).
In utero electroporation
Timed pregnant Swiss Webster female mice (Charles River Labs) were anesthetized by intraperitoneal delivery of 0.7–0.9 ml of 2.5% avertin. The abdomen was opened to expose the uterine horns. The DNA solution (1 μg/μl experimental plasmid DNA+0.025% fast green) was injected into the lateral ventricle of embryonic brains at E15.5 using a pulled glass micropipette. After injection, the head of each embryo was placed between tweezer-type electrodes (BTX Harvard Apparatus) and five square electric pulses (37 V, 50 ms) were delivered with 950 ms intervals using a pulse generator ECM 830 (BTX Harvard Apparatus). The wall and skin of the abdominal cavity were then sutured and closed.
Generation of transgenic mice
Digested DNA (CR5-GFP) was gel purified using Seakem GTG agarose gel. Purified DNA (3–5 pg) was introduced by microinjection into 0.5 dpc (days post coitum) fertilized F1 (C57Bl/6J x CBA, Jackson Labs) mouse embryos and transferred to pseudopregnant recipient females. Reimplanted embryos were allowed to develop in utero to a time point that recipient female were sacrificed or allowed to give birth. Skin or tail DNA was prepared following standard protocol for genotyping. The transmission of the transgene in following generations was verified by Southern blotting and/or PCR genotyping (forward primer: GCA ACG TGC TGG TTA TTG TGC TGT; reverse primer: GTG GTA TTT GTG AGC CAG GGC ATT).
Tissue harvesting, processing and immunohistochemistry
Tissues from mouse brain were harvested at various embryonic and postnatal stages, fixed in 4% paraformaldehyde overnight, and washed in PBS 3 times for 5 min at 4 °C. Tissues were cryoprotected in 30% sucrose overnight until they became submerged in solution at 4 °C; they were embedded in OCT, sectioned at 10–15 μm thickness using a cryostat (Thermo 0620E), mounted on Superfrost slides (Fisher Scientific), and air-dried for 30 min. Immunostaining was performed using a Shandon Slide Rack (Thermo Scientific, MA) as previously described (Doh et al., 2010). Sections were incubated in blocking solution (0.05% Triton X-100, 10% goat serum, 3% BSA in PBS) for 30 min at room temperature followed by an overnight incubation with primary antibodies. GFP signal was retrieved by staining with anti-GFP (1:1000 dilutions, Invitrogen; 1:500 dilution, Abcam). Other primary antibodies included anti-NeuN (1:1000 dilution, Millipore), Sox5 (1:200 dilution, Santa Cruz), NG2 (1:200 dilution, Millipore), Tbr1 (1:200 dilution, Santa Cruz), Tbr2 (1:200 dilution, abcam), Gsx1 (1:200 dilution, Santa Cruz), PDGFRα (1:1000 dilution, abcam), GFAP (1:1000 dilution, Sigma), BLBP (1:1000 dilution, Chemicon), Pax6(1:200 dilution, Millipore), Mash1(1:100 dilution, BD Biosciences), S100β (1:1000 dilution, Sigma), PH3 (1:100 dilution, abcam), and Ki67 (1:100 dilution, BD Pharmingen). Staining with anti-Olig2 antibody (a gift from Dr. Charles Stiles at Harvard University) required pre-heating of slides with 1 mM Tris-EDTA buffer (PH 8.5) at 96 °C for 12 min to retrieve the antigen. Slides were then washed with PBS. Subsequently, tissue sections were incubated with appropriate secondary antibodies conjugated to different fluorophores (donkey anti-RbIgG Alexa 488 or donkey anti-GtIgG Alexa 488, 1:300, Jackson ImmunoResearch Labs) (donkey anti-mIgG Alexa 647 or donkey anti-RbIgG Alexa647, 1:150, Jackson ImmunoResearch Labs). Secondary antibodies were prepared in blocking buffer and applied at room temperature for 1 h, followed by three 10 min washes with PBS and a 5 min rinse in distilled water to remove salt crystals. After air-drying for 5 min, slides were mounted with 40 μl of mounting media with Dapi (Vector Laboratories).
Cell counting and statistical analysis
For counting double-labeled cells, confocal images were captured using a digital Zeiss AxioCam MR camera using a Zeiss Axio Imager Z1 with ApoTome application (Optical sectioning using structured illumination), and analyzed to detect GFP+ cells and cells stained with a specific marker. The number of GFP+ cells and GFP+ cells co-stained with a cell type-specific marker was counted manually in 4–5 sections from at least 3 animals per time point. In addition, DAPI staining was used to ensure that GFP+ cells were co-labeled with a cell marker (Fig. S2). The percentage of co-labeled cells over total number of GFP+ cells was determined. Results were presented as mean ± SD. Statistical significance was determined using student’s t-test at the level of p < 0.01.
Electrophoretic mobility shift assay (EMSA)
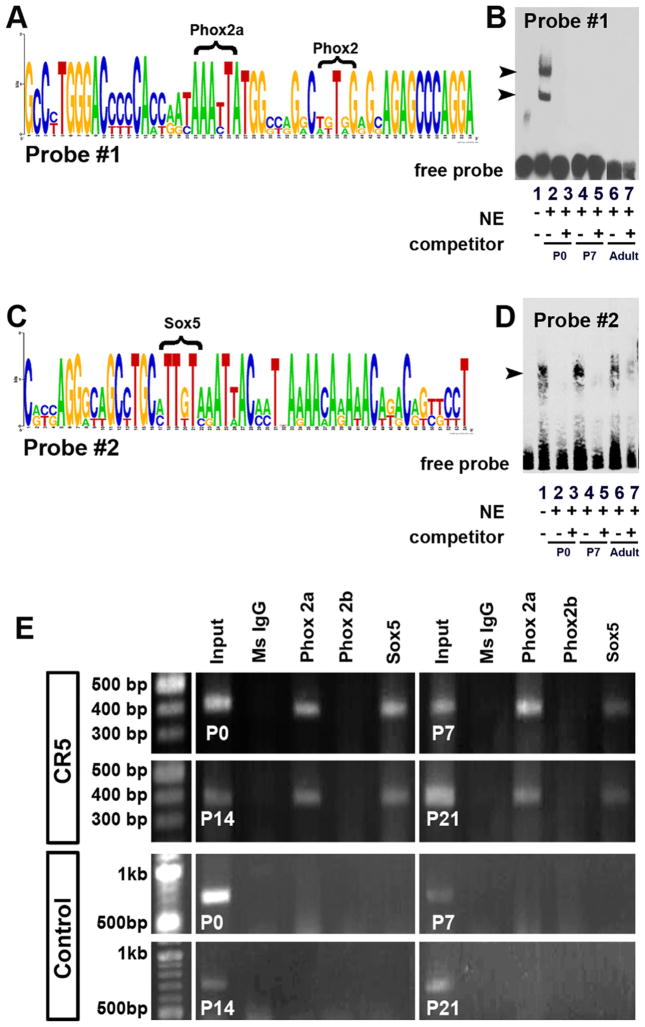
EMSA was performed to identify sub-regions of CR5 with potential binding activity with transcription factors. MatInspector, an online search tool from Genomatix (Germany) that provides potential trans-acting factor binding sites in nucleotide sequences (Cartharius et al., 2005; Quandt et al., 1995; Werner, 2000), was used to identify known sequence-specific binding sites for protein factors. Double-stranded DNA probes (40–80 bp in sequence length) were designed to span the entire conserved region (Table 2). Probes were synthesized (IDT, Piscataway, NJ) as single stranded oligonucleotides, biotinylated using the Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific) and annealed at room temperature one hour immediately prior to binding assay. Unlabeled single stranded probes were annealed and used as double-stranded competition probes. A ratio of 40:1 was used for competition probe to labeled probes. Nuclear extracts were prepared from the brain tissues (the VZ and SVZ of the cerebral cortex, the striatum and the ventral forebrain) of the Swiss Webster mice at various stages. The EMSA binding reaction and competition reaction were performed according to the LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific) protocol. The reaction mixture was loaded onto a 10% non-denaturing polyacrylamide gel containing 0.5 × TBE (40 mM Tris, 40 mM borate, 1 mM EDTA). Mini (8 × 8 × 0.1 cm) gels were run at 100 V for 220 min at 4 °C and dried under vacuum.
Table 2.
A list of EMSA probes for CR5.
| EMSA probes | Forward sequence | Potential binding transcription factors |
|---|---|---|
| Probe #1 | GCCCTGGGACCCCCACCAATAAATTATGGGTGGACATTAGGGGAGAGCCCAGGA | WT1 PHOX2a ISL2 PAX6 PIT1 EN1 PHOX2 |
| Probe #2 | CGTGAGGGCAGCCTGCATTGTAAATTACAATTAAAACAGAAACAGACAGTTCCT |
SOX5 HOXB6 MEOX1 NKX25 EN1 HOXB8 |
| Probe #3 | ATGACCAAGATGGGGACATTGTGTTTACCTACTTGAG | |
| Probe #4 | ACCTACTTGAGCAGAGGAGAAGGTGACCGTGAGGGCAGCCT | |
| Probe #5 | TCAGATAATCGCCTCCCTCCCGGCTGTCAGGGGTGCAGCCACTGCCAAT | |
| Probe #6 | CAGCCACTGCCAATTCACAGCGCCCTCCGAGAAAGTACCCTTGTCTGTGAT | |
| Probe #7 | GATGGCACACTCCATTTGATAATGGCTCTCATCTGCCTCAGATAATCGCC |
Chromatin immunoprecipitation assay (ChIP)
ChIP assays were performed to determine which transcription factors bind to the verified regulatory regions of CR5 in embryonic mouse tissue using a commercially available kit (MAGnify™ Chromatin Immunoprecipitation System, Invitrogen). The brain tissues (the VZ and SVZ of the cerebral cortex, the striatum and the ventral forebrain) were harvested from Swiss Webster mice at various stages. The brain tissue was homogenized through pipetting. Dissociated cells were cross-linked for 15 min at 37 °C with 1% formaldehyde. The cells were incubated in lysis buffer containing protease inhibitor for 5 min on ice. Sonication for 4~6 cycles of 30 s ‘ON’ and 30 s ‘OFF’ yielded 500–1000 bp fragments of sheared chromatin. The remaining steps were performed following the manufacturer’s instructions. Antibodies for immunoprecipitation included anti-Phox2a, Phox2b and Sox5 (Santa Cruz Biotechnology). ChIP DNA from individual experiments were amplified by PCR using the primers listed in Table 1.
Results
CR5 is active in the neural progenitor cells of neonatal forebrain
To identify cis-elements that regulate cell-specific gene expression, we performed comparative sequence analysis. Twelve conserved regions (CR1-CR12) surrounding the Olig2 gene locus were identified (Fig. S1A); their ability to direct tissue/cell-specific gene expression during mouse CNS development was screened by reporter assays using in vivo electroporation (IVE). The experimental DNA construct (containing a conserved region and GFP as a reporter) was co-injected with a transfection control (CAG-DsRed) into the developing mouse forebrain at postnatal day 0 (P0) and electroporated to transfect the neural progenitor cells in the ventricular zone (VZ) and/or the subventricular zone (SVZ) (Fig. S1B and C). Reporter GFP expression was examined in transfected tissues at various stages during the CNS development. Negative control experiments were performed with DNA constructs containing a minimal βGP alone without a conserved element or with a random DNA sequence comparable in size to ensure GFP reporter expression is solely due to activity of a CR. No GFP expression in transfected tissues was detected at any examined stages, indicating that the minimal βGP alone or a random sequence could not direct GFP expression. Five of the 12 CRs (CR1, 3, 4, 5 and 8; see Table 1) showed gene regulatory activity and CR5 was the strongest. Thus, its gene regulatory activities were further characterized. CR5 (Chr. 16:91082197–91082573) is a 377 bp non-coding DNA fragment located ~32 kbp upstream of Olig2 transcription start site. It is 100% conserved among various mouse strains, including C57BL/6, CBA, and Swiss Wester.
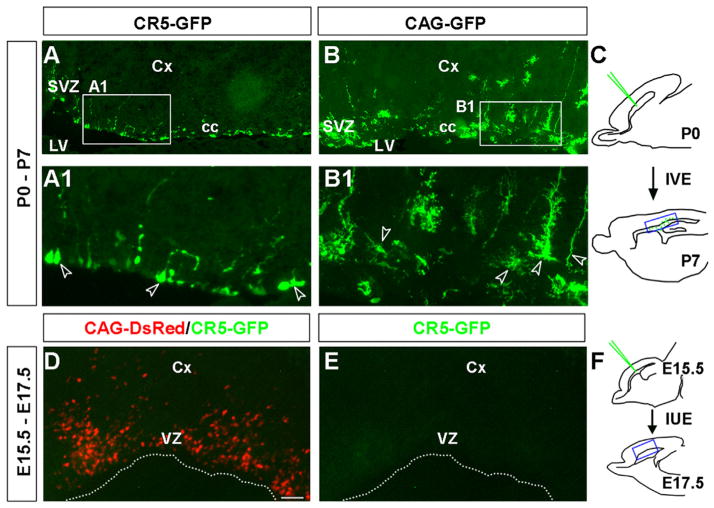
We further examined the transfection derived GFP+ cells at P7 after in vivo electroporation at P0. GFP+ cells were detected in the SVZ and the corpus callosum of the postnatal forebrain (Fig. 1A–C). The experimental CR5-GFP+ cells (Fig. 1A) and the control CAG-GFP+ cells (Fig. 1B) had dramatically different morphologies in the P7 forebrain. For example, the control CAG-GFP+ cells contain various morphologically different cells resemble astrocyte, oligodendrocyte, and radial glia (Fig. 1B,B1); while the majority of CR5-GFP+ cells were with a radial glia or a undifferentiated cell morphology (Fig. 1A,A1).
Fig. 1.
CR5 exhibits activity in multipotent cells at P0, but not during embryonic stages. Gene regulatory activity of CR5 was tested during both embryonic and postnatal forebrain development. For postnatal experiments, P0 SVZ was injected and electroporated with the experimental construct CR5-GFP or the control construct CAG-GFP individually. Brain tissues were harvested at P7 seven days after electroporation. Reporter expression was examined on sagittal sections. CR5-GFP+ cells were detected in the SVZ and “cc” areas of the postnatal forebrain (A). The control CAG-GFP+ cells were found in the SVZ, cc, and Cx areas and showed morphology of various cell types (B). Higher magnifications of the boxed areas in (A and B) are shown in (A1 and B1) and cells with specific morphology were indicated by arrowheads. (C) Diagrams of a mouse brain in sagittal plane depicting targeted cells by IVE at P0 and detection of reporter gene expression at P7. The location of A and B is shown in C indicated by a blue box. For embryonic experiments, developing VZ were co-electroporated with the experimental construct CR5-GFP and the control construct CAG-DsRed at E15.5. Brain tissues were harvested at E17.5 two days after electroporation. Reporter expression (GFP and DsRed) was examined on sagittal sections in embryonic forebrain at E17.5. No CR5-GFP expression was detected (D and E), while large amount of CAG-DsRed+ cells were observed in all transfected brains (red cells in D). (F) Diagrams of a mouse brain in sagittal plane depicting targeted cells by IUE at E15.5 and detection of reporter gene expression at E17.5. Blue boxed area in F is the location of D and E. Cx, cortex; cc, corpus callosum; LV, lateral ventricle; VZ, ventricular zone; SVZ, subventricular zone. IUE, in utero electroporation; IVE, in vivo electroporation. Scale bar=50 μm.
Since CR5 possess gene regulatory activity in neonatal stages, its activity during embryonic development was then examined using in utero electroporation (IUE). The lateral ventricles at embryonic day 15.5 (E15.5) were injected and electroporated with the experimental construct (CR5-GFP) to transfect the neural progenitors lining the VZ/SVZ. No CR5-GFP expression was detected in the transfected tissues at E17.5 two days after IUE (Fig. 1D–F), indicating that CR5 may not be active at this embryonic stage. To rule out the possibility that the lack of CR5-GFP+ expression might be caused by the failure of electroporation, a control construct CAG-DsRed was co-electroporated with CR5-GFP construct. Strong CAG-DsRed+ cells (red) were observed in all transfected brain tissues (Fig. 1D), confirming that CR5 activity was not detected in the embryonic stage.
CR5 activity is preferentially in the Sox5+/NG2+ progenitors of the neonatal forebrain
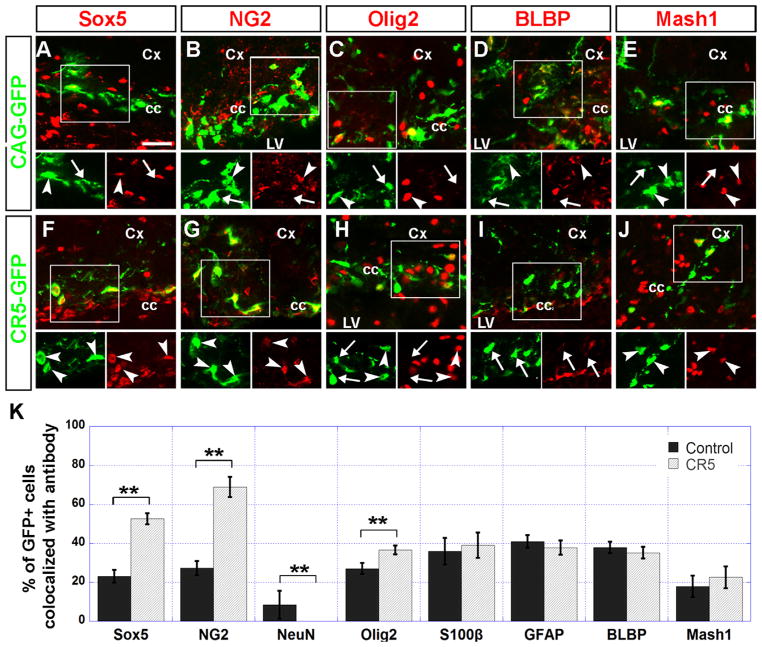
We next determined the cellular identity of transfection-derived CR5-GFP+ cells by immunostaining with neural stem/progenitor markers of Sox5 (Fig. 2A and F), NG2 (Fig. 2B and G), Olig2 (Fig. 2C and H), BLBP (Fig. 2D and I), Mash1 (Fig. 2E and J), GFAP (Fig. S3 A and B); astrocyte marker S100β (Fig. S3 C and D), and neuronal marker NeuN (Fig. S3 E and F). Compared with the control CAG-GFP+ cells, a significantly higher percentage of CR5-GFP+ cells were co-labeled with Sox5 (52.6% vs 23.1%) and NG2 (68.9% vs 27.3%) (Fig. 2K). We found no CR5-GFP+ cells were co-labeled with NeuN (Fig. S3E and F and Fig. 2K), indicating that CR5 activity is not in the differentiated neurons. The fact that the majority of the CR5-GFP+ cells were co-labeled with Sox5 and NG2 indicates that CR5 was preferentially active in Sox5+/NG2+ progenitors during neonatal forebrain development. To our surprise, only 37.8% of CR5-GFP+ cells were co-labeled with Olig2 (Fig. 2C, H and K), suggesting that CR5 may not be a cis-element/enhancer for Olig2 expression (Fig. 2C and H and Table S1).
Fig. 2.
CR5 activity is preferentially in the Sox5+ and NG2+ progenitors. Sagittal sections of the P7 mouse forebrain were immunostained with anti-GFP (green) antibody and specific cell markers (red). GFP+ cells observed in the SVZ, corpus callosum, and neocortical areas after in vivo electroporation at P0 were further examined for the expression pattern of progenitor markers including Sox5 (A and F), NG2 (B and G); Olig2 (C and H); radial glia marker BLBP (D and I); and type C cell marker Mash1 (E and J). Boxed areas were shown in green and red channel separately. Double-labeled cells were indicated by arrowheads. Cells only express GFP but not labeled with a cell marker were indicated by arrows. (K) A histograph showing the percentage of double labeled cells (GFP+/cell marker+) over GFP+ cells. Error bars indicate values of the standard deviation. The histograph also includes data of immunostaining with astrocyte markers GFAP, S100β, and neuronal marker NeuN (see Fig. S3). Compare with the control group, a higher percentage of the CR5-GFP+ cells were co-stained with Sox5, NG2. No significant difference was observed with the S100β, GFAP, BLBP, and Mash1 co-labeling. The significance of difference between CR5 and the control group was assessed by student’s t-test (see Table S1). (**p < 0.01; n=3). Cx, cortex; cc, corpus callosum; LV, lateral ventricle. Scale bar=20 μm.
CR5 activity is exclusively in a subset of Sox5+ cells in the neonatal ventral forebrain
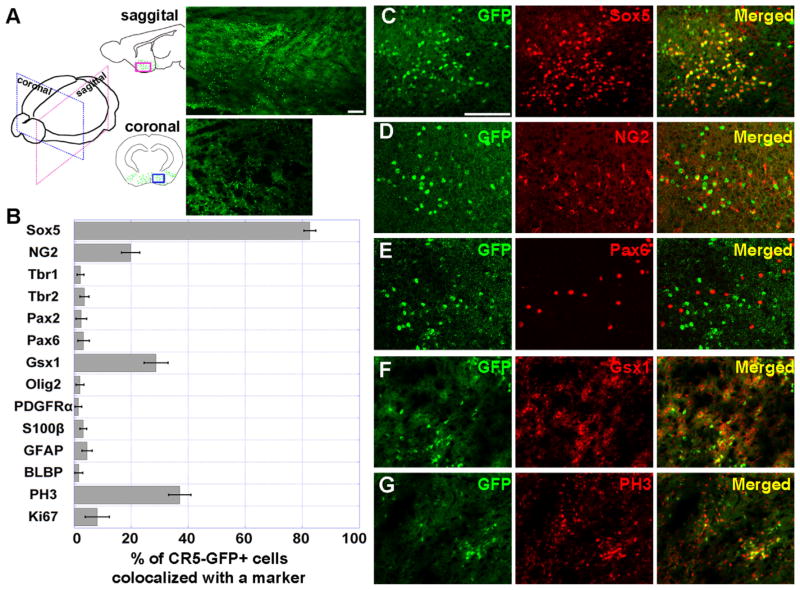
The electroporation targets only a limited regional cell population and results in a transient episomal gene expression. To determine the spatiotemporal gene regulatory activity of CR5, we thus generated transgenic mice containing the CR5-GFP construct. The transgene CR5-GFP expression was examined at various embryonic (E11.5, E13.5, E15.5, E17.5, E19.5) and postnatal stages (P0, P7, P14, and P21). Consistent with results obtained from the in vivo/utero electroporation, CR5-GFP+ cells in transgenic mice were detected at a neonatal stage, E19.5 (n=2)/P0 (n=9); and interestingly, they were predominately located in the ventral forebrain (Fig. 3A). Even with extensive examinations, no GFP expression was detected at embryonic stages and other postnatal stages we examined (data not shown). Thus, CR5 activity is exclusively in the neonatal ventral forebrain.
Fig. 3.
CR5 is preferentially active in Sox5 expressing cells of the ventral forebrain around postnatal day 0. (A) Sagittal and coronal brain sections of P0 transgenic mouse show that CR5-GFP+ cells were primarily located in ventral forebrain. Diagrams of the sagittal and coronal planes show the distribution pattern of CR5-GFP+ cells indicated by green dots. Immunostaining results with anti-GFP antibody of the boxed area were displayed on the right images. CR5-GFP+ cells in the ventral forebrain from sagittal samples were immunostained with specific cell marker Sox5 (C), NG2 (D), Pax6 (E), Gsx1 (F), and PH3 (G). A histograph showing the percentage of double labeled cells (GFP+/cell marker+) over GFP+ cells (B), including other markers, e.g., neuronal marker Tbr1 and Tbr2; oligodendrocyte marker Olig2 and PDGFRα, radial glia marker GFAP and BLBP; and astrocyte marker S100β (Fig. S4). Error bars indicate values of the standard deviation (see Table S2). Scale bar=50 μm.
We then determined the cellular identities of transgenic CR5-GFP+ cells by immunostaining the sagittal sections of P0 samples. We found that the vast majority of CR5-GFP+ cells were co-labeled with Sox5 (82.6%, n=3; p < 0.01) (Fig. 3B and G). Although some CR5-GFP+ cells also co-labeled with other progenitor markers, the percentages of co-labeled cells were relatively low, e.g., NG2 (19.8%, n=3, Fig. 3B and C), Gsx1 (28.7%, n=3, Fig. 3B and F), and mitotic cell marker PH3 (37%, n=3, Fig. 3B and G). Thus, CR5 activity is predominantly in the Sox5+ cells.
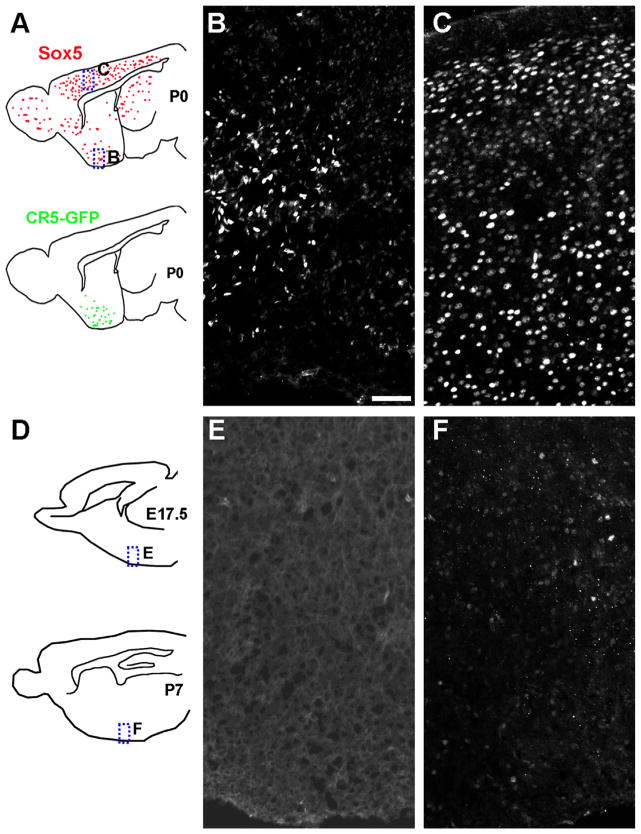
Sox5 is known to be expressed in neural progenitors and controls neocortical neuron differentiation (Azim et al., 2009; Greig et al., 2013; Perez-Alcala et al., 2004). However, its role in the ventral forebrain development is not clear. We then determined the pattern of Sox5 expression by immunohistochemistry. A peak Sox5 expression was detected in both the dorsal and ventral forebrain at P0 (Fig. 4A–C). Sox5 expression was not detected at E17.5 (Fig. 4D and E) and dramatically reduced by P7 (Fig. 4D and F) in the ventral forebrain. This ventral forebrain expression pattern of Sox5 is well correlated with CR5 activity in this brain region.
Fig. 4.
Sox5 expression is in the neonatal ventral forebrain. Sagittal sections from mouse brain at P0 (A–C), E17.5 (D and E), and P7 (D and F) were immunostained with anti-Sox5 antibody. Strong Sox5 expression was detected in both the ventral forebrain (B) and dorsal cortex (C) at P0, but not detected in the E17.5 ventral forebrain (E). Weak Sox5 expression was detected in the P7 ventral forebrain (F). Diagram of sagittal brain are shown in A and D. Dashed boxed area in the diagram are shown in the right images with corresponding labels. Sox5 expression pattern at P0 brain is indicated by red dots while CR5-GFP expression is indicated by green dots. VB, ventral forebrain; CX: cortex. Scale bar=50 μm.
Similar to electroporation results, a small percentage of CR5-GFP+ cells were co-labeled with oligodendrocyte-lineage markers Olig2 (2%) and PDGFRα (1.5%), astrocyte marker S100β (3.2%), radial glia markers GFAP (4.5%) and BLBP (1.6%), intermediate progenitor markers Tbr1 (2.2%), Tbr2 (3.6%), and Pax6 (3.3%) (Fig. 3 and Fig. S4). These results suggest that CR5 activity is time-specific, brain region-restricted and is highly induced in a subset of Sox5+ progenitor cells in the ventral forebrain.
Specific nuclear factors bind to CR5
Activation of CR5 requires the binding of transcription factor(s). To determine which specific nuclear factors may bind to CR5, we performed the electrophoretic mobility shift assays (EMSA). A total of 7 probes were designed to span the whole length of CR5 (Table 2). EMSA results showed that two sub-regions within CR5 corresponding to probe #1 (Fig. 5A and B) and probe #2 (Fig. 5C and D) have specific nuclear protein-binding activity. We analyzed the transcription factor binding sites within these two sub-regions using MatInspector (Genomatix, Germany) (Cartharius et al., 2005; Quandt et al., 1995) to identify candidate binding sites for the transcription factors (the last column in Table 2). After further literature search, we narrowed down the number of candidate transcription factor binding sites to Sox5, Phox2a and Phox2b. To determine whether these three factors bind with CR5 in vivo, we performed the chromatin immunoprecipitation (ChIP) assays using chromatin obtained from brain tissues at various developmental stages (P0, P7, P14, P21 and adult) and immunoprecipitated using antibodies against the three protein factors individually. Results from ChIP assays showed that Sox5 and Phox2a (but not Phox2b) bound with CR5 (Fig. 5E), suggesting that binding of Sox5 and Phox2a is important for the activation of CR5 and its gene regulatory activity.
Fig. 5.
CR5 interacts with specific nuclear protein factors. Analysis of homologous CR5 sequences from 8 species by a web based sequence analysis tool Weblogo revealed two highly conserved motifs (A and C). Electrophoretic mobility shift assays (EMSA) were performed to identify the in vitro binding activity of nuclear proteins with CR5. Among a total number of seven probes that cover the entire CR5, probe #1 (A and B) and probe #2 (C and D) showed binding activity. The competition assay was carried out using unlabeled probes at 40-fold higher concentration. The nuclear extracts from developing mouse brain at various stages were obtained (P0: lanes 2 and 3; P7: lanes 4 and 5; Adult: lanes 6 and 7). For probe #1, the arrowheads indicate the retarded band obtained using the P0 brain nuclear extracts (lane 2). The shift disappeared when competitor was added (lane 3). No shift was observed in the assays using P7 and adult mouse brain nuclear extracts (lanes 4–7). For Probe #2, the retarded bands were observed for all the stages (arrowhead, lane 2, 4, 6). The shift diminished when competitor was added (arrowhead, lane 3, 5, 7). Chromatin immunoprecipitation (ChIP) assays detected in vivo binding of protein factors (Phox2a, Phox2b, and Sox5) to CR5 (E). Chromatin obtained from P0, P7, P14, and P21 brain tissues of mouse pups was immunoprecipitated using antibodies against individual protein factor. The pure mouse IgG antibody was used as a negative control. The input represented 1% of the total chromatin extract. The precipitated DNA fragments were amplified by a set of primers flanking CR5 sequences. The PCR products of the expected size of 416 bp were obtained. As a negative control, primers flanking a random sequence of ~700 bp which does not include specific binding sites were tested and no PCR product was detected.
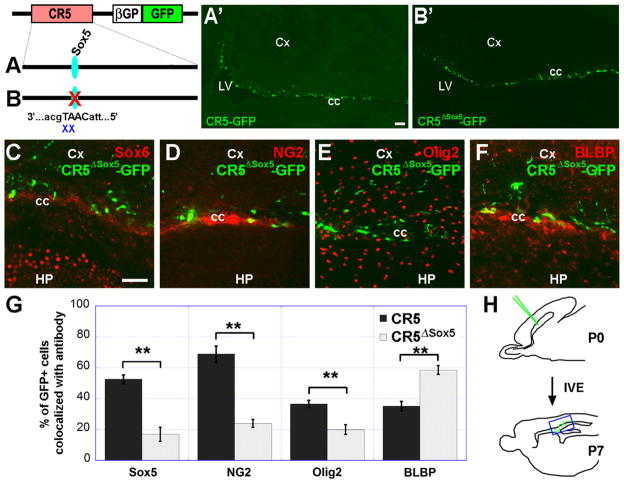
Lack of Sox5 binding site in CR5 alters gene regulatory activity
Since the majority of CR5-GFP+ were co-labeled with Sox5 (Fig. 3B and G) and Sox5 is known to control cell cycle progression in neural progenitors and production of distinct neuronal cell types (Martinez-Morales et al., 2010; Perez-Alcala et al., 2004), we thus focused our further investigation on the role of Sox5 in CR5 activation. We next determined whether the Sox5 binding site is required for gene regulatory activity using site-directed mutagenesis assay. Mutant CR5ΔSox5-GFP construct (deletion of AT from CAAT) (Fig. 6A and B) was injected into the P0 mouse brain (for all data points, n ≥ 3) followed by electroporation to transfect the neural progenitors in the SVZ. To our surprise, GFP+ cells were observed from the mutant construct CR5ΔSox5-GFP, comparable to the wild-type CR5-GFP+ cells (Fig. 6A′) with no obvious difference (Fig. 6B′), suggesting that Sox5 binding site is not essential for CR5 activation. However, after further analysis of the molecular identities of the resulting GFP+ cells, we found that CR5ΔSox5-GFP+ cells and CR5-GFP+ cells have a different distribution (Fig. 6G and Table S2). Compared with the CR5-GFP+ cell population, a significant lower percentage of CR5ΔSox5-GFP+ cells were co-localized with Sox5 (52.6% vs 16.98%; Fig. 6C), NG2 (68.9% vs 23.98%; Fig. 6D) and Olig2 (36.6% vs 19.98%; Fig. 6E); while a higher percentage of CR5ΔSox5-GFP+ cells were co-labeled with BLBP (35.2% vs 58.58%; Fig. 6F). These findings suggest that although Sox5 binding site is not required for the activation of CR5, it affects the cellular specificity of CR5 in its gene regulatory activity.
Fig. 6.
Sox5 binding site affects gene regulatory activity of CR5. Schematic representation of plasmid constructs of CR5-GFP (A) and CR5ΔSox5-GFP with deletion of “AT” from Sox5 binding site “CAAT” (B). Mutated Sox5 binding site is indicated by a red X. Sagittal sections of the P7 mouse forebrain after in vivo electroporation at P0 showing GFP+ cells in transfected brain tissues with CR5-GFP (A′) or CR5ΔSox5-GFP (B′) constructs. The location of A′–B′ is indicated by blue box in H. GFP+ cells were further examined for the expression of neural stem/progenitor markers: Sox5 (C), NG2 (D), Olig2 (E), and BLBP (F). A histograph showing the percentage of double labeled cells (GFP+/cell marker+) over GFP+ cells (G). Error bars indicate values of the mean of standard deviation. Compare with the wild-type CR5 group, a lower percentage of CR5ΔSox5-GFP+ cells express neural stem/progenitor markers Sox5 and NG2; and a higher percentage of CR5ΔSox5-GFP+ cells express BLBP. (H) Diagrams of a mouse brain in sagittal plane depicting targeted cells by IVE at P0 and detection of reporter gene expression at P7. The significance of difference was assessed by student’s t-test. (**p < 0.01; n=3). Cx, cortex; cc, corpus callosum; LV, HP, Hippocampus; lateral ventricle. Scale bar=50 μm.
Discussion
The molecular mechanism underlying lineage-specific gene expression is a key to the understanding of how cell diversity is generated during the development of the CNS. In this study, we report the identification of a novel cis-element, CR5, an evolutionarily conserved non-coding DNA fragment located upstream of Olig2 locus, capable of regulating gene expression especially in a subpopulation of Sox5+ progenitor cells in the ventral forebrain during neonatal development. We demonstrated that Sox5 binding site in CR5 is important for cell-specific gene regulatory activity.
The VZ/SVZ progenitors in the embryonic and postnatal mammalian forebrain are known to generate olfactory interneurons, astrocytes, and oligodendrocytes (Marshall et al., 2005; Marshall et al., 2003; Menn et al., 2006). The SVZ population also contains radial glia that serves as neural progenitors (Middeldorp et al., 2010). Consistent with previous observations, our electroporation experiments showed that CAG driven reporter GFP or DsRed expression was detected in all above mentioned cell types in both embryonic and neonatal stages, while CR5 driven GFP expression was only detected in the SVZ at a stage between P0 to P7, but not in the embryonic stages (Figs. 1 and 2 and Fig. S3). Together with data from the transgenic mice (Fig. 3), we demonstrated that CR5 activity exists only in the neonatal developing brain.
In both the in vivo electroporation and transgenic mouse studies, the majority of CR5-GFP+ cells were co-localized with Sox5 (82.6% in transgenic animals and 52.6% in electroporation experiments) indicating that CR5 activity is preferentially in a subpopulation of Sox5+ progenitors. The difference in the percentage of CR5-GFP+/Sox5+ cells observed in the two experiments may be due to the fact that CR5-GFP construct targets only the SVZ progenitors at P0 in the electroporation experiment and resulted in a transient episomal GFP expression; while in transgenic animals, CR5-GFP construct targets the embryonic stem cells, CR5-GFP was integrated into the genome, and resulted in a spatiotemporal regulated GFP expression. We noticed that there was also a large difference in the percentages of CR5-GFP+/NG2+ cells, i.e., 19.8% in transgenic animals and 68.9% in electroporation experiments. This can be explained again by the difference of the two methods targeting different cell population, i.e., embryonic neural stem cells in transgenic mouse study vs SVZ progenitors at neonatal stage (P0) in electroporation experiments.
It is interesting that CR5-GFP+ cells were not detected in the postnatal SVZ of the transgenic animals (Figs. 1–3), indicating that CR5 activity may not be in the SVZ region of the cerebral cortex. Alternatively, it is also possible that the CR5-GFP level was too low to be detected. Although, CR5 activity may not be in the cerebral cortex, the electroporation derived GFP+ cells may be a result of the electrical stimulation to the SVZ progenitors. The transgenic experiments reveal the spatiotemporal activity of CR5 during CNS development, while the in vivo electroporation experiments provide a rapid screen method for functional cis-elements. Our results from the CR5-GFP transgenic mouse study provide not only evidence to support the conclusion that CR5 activity is in a subpopulation of Sox5+ progenitors but also further determined that this progenitor population is located in the neonatal ventral forebrain (Figs. 3 and 4). Thus, CR5 activity is in a spatiotemporally restricted manner.
Sox5 is well known for its function in controlling cell cycle and sequential generation of distinct corticofugal neuron subtypes. Our analysis revealed a novel role of Sox5 in neural progenitors of the neonatal ventral forebrain regulated by CR5 activity (Fig. 3). To support this, a transient Sox5 expression was detected in the P0 ventral forebrain (Fig. 4). By P7, Sox5 expression was dramatically decreased and barely detectable (Fig. 4). The observation that no CR5-GFP+ cells were detected in the transgenic neocortex where Sox5 is highly expressed implies that CR5 was not active in this brain region. Thus, Sox5 expression in different brain areas may be regulated by different mechanisms.
Despite its location upstream of the Olig2 gene, CR5 may not directly regulate Olig2 expression as the majority of CR5-GFP+ cells were not co-localized with Olig2 or oligodendrocyte progenitor marker PDGFRα (Figs. 2–3 and Fig. S3). Thus, CR5 activity may not be in the oligodendrocyte lineage. Since CR5-GFP+ cells were co-localized with Sox5 and not with Olig2, thus, Sox5 may not directly regulate Olig2 expression. This is consistent with the role of Sox5 in suppressing myelin gene expression in oligodendrocytes (Stolt et al., 2006). Sox5 expression is found in VZ cells, astroglia, and specific neuronal populations (Batista-Brito et al., 2009; Lai et al., 2008) and not in differentiating oligodendrocytes (Stolt et al., 2006).
The diversity of neuronal progeny in the early postnatal brain contributes to the anatomical organization and cell specification (Lledo et al., 2008). Recent studies have revealed that distinct molecules mobilize stem cells toward neurogenesis in different regions (Lopez-Juarez et al., 2013). Many molecules exerts regulatory function in a region-specific manner (Brill et al., 2008; Lledo et al., 2008; Lopez-Juarez et al., 2013; Merkle et al., 2007). Given the regionally restricted expression of CR5-GFP, our results suggest that CR5 is an important regulatory element for proper lineage progression of Sox5+/NG2+ subpopulation. Previous studies demonstrated that Olig2 is not co-expressed with NG2 in the ventral forebrain; NG2+/Olig2− cells in this area differentiated into astrocytes, but not oligodendrocytes (Zhu et al., 2012). A subpopulation of CR5-GFP+/NG2+ in the transgenic ventral brain indicates that CR5 might play a role in controlling cell diversity in this brain region. In addition, a recent study suggests that Gsx1 is likely to be a regulator in the development of lateral and ventral neural stem cells (Lopez-Juarez et al., 2013). Our finding that a subpopulation of CR5-GFP+ cells in transgenic animals were co-localized with Gsx1 (Fig. 3F) further supports the notion that CR5 is involved in the generation of cell diversity in the forebrain.
Gene regulatory ability of CR5 is attributed to the binding activities with specific protein factors. We identified Sox5 as a CR5 binding trans-acting factor, which was confirmed by EMSA, ChIP (Fig. 5), and site-directed mutagenesis and in vivo electroporation assays (Fig. 6). Although no obvious changes in the level of GFP expression was detected with CR5ΔSox5-GFP construct, the mutant Sox5 binding site did alter the composition of reporter gene expression in the resulting GFP+ cell population: a dramatic decrease of the percentage of CR5ΔSox5-GFP+ cells co-labeled with Sox5 and NG2, and a significant increase in the percentage of CR5ΔSox5-GFP+ cells co-labeled with BLBP (Fig. 6). These findings suggest that although Sox5 binding site is not required for the activation of CR5, it does affect the specificity of CR5 in Sox5 cell population.
Sox5 belongs to SoxD group, which contains two other factors Sox6 and Sox13 (Guth and Wegner, 2008). Our analysis showed that CR5 contains binding sites for Sox5, but not for Sox6 or Sox13. For this reason, the role of Sox6 or Sox13 in regulating CR5 activity was not examined. However, the possibility that Sox6 and/or Sox13 may interact with Sox5 and participate in CR5 activation cannot be ruled out.
A major challenge in understanding how various cell types in the CNS are generated is to elucidate the mechanism of cell-specific gene expression. The identification of the novel gene regulatory element in this study represents one step in this effort. Based on our findings, we propose that the cis-element CR5 and its binding factors participate in the regulation of cell-specific gene expression in a spatiotemporal manner. Our study provides an example of such cell-specific gene expression mediated by a direct interaction of trans-acting factors (e.g., Sox5 and possibly Phox2a) with a cis-element (e.g., CR5). This study also represents a useful and effective method for the functional study of non-coding regulatory sequences and their binding protein factors in cell lineage development.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Connie Cepko for a reporter construct of plasmid DNA pCAG-GFP, Dr. Charles Stiles for Olig2 antibody, Chris Ricupero in Dr. Ronald Hart’s lab for helping with ChIP experiments, and the members of the Cai lab for helpful discussion and proof-reading. We would like to acknowledge the assistance of the Transgenic and Knockout Mouse Shared Resource of the Rutgers Cancer Institute of New Jersey -NCI/CCSG (P30CA072720) and Robert Wood Johnson Foundation support of the Child Health Institute of New Jersey. This work was supported in part by the grant EY018738 from the National Institute of Health; the New Jersey Commission on Spinal Cord Research Grants (08-3074-SCR-E-0 and 10A-003-SCR1) and the Busch Biomedical Research Awards (6-49121).
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2014.06.010.
References
- Akagi T, Inoue T, Miyoshi G, Bessho Y, Takahashi M, Lee JE, Guillemot F, Kageyama R. Requirement of multiple basic helix-loop-helix genes for retinal neuronal subtype specification. J Biol Chem. 2004;279:28492–28498. doi: 10.1074/jbc.M400871200. [DOI] [PubMed] [Google Scholar]
- Azim E, Jabaudon D, Fame RM, Macklis JD. SOX6 controls dorsal progenitor identity and interneuron diversity during neocortical development. Nat Neurosci. 2009;12:1238–1247. doi: 10.1038/nn.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista-Brito R, Rossignol E, Hjerling-Leffler J, Denaxa M, Wegner M, Lefebvre V, Pachnis V, Fishell G. The cell-intrinsic requirement of Sox6 for cortical interneuron development. Neuron. 2009;63:466–481. doi: 10.1016/j.neuron.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand N, Castro DS, Guillemot F. Proneural genes and the specification of neural cell types. Nat Rev Neurosci. 2002;3:517–530. doi: 10.1038/nrn874. [DOI] [PubMed] [Google Scholar]
- Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- Brill MS, Snapyan M, Wohlfrom H, Ninkovic J, Jawerka M, Mastick GS, Ashery-Padan R, Saghatelyan A, Berninger B, Gotz M. A dlx2- and pax6-dependent transcriptional code for periglomerular neuron specification in the adult olfactory bulb. J Neurosci: Off J Soc Neurosci. 2008;28:6439–6452. doi: 10.1523/JNEUROSCI.0700-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt SJ, Fuccillo M, Nery S, Noctor S, Kriegstein A, Corbin JG, Fishell G. The temporal and spatial origins of cortical interneurons predict their physiological subtype. Neuron. 2005;48:591–604. doi: 10.1016/j.neuron.2005.09.034. [DOI] [PubMed] [Google Scholar]
- Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- Dessaud E, McMahon AP, Briscoe J. Pattern formation in the vertebrate neural tube: a sonic hedgehog morphogen-regulated transcriptional network. Development. 2008;135:2489–2503. doi: 10.1242/dev.009324. [DOI] [PubMed] [Google Scholar]
- Doh ST, Hao H, Loh SC, Patel T, Tawil HY, Chen DK, Pashkova A, Shen A, Wang H, Cai L. Analysis of retinal cell development in chick embryo by immunohistochemistry and in ovo electroporation techniques. BMC Dev Biol. 2010;10:8. doi: 10.1186/1471-213X-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh ST, Zhang Y, Temple MH, Cai L. Non-coding sequence retrieval system for comparative genomic analysis of gene regulatory elements. BMC Bioinform. 2007;8:94. doi: 10.1186/1471-2105-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BL, Darlington RB. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucl Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedli M, Barde I, Arcangeli M, Verp S, Quazzola A, Zakany J, Lin-Marq N, Robyr D, Attanasio C, Spitz F, Duboule D, Trono D, Antonarakis SE. A systematic enhancer screen using lentivector transgenesis identifies conserved and non-conserved functional elements at the Olig1 and Olig2 locus. PLoS One. 2010;5:e15741. doi: 10.1371/journal.pone.0015741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Lenhard B, Becker TS. New technologies, new findings, and new concepts in the study of vertebrate cis-regulatory sequences. Dev Dyn. 2006;235:870–885. doi: 10.1002/dvdy.20659. [DOI] [PubMed] [Google Scholar]
- Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci. 2013;14:755–769. doi: 10.1038/nrn3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guth SI, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci: CMLS. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M. Neuronal subtype specification in the cerebellum and dorsal hindbrain. Dev Growth Differ. 2012;54:317–326. doi: 10.1111/j.1440-169X.2012.01330.x. [DOI] [PubMed] [Google Scholar]
- Jeziorska DM, Jordan KW, Vance KW. A systems biology approach to understanding cis-regulatory module function. Semin Cell Dev Biol. 2009;20:856–862. doi: 10.1016/j.semcdb.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Lai T, Jabaudon D, Molyneaux BJ, Azim E, Arlotta P, Menezes JR, Macklis JD. SOX5 controls the sequential generation of distinct corticofugal neuron subtypes. Neuron. 2008;57:232–247. doi: 10.1016/j.neuron.2007.12.023. [DOI] [PubMed] [Google Scholar]
- Lledo PM, Merkle FT, Alvarez-Buylla A. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31:392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo L, Dormand E, Greenwood A, Anderson DJ. Comparison of the generic neuronal differentiation and neuron subtype specification functions of mammalian achaetescute and atonal homologs in cultured neural progenitor cells. Development. 2002;129:1553–1567. doi: 10.1242/dev.129.7.1553. [DOI] [PubMed] [Google Scholar]
- Long X, Miano JM. Remote control of gene expression. J Biol Chem. 2007;282:15941–15945. doi: 10.1074/jbc.R700010200. [DOI] [PubMed] [Google Scholar]
- Lopez-Juarez A, Howard J, Ullom K, Howard L, Grande A, Pardo A, Waclaw R, Sun YY, Yang D, Kuan CY, Campbell K, Nakafuku M. Gsx2 controls region-specific activation of neural stem cells and injury-induced neurogenesis in the adult subventricular zone. Genes Dev. 2013;27:1272–1287. doi: 10.1101/gad.217539.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu QR, Cai L, Rowitch D, Cepko CL, Stiles CD. Ectopic expression of Olig1 promotes oligodendrocyte formation and reduces neuronal survival in developing mouse cortex. Nat Neurosci. 2001;4:973–974. doi: 10.1038/nn718. [DOI] [PubMed] [Google Scholar]
- Lu QR, Sun T, Zhu Z, Ma N, Garcia M, Stiles CD, Rowitch DH. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Lu QR, Yuk D, Alberta JA, Zhu Z, Pawlitzky I, Chan J, McMahon AP, Stiles CD, Rowitch DH. Sonic hedgehog–regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Marin O, Rubenstein JL. A long, remarkable journey: tangential migration in the telencephalon. Nat Rev Neurosci. 2001;2:780–790. doi: 10.1038/35097509. [DOI] [PubMed] [Google Scholar]
- Marshall CA, Novitch BG, Goldman JE. Olig2 directs astrocyte and oligodendrocyte formation in postnatal subventricular zone cells. J Neuroscin: Off J Soc Neurosci. 2005;25:7289–7298. doi: 10.1523/JNEUROSCI.1924-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CA, Suzuki SO, Goldman JE. Gliogenic and neurogenic progenitors of the subventricular zone: who are they, where did they come from, and where are they going? Glia. 2003;43:52–61. doi: 10.1002/glia.10213. [DOI] [PubMed] [Google Scholar]
- Martinez-Morales PL, Quiroga AC, Barbas JA, Morales AV. SOX5 controls cell cycle progression in neural progenitors by interfering with the WNT-beta-catenin pathway. EMBO Rep. 2010;11:466–472. doi: 10.1038/embor.2010.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menn B, Garcia-Verdugo JM, Yaschine C, Gonzalez-Perez O, Rowitch D, Alvarez-Buylla A. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci: Off J Soc Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Mirzadeh Z, Alvarez-Buylla A. Mosaic organization of neural stem cells in the adult brain. Science. 2007;317:381–384. doi: 10.1126/science.1144914. [DOI] [PubMed] [Google Scholar]
- Middeldorp J, Boer K, Sluijs JA, De Filippis L, Encha-Razavi F, Vescovi AL, Swaab DF, Aronica E, Hol EM. GFAPdelta in radial glia and subventricular zone progenitors in the developing human cortex. Development. 2010;137:313–321. doi: 10.1242/dev.041632. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat Rev Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Panman L, Andersson E, Alekseenko Z, Hedlund E, Kee N, Mong J, Uhde CW, Deng Q, Sandberg R, Stanton LW, Ericson J, Perlmann T. Transcription factor-induced lineage selection of stem-cell-derived neural progenitor cells. Cell Stem Cell. 2011;8:663–675. doi: 10.1016/j.stem.2011.04.001. [DOI] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Alcala S, Nieto MA, Barbas JA. LSox5 regulates RhoB expression in the neural tube and promotes generation of the neural crest. Development. 2004;131:4455–4465. doi: 10.1242/dev.01329. [DOI] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucl Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Sun T, Hafler BP, Kaing S, Kitada M, Ligon KL, Widlund HR, Yuk DI, Stiles CD, Rowitch DH. Evidence for motoneuron lineage-specific regulation of Olig2 in the vertebrate neural tube. Dev Biol. 2006;292:152–164. doi: 10.1016/j.ydbio.2005.12.047. [DOI] [PubMed] [Google Scholar]
- Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist’s view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Werner T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol Biol. 2000;132:337–349. doi: 10.1385/1-59259-192-2:337. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chen Y, Kim B, Wang H, Zhao C, He X, Liu L, Liu W, Wu LM, Mao M, Chan JR, Wu J, Lu QR. Olig2 targets chromatin remodelers to enhancers to initiate oligodendrocyte differentiation. Cell. 2013;152:248–261. doi: 10.1016/j.cell.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson DJ. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhu X, Zuo H, Maher BJ, Serwanski DR, LoTurco JJ, Lu QR, Nishiyama A. Olig2-dependent developmental fate switch of NG2 cells. Development. 2012;139:2299–2307. doi: 10.1242/dev.078873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.