Abstract
Both classical brown adipocytes and brown-like beige adipocytes are considered as promising therapeutic targets for obesity; however, their development, relative importance, and functional coordination are not well understood. Here we show that a modest expression of miR-378/378* in adipose tissue specifically increases classical brown fat (BAT) mass, but not white fat (WAT) mass. Remarkably, BAT expansion, rather than miR-378 per se, suppresses formation of beige adipocytes in subcutaneous WAT. Despite this negative feedback, the expanded BAT depot is sufficient to prevent both genetic and high fat diet-induced obesity. At the molecular level, we find that miR-378 targets phosphodiesterase Pde1b in BAT, but not in WAT. Indeed, miR-378 and Pde1b inversely regulate brown adipogenesis in vitro in the absence of phosphodiesterase inhibitor IBMX. Our work identifies miR-378 as a key regulatory component underlying classical BAT-specific expansion and obesity resistance, and adds novel insights into the physiological cross-talk between BAT and WAT.
INTRODUCTION
Classical BAT, primary located in the interscapular region of many mammals, has long been well recognized as an organ specialized for heat production, a property that is attributed to its extensive vascularization and sympathetic innervation, high mitochondrial density and oxidative capacity, and abundant expression of uncoupling protein-1 (Ucp1)1. In addition, certain WAT depots, such as subcutaneous inguinal WAT (iWAT), possess a subpopulation of adipocytes called beige or brite cells that confer plasticity and can be induced to brown-like adipocytes in response to various stimuli2–6. Both classical BAT and beige adipocytes are found in adult humans and their mass and/or activity are inversely correlated with body mass index6–16. These findings lead to the proposal that increasing BAT mass and/or activity or converting WAT adipocytes to beige adipocytes might be a promising therapeutic strategy for obesity and metabolic diseases, an idea that has gained strong support from numerous studies performed in rodents17–19. However, in many of these studies, a concomitant increase of activities of both classical BAT and beige adipocytes has been observed; hence, their relative contributions to obesity resistance are not fully understood17,18,20, and it remains to be determined whether and to what extent of expansion of classical BAT alone is sufficient to prevent obesity. Moreover, how the functions of classical BAT and beige adipocytes are coordinated are largely unclear.
Adipogenesis of both WAT and BAT is driven by a Pparγ transcriptional cascade, and many signaling molecules impinge upon the Pparγ pathway to modulate adipogenesis21–24. Among them is cAMP second messenger. cAMP is turned over by members in the cyclic nucleotide phosphodiesterase (Pde) superfamily25, and for this reason, a nonselective PDE enzyme inhibitor isobutylmethylxanthine (IBMX) has been widely used to promote fat cell differentiation in vitro. However, how the activities of Pde enzymes are naturally attenuated during adipocyte differentiation is unknown.
Brown and white adipogenesis are also specified by separate genetic entities. For example, a few genes have been shown to selectively operate in brown adipocytes to promote their adipogenesis26–32. For some of these genes such as C/EBPβ, Ebf2 and Orexin, the lack of adipogenic effects on white adipocytes is due to either their low expression levels or absence of intact signaling transduction pathways, which ultimately fail to activate Pparγ. On the other hand, co-factor Tle3 facilitates white adipogenesis by competing with Prdm16 for its interaction with PPARγ33. Clearly, identifying additional regulatory components governing fat type-specific development and understanding their mechanisms of action are of great biomedical importance.
MicroRNAs (miRNAs) are endogenous, non-coding small RNAs that control multiple developmental and cellular processes. miRNAs post-transcriptionally regulate gene expression through complementary base-pairing between its seed sequence and 3’ UTR of target mRNAs, triggering mRNA destabilization or translational repression34,35. miRNA targets are highly tissue- or cell type-dependent and one miRNA can have different functions by targeting different mRNAs in different tissues34,36,37. Several groups have investigated the importance of BAT-selectively expressed miRNAs in brown adipogenesis. These studies reveal that miR-155 and miR-133a/b inhibit brown adipogenesis38–41, and miR-193b-365 cluster enhances both brown and white adipogenesis42, suggesting diverse roles of miRNAs in BAT development. Despite these progresses, to date, miRNAs that positively control BAT-specific development have not been identified.
Here we report that a modest expression of miR-378, at least in part by targeting phosphodiesterase Pde1b, promotes BAT-specific expansion, which offers complete resistance to both genetic and high fat diet-induced obesity. Surprisingly, BAT expansion, but not miR-378 per se, leads to suppression of development of beige adipocytes. Our work reveals a fat type-specific action of miR-378 in control of adipose development and obesity resistance, and provides insights into the physiological cross-talk between BAT and WAT.
RESULTS
miR-378 transgene in adipose tissue expands classical BAT
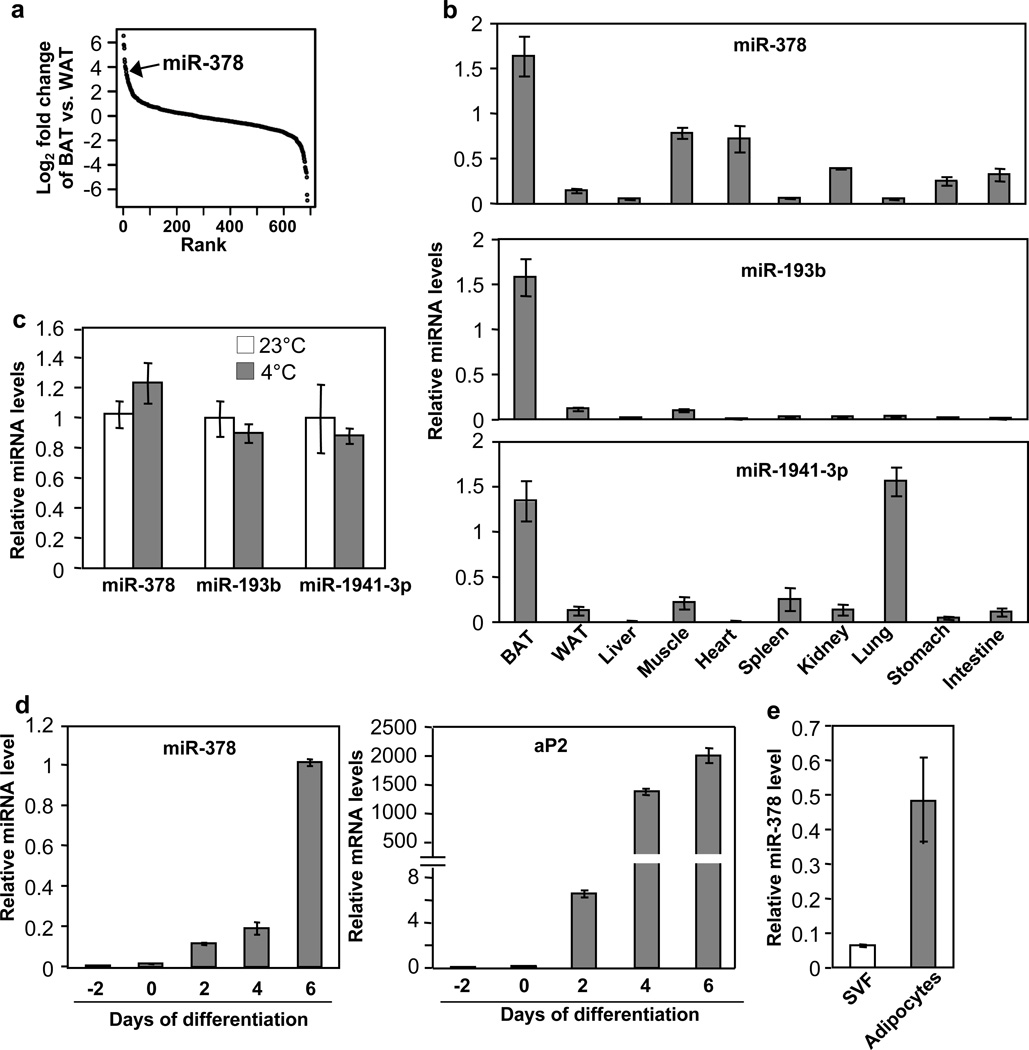
To begin investigating potential functions of miRNAs in BAT, we measured the expression of a total of 688 miRNAs in BAT and gonadal WAT by RT-qPCR (Fig. 1a and Supplementary Data 1). miRNAs that displayed significantly higher level in BAT were selected and further examined for their expression in other tissues. While, as noted previously43, most of our selected miRNAs are in fact skeletal muscle-specific miRNAs, three miRNAs, miR-378, miR-193b and miR-1941-3p, were highly enriched in BAT (Fig. 1b)42. Interestingly, none of these three miRNAs was induced by acute cold exposure (Fig. 1c). miR-378 was of particular interest, as it is induced during brown adipogenesis (Fig. 1d and 1e) and resides in the sense strand of the first intron of Pgc-1β, which is a transcriptional co-activator that regulates mitochondrial biogenesis and brown fat-specific gene expression44. In addition to miR-378, miR-378*, produced from the passenger strand of the 22 nucleotide RNA duplex, was detected in BAT at a very low level.
Fig. 1. Screening for BAT-selective miRNAs.
(a) miRNAs were ranked in order of differential expression between BAT and gonadal WAT (n=5 mice, 10-week-old male mice).
(b) miRNA tissue expression profile analyzed by quantitative RT-PCR. n=5 mice (10-week-old male mice).
(c) miRNA levels in BAT at 23°C and at 4°C (6 hr). n=5 mice per group (10-week-old male mice).
(d) miR-378 (left) and aP2 mRNA (right) levels during brown fat cells differentiation.
(e) miR-378 level in SVF and mature adipocytes isolated from BAT of wild type mice.
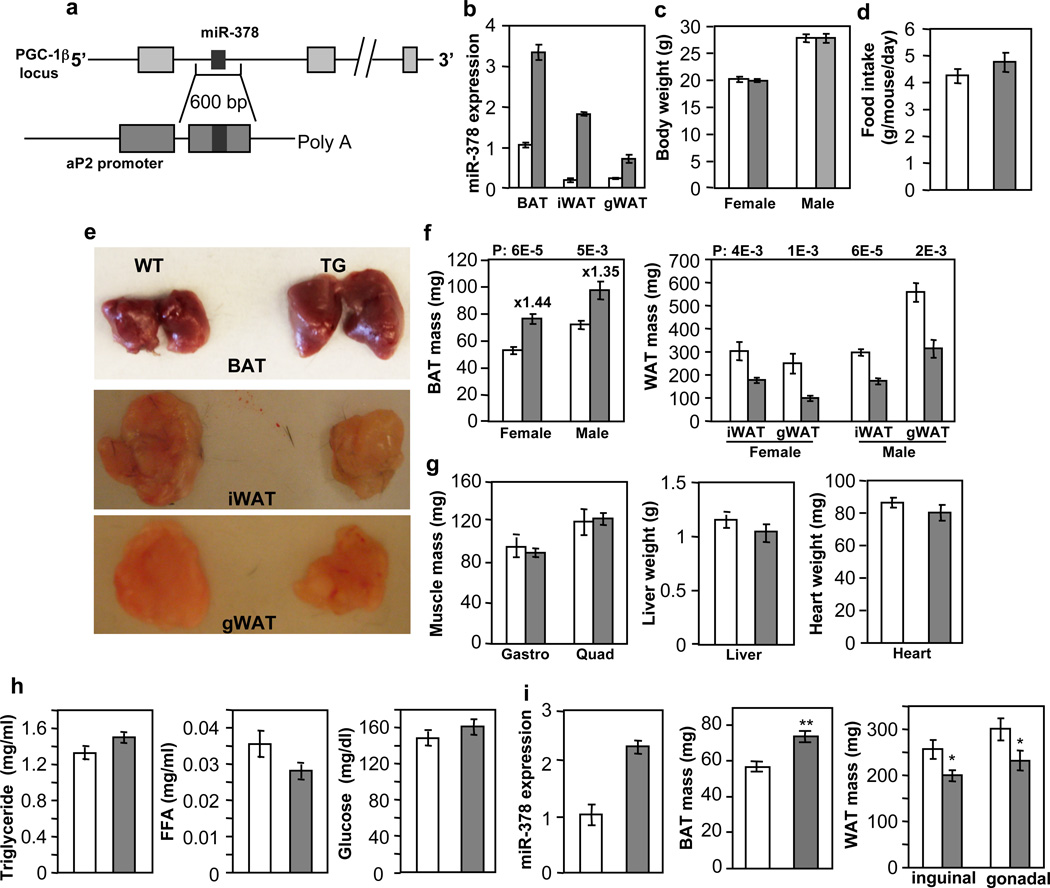
To pinpoint the physiological role of miR-378, we generated transgenic mice ectopically expressing the miR-378 locus under the control of the aP2 promoter45 (Fig. 2a), given that endogenous aP2 and miR-378 displayed a similar expression pattern during brown adipogenesis (Fig. 1d). The levels of miR-378 in the transgenic mice were about 3.3-fold in BAT and 3 to 10-fold in WAT, compared to the control littermates (Fig. 2b). Similar folds of miR-378* expression were observed (Supplementary Fig. 1), thus total level of miR-378* remained extremely low. On a normal chow diet, the transgenic mice did not show any noticeable difference in outward appearance. Their body weights as well as food intakes were similar (Fig. 2c and 2d). However, upon dissection, it was evident that BAT was enlarged (Fig. 2e). We observed a 44% increase of BAT mass in female and a 35% increase in male transgenic mice (Fig. 2f). Surprisingly, both inguinal and gonadal WAT depots were significantly reduced (Fig. 2e and 2f). Other organs such as liver, skeletal muscle and heart showed no weight difference (Fig. 2g), and levels of circulating triglycerides, free fatty acids and glucose were normal (Fig. 2h). We also obtained a second miR-378 transgenic line, which displayed a similar BAT expansion phenotype and other phenotypes examined, albeit to a less extent, presumably due to a lower transgene expression (Fig. 2i). We thus characterized the first transgenic line in detail. In addition, we have generated aP2 promoter-driven miR-1941-3p transgenic mice and no BAT expansion phenotype was found. Together, we conclude that a very modest ectopic expression of miR-378 locus in adipose tissue results in a BAT-specific expansion accompanied with diminished WAT depots.
Fig. 2. Modest transgenic expression of miR-378 in adipose tissue produces a BAT-specific expansion.
Empty bar, wild type mice (WT); Grey bar, transgenic mice (TG). Mice were fed a normal chow diet.
(a) Outline of miR-378/378* transgenic construct.
(b) qPCR analysis of miR-378 levels in 9-week-old male WT and TG mice (n=3 mice per group). iWAT, subcutaneous inguinal WAT; gWAT, gonadal WAT.
(c) Body weights. Female, n=10–12 mice per group (9-week-old mice); male, n=6 mice per group (14-week-old mice).
(d) Food intake. n=5 male mice per group (10-week-old mice).
(e and f) BAT and WAT mass. Female, n=10–12 mice per group (9-week-old mice); male, n=6 mice per group (14-week-old mice). P values (TG vs. WT, two-tailed Student’s t-test) are indicated on top of the panels.
(g) Liver, skeletal muscle and heart weights from mice in (d). Gastro, gastrocnemius muscle; Quad, quadriceps muscle.
(h) Circulating triglycerides, free fatty acids and glucose levels. n=6–8 mice per group (14-week-old male mice).
(i) miR-378 expression and BAT expansion in a second miR-378 transgenic line. n= 6 mice per group (9-week-old male mice). *P<0.05, **P<0.005 (two-tailed Student’s t-test). All data are mean ± SE.
We performed Ki67 immunohistochemical staining on section of BAT at E16.5 or postnatal day 21. In either case, we found no apparent difference in preadipocyte proliferation between transgenic and control mice (Supplementary Fig. 2A and 2B). Overexpression of miR-378 in immortalized brown preadipocytes also had no effect on proliferation (Supplementary Fig. 2C). Our data suggest that BAT expansion is not due to an increase in preadipocyte proliferation. Instead, we hypothesized that miR-378 transgene promotes more preadipocytes to differentiate into adipocytes, and the preadipocyte pool is then rapidly replenished and maintained.
miR-378 transgene enhances brown adipogenesis in vivo
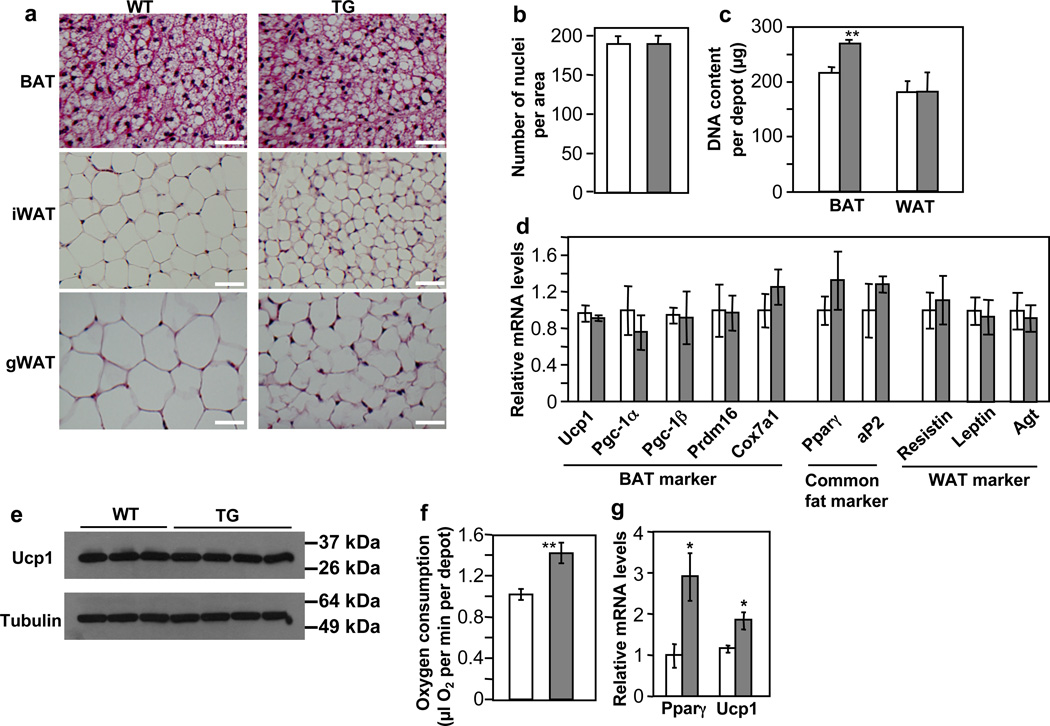
An increase of BAT mass can be attributed to cell number, cell size, or both. Inspection of hematoxylin and eosin stained BAT images (Fig. 3a) and estimation of nuclei densities (number of nuclei per unit area) (Fig. 3b) from these images suggested similar cell size between the transgenic and control mice, though the transgenic mice appeared to contain a slightly higher amount of lipid in BAT (Fig. 3a). Measurements of genomic DNA content revealed that BAT depots from the transgenic mice had a significantly higher cellularity (Fig. 3c). In contrast, the WAT adipocyte size in the transgenic mice was clearly smaller (Fig. 3a), and genomic DNA content per depot was similar compared to control mice (Fig. 3c). These results suggest that the expansion of BAT mass in the miR-378 transgenic mice is likely due to cell number, not cell size, whereas diminished WAT mass is likely due to cell size. Thus, ectopic expression of miR-378 in adipose promotes the formation of brown adipocytes but not white adipocytes, despite that both fat types express the miR-378 transgene. Furthermore, the shrinkage of white adipocyte might be secondary, caused by BAT expansion, which shifts the energy balance from storage to burning. Of note, the gradual increase of miR-378 level during adipogenesis (Fig. 1d) is consistent with its function, as similar expression profile has been observed for key adipogenic regulators such as Pparγ.
Fig. 3. miR-378 promotes brown adipogenesis but not white adipogenesis in vivo.
(a) Hematoxyln and Eosin staining for adipose tissue from 9-week-old male WT and TG mice. Scale bar, 50 µm.
(b) Number of nuclei per BAT image. 20 random images from 3 mice per group were counted, mean ± SE.
(c) Genomic DNA content per fat depot. n=6 mice per group (11-week-old female mice), mean ± SE, **P=0.01 (two-tailed Student’s t-test).
(d) Expression of fat markers determined by real-time RT-qPCR in BAT. n=4–6 mice per group (9-week-old male mice), mean ± SE.
(e) Immunoblotting showing Ucp1 protein levels in BAT (5-month-old female mice).
(f) Measurement of oxygen consumption per BAT depot with a Clark-type oxygen electrode. n=4 mice per group (9-week-old female mice), mean ± SE, **P=0.01 (two-tailed Student’s t-test).
(g) Gene expression in BAT of neonatal mice determined by real-time RT-qPCR. n=3–6 mice per group, mean ± SE, *P<0.05 (two-tailed Student’s t-test).
To ascertain that the expanded BAT is “true” brown fat, not white fat-like, we analyzed the expression of a number of fat markers in adult mice by RT-qPCR, which in principle measures relative expression level per cell. The mRNA levels of BAT-specific markers, common fat markers, and WAT-specific marker were comparable between transgenic and control mice (Fig. 3d). As expected, Ucp1 protein levels remained unchanged as well (Fig. 3e). Moreover, the expanded BAT is fully functional, as indicated by ex vivo oxygen consumption assays (Fig. 3f). Our observation of no changes of common fat gene expression or BAT-specific gene expression is not unexpected; similar scenarios have been seen in BMP receptor (Bmpr1a) knockout46 and aP2-Prdm16 transgenic mice47, respectively. It is possible that the normal differentiation process in vivo has already allowed a great extent of their expression that cannot be pushed further by the miR-378 transgene. Since brown adipocytes in neonatal mice are not as fully differentiated as those of adult mice, we wondered whether an effect of miR-378 on adipogenic gene expression would be observed at this early stage. Indeed, neonatal miR-378 transgenic mice displayed a higher expression of Pparγ and Ucp1 (Fig. 3g). Taken together, our data strongly suggest that the expanded BAT is authentic and most likely retains its full range of molecular characteristics.
BAT expansion suppresses formation of beige adipocytes
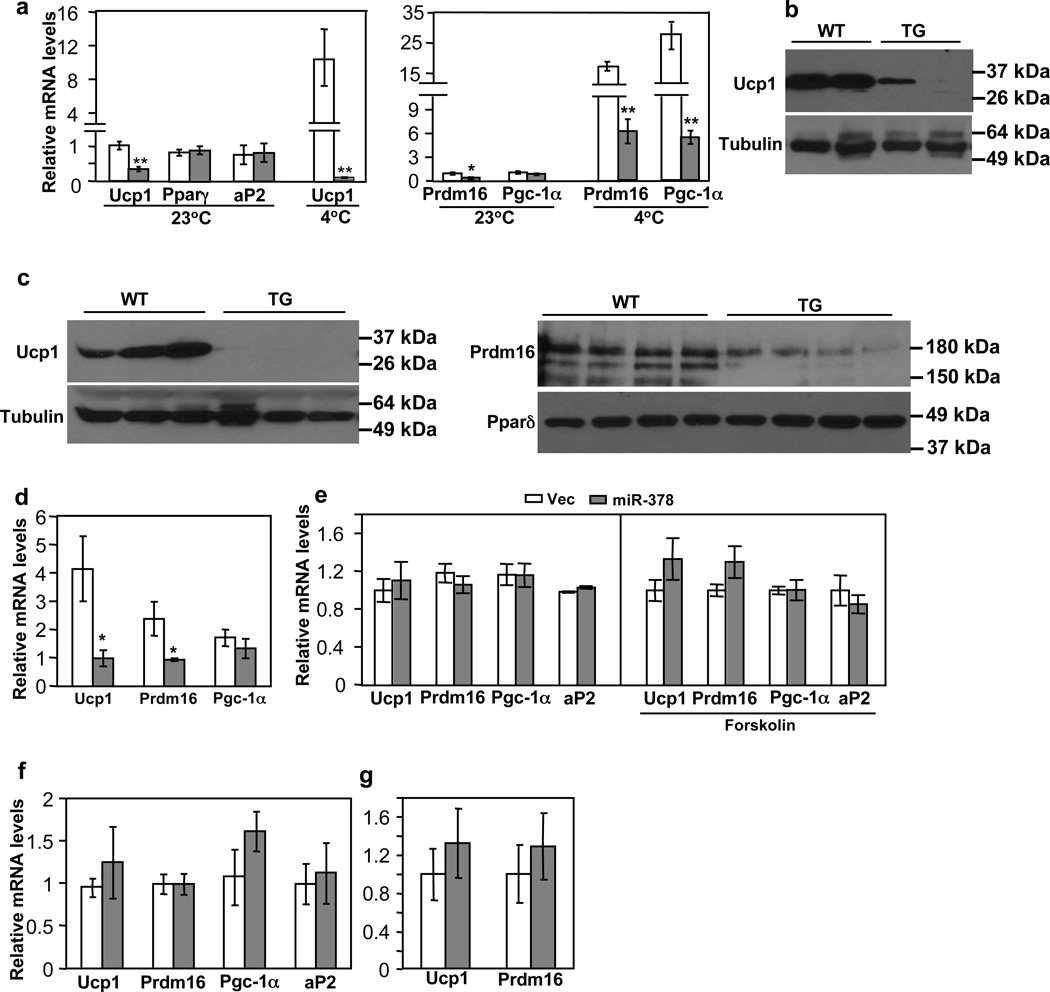
iWAT expresses a considerable basal level of Ucp1 and displays high plasticity that is amendable to browning. We determined whether ectopic expression of miR-378 in this depot promotes the formation of beige adipocytes. In contrast to what we expect, basal levels of Ucp1 mRNA and protein, as well as expression of Prdm16, which is a major regulator for BAT determination and beige adipocyte development47,48, were significantly decreased at room temperature (Fig. 4a and 4b), whereas levels of common adipogenic markers (Pparγ and aP2) remained normal. No change of Ucp1 in gonadal WAT was observed (Supplementary Fig. 3A). We envisioned that the expansion of BAT might cause a negative feedback response in iWAT. To investigate this further, we challenged the animals at 4°C for 6 hr. As expected, cold exposure induced Ucp1 expression in BAT of both wild type and transgenic mice (Supplementary Fig. 3B) and in iWAT of wild type mice (Fig. 4a). Strikingly, thermogenic pathway was completely blocked in iWAT of the transgenic mice, and Ucp1 expression was further suppressed at 4°C (Fig. 4a and 4c). Consistent with this, we observed that Prdm16 level remained significantly lower and cold-induction of Pgc-1α was severely impaired (Fig. 4a and 4c). We interpreted that high Ucp1 induction by cold in the expanded BAT somehow signals iWAT to curb its Ucp1 expression in order to maintain a normal and perhaps intrinsic cold response. Indeed, core body temperature was similar between the transgenic and control mice when shifted to 4°C for 6 hr (Supplementary Fig. 3C).
Fig. 4. BAT expansion, not miR-378 per se, suppresses browning of subcutaneous iWAT.
Empty bar, wild type mice; Grey bar, transgenic mice.
(a) Ucp1, Prdm16 and Pgc-1α mRNA levels in iWAT from mice maintained at 23°C or shifted to 4°C for 6 hr. n=4–6 mice per group (10-week-old male mice). WT vs. TG: *P<0.05, **P<0.01 (two-tailed Student’s t-test). Ucp1 expression at 23°C vs. at 4°C: P=0.009 for WT, and P=0.001 for TG. Data are mean ± SE.
(b) Immunoblotting of Ucp1 protein in iWAT from mice maintained at 23°C (5-month-old female mice).
(c) Immunoblotting of Ucp1 (left) and Prdm16 (right) protein in iWAT from 8-week-old male mice exposed to 4°C for 6 hr.
(d) Ucp1, Prdm16 and Pgc-1α mRNA levels in iWAT from 8-week-old male mice treated with CL-316,243 for 5 days. Data were normalized to those of untreated wild type mice. n=5–6 mice per group, mean ± SE, *P<0.05 (two-tailed Student’s t-test).
(e) iWAT preadipocytes from wild type mice were infected with miR-378 or control lentiviruses and then differentiated. At Day 6, adipocytes were either untreated or treated with forskolin (10 µM) for 5 hr before harvesting (n=3, mean ± SE). All data were relative to their respective controls. We typically observed that Ucp1 was induced to ~ 20-fold by forskolin relative to untreated adipocytes.
(f) iWAT preadipocytes from wild type mice and transgenic mice were differentiated. At Day 6, adipocytes were harvested. n=4–6 mice per group, mean ± SE.
(g) Real-time quantification of Ucp1 and Prdm16 mRNA levels in iWAT after interscapular BAT was removed. n=4 mice per group (9-week-old female mice), mean ± SE.
This negative response could be due to either suppression of sympathetic tone into iWAT or due to suppression of β3-adrenergic signal transduction within iWAT adipocytes. To distinguish these possibilities, we intraperitoneally injected agonist CL-316,243 daily for 5 days into miR-378 transgenic and littermate control mice. While levels of Ucp1 and Prdm16 were induced to 4-fold and 2-fold, respectively, in the iWAT of control mice compared to untreated control mice, β3-adrenergic agonist treatment failed to rescue their expression in the transgenic mice (Fig. 4d). These results suggest that the recipient site responsive to BAT expansion is likely to be located within the iWAT adipocytes.
To rigorously test that the above observed phenotype was indeed through a cross-talk between BAT and iWAT, not caused by a cell autonomous effect of miR-378, we overexpressed miR-378 through lentiviral infection in primary iWAT preadipocytes isolated from wild type C57BL/6 mice. Cells were differentiated into mature adipocytes under a standard differentiation protocol. Microscopic examination found no effect of miR-378 on adipogenesis (see below). We found that expression of Ucp1, Prdm16 and Pgc-1α was not affected by miR-378 expression (Fig. 4e). Similar data were obtained when cells were treated with forskolin (Fig. 4e); forskolin induced Ucp1 expression by ~ 20-fold relative to untreated adipocytes. In addition, we isolated iWAT preadipocytes from both miR-378 transgenic and control littermates and differentiated them into mature adipocytes. We found that Ucp1, Prdm16 and Pgc-1α were not downregulated in adipocytes of the transgenic mice (Fig. 4f). These results are in striking contrast to what observed in vivo, strongly supporting the idea that suppression of beige adipocyte development in vivo can not be attributed to overexpression of miR-378 within iWAT.
Finally, we surgically removed BAT from interscapular regions of transgenic and control mice, and then allowed the animals to recover for 10 days. We found that, after BAT removal, levels of Ucp1 and Prdm16 gene expression were no longer lower in the transgenic mice (Fig. 4g). Taken together, the data presented in Fig. 4 reveal a cross-talk between BAT and iWAT in which BAT expansion robustly suppresses Ucp1 expression and formation of beige adipocytes in iWAT. This negative response is at least in part mediated by downregulation of Prdm16.
BAT expansion prevents both genetic and diet-induced obesity
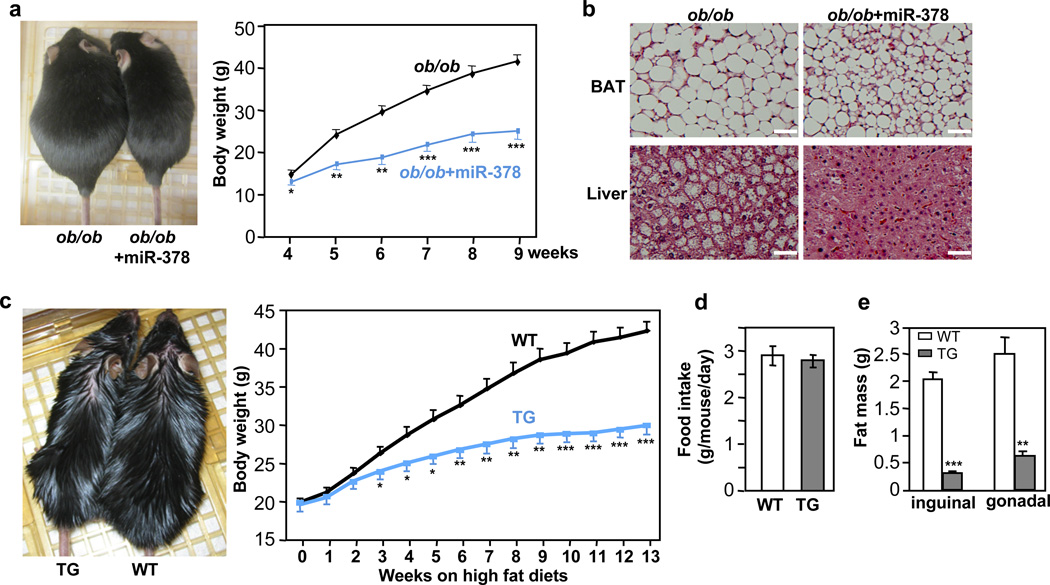
One important question is whether the expanded BAT is sufficient to prevent obesity, in particular in the face of a decreased basal Ucp1 expression in the iWAT. We first addressed this question using ob/ob mice, since these mice have low energy expenditure and their BAT depots become impaired with time. We crossed the miR-378 transgenic mice with the ob/ob mice. As shown in Fig. 5a, the miR-378 transgene completely reversed the obese phenotype. In fact, the body weights of the ob/ob mice with the miR-378 transgene were similar to those of age-matched wild type mice. The miR-378 transgene also improved BAT morphology and rescued hepatic steatosis in the ob/ob mice (Fig. 5b). Next, we fed the miR-378 transgenic mice and wild type littermates with a high fat diet. Their body weights, which were similar before high fat diet, started to be significantly segregated at week 3 of high fat diet feeding (Fig. 5c). At week 13, the control mice gained 22.4 0.9 g of body weight, while the transgenic mice only gained 10.3 0.8 g, which is equivalent to a typical body weight gain under a chow diet, although similar amounts of food were consumed (Fig. 5d). The body weight difference was due to massive reduction of WAT mass in the transgenic mice (Fig. 5e) that are associated with amelioration of adipocyte hypertrophy (Supplementary Fig. 4). Taken together, we conclude that BAT expansion in the transgenic mice not only overcomes the negative response in iWAT, but also expends far more energy, leading to a complete prevention of both genetic and high fat diet-induced obesity.
Fig. 5. miR-378-directed BAT expansion is sufficient to protect against both genetic and high fat diet-induced obesity.
(a) Left, representatives of ob/ob mice and ob/ob mice carrying the miR-378 transgene at nine-week-old; Right, body weights of ob/ob mice (n=7 mice) and ob/ob mice with the miR-378 transgene (n=4 mice) fed a normal chow diet. Mean ± SE, *P<0.05, **P<10−4, ***P<10−5 (two-tailed Student’s t-test).
(b) Amelioration of liver steatosis and reduction of BAT lipid accumulation by miR-378 in ob/ob mice. Scale bar, 50 µm.
(c) 6-week-old male mice were fed a high fat diet for 13 weeks. Left, representatives of wild type (WT) and transgenic (TG) mice after a 13-week high fat diet; Right, body weights were measured weekly. n=8–9 mice per group, mean ± SE, *P<10−3, **P<10−4, ***P<10−6 (two-tailed Student’s t-test).
(d) Consumption of high fat diet by wild type (WT) and transgenic (TG) mice. n=5 mice per group (10-week-old male mice), mean ± SE.
(e) Inguinal and gonadal WAT mass of mice in (d). Mean ± SE, **P<10−4, ***P<10−6 (two-tailed Student’s t-test).
We performed indirect calorimetry measurements in adult mice fed a normal chow diet or a 3-day high fat diet, conditions that transgenic and control mice had similar body weights. These studies confirmed our earlier results of no difference in food intake, and showed transgenic mice tended to be less active at night phase, although no statistical significance reached (Supplementary Fig. 5), suggesting that resistance to obesity is not due to potential decreased food intake or increased activity. Interestingly, we did not observe any difference in oxygen consumption or respiratory exchange ratio (RER) (Supplementary Fig. 5), despite that BAT depot ex vivo on a per mouse basis had a higher oxygen consumption (Fig. 3f). Other mouse models that display changes in oxidative metabolism in adipose tissue or skeletal muscle but no difference in oxygen consumption and/or RER have been reported49–51. It has been discussed that a small difference in energy expenditure that is beyond the detection limit of indirect calorimetry could result in body weight difference over a long term52.
miR-378 targets Pde1b in BAT but not in WAT
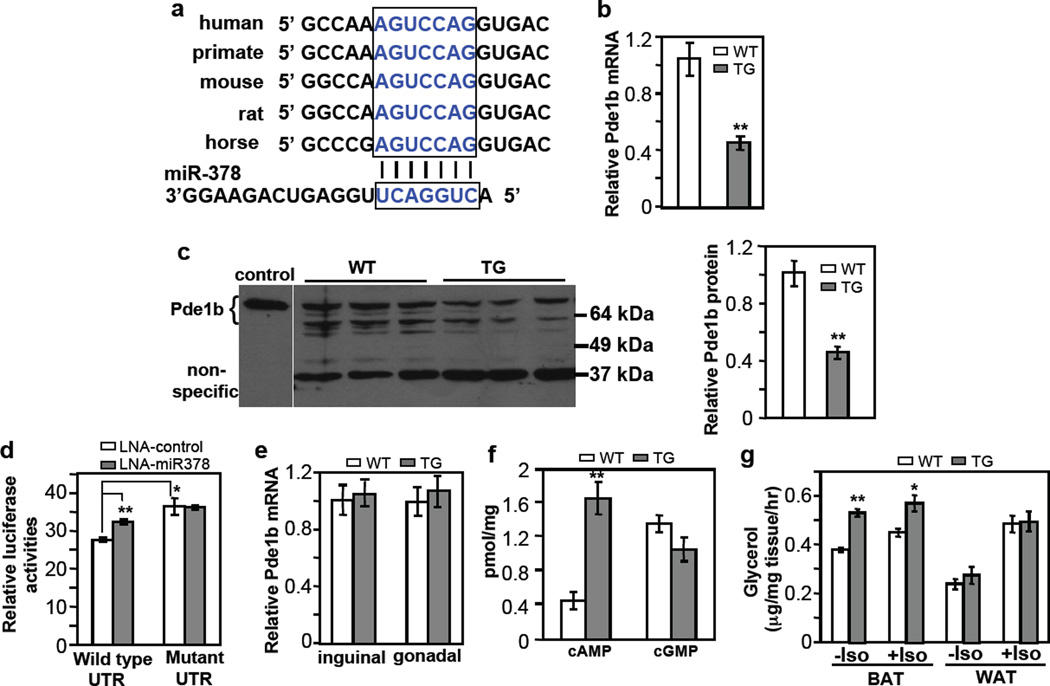
We sought to understand the molecular mechanism by which miR-378 promotes BAT expansion. Targetscan (www.targetscan.org) predicts Pde1b, a phosphodiesterase that catalyzes the turnover of cAMP and cGMP25, is a potential miR-378 target. cAMP and cGMP are signaling molecules that potentiate adipocyte differentiation21–23,53. Pde1b appeared to be ubiquitously expressed with a higher level in WAT than in BAT (Supplementary Fig. 6A). Pde1b level was initially decreased and then increased during brown adipogenesis (Supplementary Fig. 6B). Interestingly, single nucleotide polymorphisms in the Pde1b gene were found to be associated with cattle backfat thickness54,55. The target sequence at the 3’UTR of Pde1b is well conserved in mammals (Fig. 6a). We found that both endogenous Pde1b mRNA and protein in the BAT of transgenic mice were significantly decreased (Fig. 6b and 6c). To test whether Pde1b is a direct target of miR-378, we generated a luciferase reporter containing, at its downstream, either a wild type 3’UTR of mouse Pde1b or a variant in which the target sequence was mutated. We transfected these reporters into HEK293 cells. We found that the mutant reporter had a significantly higher activity than the wild type reporter (Fig. 6d), most likely due to the presence of endogenous miR-378, which has a well-conserved sequence in human genome. Indeed, transfection of HEK293 cells with the miR-378 inhibitor increased the activity of the wild type reporter but had no effect on the mutant reporter (Fig. 6d). These results demonstrated that Pde1b is a direct target of miR-378 in BAT.
Fig. 6. BAT-specific suppression of Pde1b by miR-378.
(a) Conserved miR-378 targeting sequence at Pde1b 3’ UTR.
(b) Pde1b mRNA is downregulated in BAT of miR-378 transgenic mice. n=5 mice per group (9-week-old male mice), mean ± SE, **P<0.001 (two-tailed Student’s t-test).
(c) Pde1b protein levels in BAT from 5-week-old female WT and TG mice. Cell extracts from HEK293 cells transfected with a full-length Pde1b1 plasmid were used as a control to indicate the position of endogenous mouse Pde1b. BAT produces two expected major splice variants of Pde1b (Pde1b1 and Pde1b2) differing in their amino-termini70. Right panel shows quantification of the western blot. Mean ± SE, **P=0.005 (two-tailed Student’s t-test).
(d) HEK293 cells were transfected with locked nucleic acid (LNA) miR-378 inhibitor and a luciferase reporter carrying 3’ UTR of Pde1b as described in the online methods. Luciferase activity was measured. n=3, mean ± SE, *P<0.05, **P<0.001 (two-tailed Student’s t-test).
(e) Relative Pde1b mRNA level in inguinal and gonadal WAT of the transgenic mice. n=5 mice per group (9-week-old male mice), mean ± SE.
(f) cAMP and cGMP levels in BAT. n=7 mice per group (9-week-old male mice), mean ± SE, **P<0.001 (two-tailed Student’s t-test).
(g) BAT and WAT lipolysis with or without 10 µM isoproterenol. n=5 mice per group (5-month-old male mice), mean ± SE, **P<0.0005, *P=0.012 (two-tailed Student’s t-test).
Interestingly, Pde1b mRNA expression remained unchanged in both inguinal and gonadal WAT of miR-378 transgenic mice (Fig. 6e). These results might in part explain the lack of WAT expansion, and suggest that targets of miR-378 are highly tissue- or cell type-specific. In support this notion, we found that none of other known miR-378/378* targets identified in other tissues, including Igf1r (cardiomyocytes)56, Crat and Med13 (liver)57, and ERRγ (cancer cells)58, was downregulated by the miR-378/378* transgene in BAT (Supplementary Fig. 7), consistent with the idea that one microRNA can have distinct and tissue-specific functions through targeting different mRNAs34,36,37.
We next determined whether reduction of Pde1b modulates cAMP and cGMP levels in BAT. We found that cAMP level was increased to more than 3.6-fold in the miR-378 transgenic mice, whereas cGMP level was not significantly different (Fig. 6f). Thus, Pde1b appears to predominantly regulate cAMP turnover in BAT, and cGMP level is refractory to changes of Pde1b. Our results suggest that suppression of Pde1b by miR-378 and hence elevation of cAMP level might be part of the mechanism underlying BAT expansion.
An elevation of cAMP level is expected to lead to an increase of lipolysis in BAT. Since the process of isolation of single mature adipocytes might disturb intracellular cAMP content, we tested this idea in BAT tissue samples ex vivo. We found that lipolysis was indeed significantly higher in BAT of transgenic mice relative to control littermates (Fig. 6g). No difference was observed in WAT, consistent with the data that Pde1b was not downregulated in this adipose type. Interestingly, application of isoproterenol produced little and a slight increase of lipolysis in BAT of transgenic and control mice, respectively, but strongly enhanced lipolysis of WAT in both genotypes, which might be related to their respective preexisting basal β-adrenergic activation state. Increased lipolysis in BAT of transgenic mice did not cause an increase of plasma free fatty acids under both normal chow (Fig. 2h) and high fat diet conditions (Supplementary Fig. 8), indicating that released lipids are probably oxidized locally.
miR-378 and Pde1b inversely regulates adipogenesis in vitro
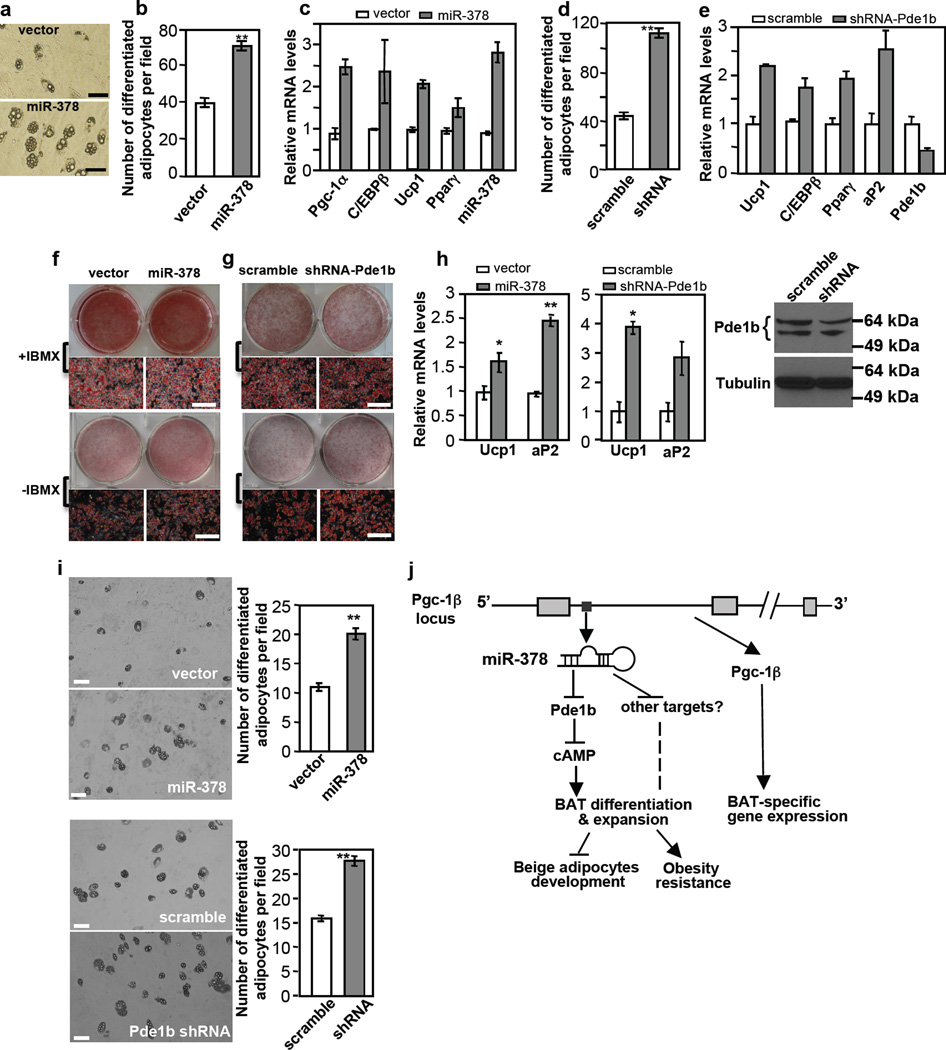
Given that miR-378 expression promotes BAT adipogenesis in vivo, we determined whether this effect can be recapitulated in vitro. We first performed our experiments in C3H10T1/2 mesenchymal stem cells. In a standard adipogenic assay condition that contains the general phosphodiesterase inhibitor IBMX, we found that ectopic expression of miR-378/378* had little stimulatory effect on adipogenesis (Supplementary Fig. 9A). Removal of IBMX abrogated adipogenesis (Supplementary Fig. 9B). Importantly, expression of miR-378/378* strongly promoted the formation of adipocytes in the absence of IBMX, that is, a much higher percentage of cells were differentiated into adipocytes (Fig. 7a and 7b). Because of increased number of adipocytes, levels of adipogenic markers, when assayed in the C3H10T1/2 population, were increased as well (Fig. 7c). Thus, miR-378 does positively regulate adipocyte differentiation in vitro, an effect that is masked by IBMX, further linking miR-378 to Pde1b. Accordingly, we infected C3H10T1/2 cells with Pde1b shRNA or scramble lentiviruses and differentiated them into adipocytes in the absence of IBMX. As shown in Fig. 7d and 7e, knockdown of Pde1b phenocopied miR-378 expression.
Fig. 7. miR-378 and Pde1b inversely regulate adipogenesis in vitro.
(a–c) Ectopic expression of miR-378 promotes adipogenesis in C3H10T1/2 cells in the absence of Pde enzyme inhibitor IBMX. Experiments were repeated 3 times. Scale bar, 100 µm. In (b), number of differentiated adipocytes per field. 20 fields were counted per condition. Mean ± SE, **P<10−5 (two-tailed Student’s t-test). In (c), real-time qPCR was used to detect gene expression of differentiation-dependent adipogenic markers.
(d, e) Knockdown of Pde1b phenocopies the effect of miR-378 overexpression in the absence of IBMX. In (d), number of differentiated adipocytes per field. 20 fields were counted per condition. Mean ± SE **P<10−5 (two-tailed Student’s t-test). In (e), expression of differentiation-dependent adipogenic markers.
(f) Adipogenesis of immortalized brown preadipocytes expressing miR-378. Experiments were repeated 3 times. Scale bar, 50 µm.
(g) Adipogenesis of immortalized brown preadipocytes with Pde1b knockdown. Experiments were repeated 3 times. In both (f) and (g), dark areas in microscopic images are undifferentiated preadipocytes. Scale bar, 50 µm.
(h) Relative Ucp1 and aP2 mRNA levels in brown adipocytes generated as described in (f) and (g). n=3, mean ± SE, *P<0.05, **P<0.01 (two-tailed Student’s t-test). Pde1b protein level was shown in brown adipocytes differentiated at a standard condition.
(i) Ectopic expression of miR-378 or knockdown of Pde1b in primary brown preadipocytes increases the number of adipocytes during differentiation in the absence of IBMX. Scale bar, 50 µm. Mean ± SE, **P<10−6 (two-tailed Student’s t-test).
(j) A model for miR-378 action in classical BAT. miR-378 controls brown adipogenesis and expansion in part through downregulating Pde1b and hence elevating cAMP content, whereas the host gene Pgc-1β controls BAT-specific gene expression but not adipogenesis per se44.
We next tested our model in immortalized brown preadipocytes. Again, same number of preadipocytes were plated for control and experimental conditions, and were subjected to differentiation. In agreement with what observed in C3H10T1/2 cells, neither miR-378 overexpression nor Pde1b knockdown had any effect in the presence of IBMX, while both of these two manipulations promoted formation of adipocytes in the absence of IBMX (Fig. 7f and 7g). Again, increased number of adipocytes was associated with higher adipogenic gene expression (Fig. 7h). In contrast, iWAT preadipocyte differentiation was not affected by miR-378 expression (Supplementary Fig. 9C).
Finally, we performed adipogenesis experiments in primary brown adipocytes. We initially used brown preadipocytes from transgenic and control littermates. However, we found that individual animals displayed considerable variations in terms of both proliferation and differentiation capacities in vitro, even these brown preadipocytes were isolated from equal BAT mass and same genotype, thus precluding us to critically evaluate in vitro adipogenesis. To circumvent this problem, we isolated primary brown preadipocytes from wild type C56BL6 mice, pooled them together, and cultured them. Equal numbers of preadipocytes were infected with lentiviruses, selected with puromycin and amplified. Cells were then replated at equal numbers and differentiated without IBMX. Because of this relatively long procedure and lack of IBMX, cells lost their differentiation ability considerably, but we were able to reproducibly observe a significant increase of adipocyte number in these primary cells with either miR-378 ectopic expression or Pde1b knockdown (Fig. 7i). Together, our data show that miR-378 promotes brown adipocyte formation both in vivo and in vitro, and this is likely to be mediated, at least in part, through downregulation of Pde1b.
DISCUSSION
Our studies have made the following discoveries: (1) miR-378 promotes brown adipogenesis, and as a result, transgenic expression of miR-378 in adipose tissue leads to the expansion of classical BAT but not of WAT; (2) BAT expansion is achieved at least in part through fat type-specific Pde1b downregulation and hence elevation of cAMP content; (3) BAT expansion suppresses development of beige adipocytes in iWAT; (4) BAT expansion alone is sufficient to prevent both genetic and high fat diet-induced obesity, even in the face of decreased activity of beige adipocytes. Our work identifies miR-378 and Pde1b axis as a potential major molecular node that fine-tunes BAT mass and regulates systematic metabolism, and provides new insights into our understanding of BAT development and cross-talk between BAT and WAT. It is important to note that this miR-378 and Pde1b axis appears to be physiologically relevant, as BAT expansion and associated phenotypes requires a merely 2 to 3-fold increase of miR-378 expression, and knockdown of Pde1b promotes adipocyte differentiation.
The process of adipogenesis per se is largely shared by both brown and white adipocytes, and many adipogenic regulators and signaling pathways have been found to impact differentiation of both fat cell types. Nevertheless, studies in recent years have identified a number of genes that selectively promote brown adipogenesis26–32. We now extend this group of genes to include a microRNA. Previous investigations by others have demonstrated key roles of miRNAs in inhibition of brown adipogenesis or enhancement of both brown and white adipogenesis38–42,59. To our knowledge, miR-378 is the first miRNA shown to date that selectively promotes brown adipogenesis.
We reveal that miR-378 directly targets Pde1b in BAT, but not in WAT, to increase intracellular cAMP content, providing a plausible molecular mechanism for BAT expansion. It is noteworthy that the BAT expansion phenotype driven by miR-378 is reminiscent of that of the aP2-Foxc2 transgenic mice, which sensitizes cAMP signaling in adipose tissue60. Moreover, our proposed mechanism is strongly supported by our in vitro experiments showing that effects of miR-378 and Pde1b on brown adipogenesis are only manifested when phosphodiesterase inhibitor IBMX is removed. Interestingly and similar to what we observed, miR-378 was previously not found to increase number of adipocyte formation of ST2 mesenchymal precursor cells in the presence of IBMX; however, adipogenesis was not examined in the absence of IBMX in this early study61. While IBMX is widely used to inhibit phosphodiesterase activity for efficient adipogenesis in vitro, the specific Pde enzyme involved in brown adipogenesis has not been firmly identified. Experiments performed a decade ago have led Nedergaard and co-works to postulate that Pde1 might be important for inhibition of intracellular cAMP accumulation in brown adipocytes62. Their studies, together with our results presented here, suggest a potential critical negative role of Pde1b in control of cAMP level and brown adipogenesis. Furthermore, our work elucidates an endogenous pathway attenuating Pde1b activity during brown adipogenesis.
Our observation of fat type-specific targeting of Pde1b by miR-378 is consistent with the notion that miRNAs confer target and functional specificity in different cell types or tissues34,36,37. Indeed, miR-378/378* has been shown to have several different functions, including lipogenesis in ST2 cells61, fatty acid oxidation in liver57, and apoptosis and hypertrophy in cardiomyocytes56,63. While how exactly Pde1b is a preferred target of miR-378 in BAT but not in WAT is currently unknown, this selective miR-378 and Pde1b interaction is presumably determined by the overall availability and complexity of other Pde1b-targeting miRNAs, other mRNA targets of miR-378, and competing endogenous RNAs, as alluded in other systems64. Interestingly, Pde1b protein is readily detectable by our western blot analysis and appears to be abundantly expressed in BAT. Thus, BAT-enriched miR-378 is employed to counterbalance abundantly expressed Pde1b, a mechanism likely to be important for maintaining the flexibility of BAT to expand. Of course, it is entirely possible that this process also involves other miR-378 targets that remain to be identified.
Surprisingly, we find that BAT expansion, not miR-378 per se, elicits a robust negative feedback response in subcutaneous iWAT that suppresses formation of beige adipocytes. Cross-talk between BAT and WAT is an emerging, interesting topic with two previous documentations in the literature, showing that ectopic expression of Ucp1 in WAT inhibits endogenous Ucp1 expression in BAT65, and that BAT paucity causes a compensatory induction of beige adipocytes in iWAT46. These studies together with ours suggest that functions of BAT and beige adipocytes are highly coordinated, and that brown-white fat communication is a two-way process and is capable of generating diverse responses. Moreover, depending on the nature of the response, distinct mechanisms can be utilized. For example, the induction of beige adipocytes is mediated by an increase of sympathetic input to iWAT46, while the suppression of beige adipocytes observed in our study appears to directly stem from iWAT adipocytes and Prdm16 reduction without the need to attenuate sympathetic input. Interestingly, recent work shows that WAT can cell-autonomously sense temperature to induce thermogenic gene program66. Currently, we do not know yet what signal is emanated from BAT and what molecular pathway in iWAT is responding, but our miR-378 animal model might be a useful tool to dissect this process.
We initially anticipated that the negative response in iWAT would neutralize the effect of BAT expansion on energy homeostasis. Instead, we find that the miR-378 transgenic mice are completely protected from both genetic and high fat diet-induced obesity, to an extent that is similar to a body gain by wild type mice on a normal chow diet, suggesting that energy expenditure due to BAT expansion far exceeds its negative impacts on iWAT. Notably, the net increase of BAT mass (~ 25 mg) in our transgenic mice only accounts for about 0.1% of total body weight, but can dramatically shift the energy balance, consistent with the idea that BAT possesses tremendous fuel combusting capacity67. It is important to note that our work in no way discounts the importance of beige adipocytes in energy homeostasis; in fact, their metabolic benefits have already been demonstrated47,50. Our data do suggest that expansion of classical BAT alone is sufficient to achieve obesity resistance, even in the face of compromised beige adipocytes. Given the presence of classical BAT in adult humans and the conservation of miR-378 in human genome, targeting the miR-378 and Pde1b axis to expand BAT might be a potential strategy for the treatment of obesity and metabolic diseases. Alternatively, unlocking the exact cellular and molecular mechanisms of the iWAT negative response may provide us information on how to use beige adipocytes for the treatment.
METHODS
miRNA screen
BAT and gonadal WAT were isolated form wild type male C57BL6 mice (n=5, 10-week-old mice) and total RNA was extracted using TRIzol. miRNAs were reversed transcribed using QuantiMir kit (System Biosciences, Cat#RA420A-1). A total of 688 miRNAs were screened using qPCR-based miRNA profiling kit (System Biosciences, Cat#RA670A-1). Data of their expression levels normalized with levels of U6 snoRNA were listed in Supplementary Data 1. miRNAs that displayed a high enrichment in BAT versus WAT were selected and their expression in other tissues was further examined.
Animals
A 600 bp genomic fragment containing the miR-378 locus was amplified from the first intron of Pgc-1β, sequenced and fused downstream with the 5.4 kb aP2 promoter. The transgenic DNA fragment was gel-purified and injected into fertilized embryos harvested from C57BL/6 × SJL hybrid mice. Two independent transgenic lines, which show similar phenotypes, were obtained and backcrossed with C57BL/6 for at least 3 generations. Heterozygous leptin-deficient mice were purchased from Jackson Laboratory (Stock # 000632) and crossed with miR-378 transgenic mice to generate ob/ob mice and ob/ob mice carrying the miR-378 transgene.
Normal diet containing 4% (w/w) fat and high fat diet containing 35% (w/w) fat were purchased from Harlan Teklad and Bioserv (Product# F3282), respectively. Food consumption was measured daily for individually caged mice for two weeks. Circulating triglycerides and free fatty acids were measured with commercial kits (Sigma, Cat#TR0100). We placed mice in a 4°C cold room for 6 hr for cold challenging experiments. We intraperitoneally injected mice with β3 agonist CL316,243 (Tocris, Cat#1499) daily at 1 mg per kg body weight for 5 days.
The University of Massachusetts Medical School’s Institutional Animal Care and Use Committee approved all the animal studies. Littermate controls were used in all the experiments.
Cellularity
To measure genomic DNA content, fat depots were excised, weighted, and minced. Samples with equal amounts were digested with 10 mM Tris buffer (pH 7.5) containing protease K (0.4 mg per ml), and genomic DNA was precipitated with ethanol. DNA content was calculated as per fat depot.
Gene expression and western blot analyses
Total RNA was extracted using TRIzol. Gene expression was measured by RT-qPCR, and normalized with internal controls. Primer sequences were listed in Supplementary Table 1.
For western blot analysis, adipose tissue samples with equal amounts were homogenized with a lysis buffer [100 mM NaCl, 50 mM Tris (pH7.5), 0.5% Triton X-100, 5% (w/v) glycerol]. Tissue lysates were quantified, separated by SDS-PAGE, immunoblotted with antibodies to Ucp1 (Sigma #U6382, 1:2,000), tubulin (DSHB#E7-S, 1:10,000), and Pde1b (Abgent #AP11846B, 1:500). To measure levels of Prdm16 protein, iWAT tissue were obtained from 2-month old wild type and transgenic mice that were exposed at 4C for 6 hr. The tissues were chopped into small pieces and homogenized in cold cytoplasmic lysis buffer (10 mM HEPES pH 7.9,10 mM KCl, 0.1 mM EDTA, protease inhibitors and 1 mM PMSF) with dounce homogenization until a single cell suspension was achieved. The cell suspensions were swollen for 15 min on ice and nuclei were released by vortex vigorously with supplementation of 0.625% NP-40 in lysis buffer. The nuclei were then recovered by centrifugation at 3000 g for 5 minutes at 4C, subsequently resuspended in 150µl buffer (20 mM HEPES pH 7.9, 0.4 M NaCl, 10% glycerol, 1mM EDTA, protease inhibitors and 1 mM PMSF), and rocked at 4C for 30 min. The nuclear lysate was centrifuged at 16000 g for 10 min at 4C and the supernatant was loaded to SDS-PAGE after protein quantification. Prdm16 antibody was purchased from R&D Systems (AF6295, 1:500).
Oxygen consumption assays
Brown fat depots were weighed and chopped into small pieces. Portions with equal amount (30 mg) were suspended in 5 ml DPBS supplemented with 25mM glucose, 1mM pyruvate, and 2% BSA, and respiration was measured with a biological oxygen monitor (YSI model 5300A). Oxygen consumption was calculated as per depot.
Luciferase reporter assays
Mouse Pde1b 3’ UTR (1.24 kb) was cloned from genomic DNA, and a variant was also generated in which point mutations at the miR-378 targeting sequences were introduced. Both fragments were verified by sequencing, and were placed downstream of the luciferase gene in the pMIR-REPORT vector (Invitrogen). HEK293 cells in 48-well plates were first transfected with locked nucleic acid (LNA) miR-378 inhibitor (50 nM) or control oligos (50 nM) (both from Exiqon), using lipofectamine 2000 (Invitrogen). The next day, cells were transfected with the luciferase reporters (50 ng) and CMV-β-galactosidase plasmids (150 ng). Luciferase activity was measured 48 h after the first transfection and normalized with β-galactosidase activity.
cAMP and cGMP assays
Commercial kits (KGE012 and KGE003, R&D Systems) were used and manufacture’s instructions were followed. Briefly, BAT depot was weighed, and equivalent amount (~ 50 mg) was minced with scissors and then homogenized with a 1 ml-glass dounce tissue grinder in 500 µl 0.1 N HCl. Lysates were collected into eppendorf tubes and centrifuged at 10,000 g for 10 min. 200 µl supernatant was transferred to new tubes, and mixed with 28 µl 1N NaOH and 228 µl calibrator diluent RD5–55. Samples were centrifuged again at 10,000 g for 10 min, and 100 µl supernatant was used for assay. cAMP and cGMP levels were expressed as pmol/mg BAT‥
Lipolysis assays ex vivo
BAT and iWAT depots were dissected from 4-months old wild type and transgenic mice. Small pieces of adipose tissue with similar weights (~ 50 mg) obtained from similar locations were incubated for 1 hr at 37°C without shaking in 500 µl of KRB buffer (12 mM HEPES pH 7.4, 121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4 and 0.33 mM CaCl2) containing 2% FA free BSA and 0.1% glucose with or without 10 µM isoproterenol. Glycerol release was measured from incubation buffer using Free Glycerol Reagent (Sigma) and calculated as µg per mg tissue per hr‥
Adipogenesis in vitro
C3H10T1/2 cells were obtained from ATCC. Immortalized brown preadipocytes were generated previously68. miR-378 overexpression and Pde1b knockdown plasmids along with control plasmids were packaged in lentiviral vector plenti-CMV/puro69 and pSP108 (Addgene), respectively. Viruses were first titered in C3H10T1/2 cells and brown preadipocytes. Cells were then infected with equal number of viral particles, and selected with puromycin to generate pooled stable cells. Equal number of cells were replated at 70% confluency. The next day (Day -2), cells were changed to medium containing 20 nM insulin and 1 nM 3,3’,5-triiodo-L-thyronine (differentiation medium). At Day 0, differentiation was induced by culturing cells in differentiation medium supplemented with 0.5 µM dexamethasone, and 0.125 mM indomethacin, and with or without 0.5 mM isobutylmethylxanthine (IBMX) as indicated, for 48 hr. After this induction period, cells were replenished with new differentiation medium every other day.
Primary iWAT preadipocytes were isolated from 2-week old C57BL6/J mice through collagenase digestion. iWAT adipogenesis was performed by culturing confluent cells in medium containing 850 nM insulin, 1 nM 3,3’,5-triiodo-L-thyronine, 0.5 µM dexamethasone, 0.125 mM indomethacin and with or without 0.5mM IBMX. Differentiated iWAT adipocytes were either harvested for gene expression analysis or fixed for Oil Red O staining.
Primary BAT preadipocytes were isolated from neonatal (Day 1) wild type C56BL6 mice through collagenase digestion. Cells were pooled together and plated. When cells were reached 70% confluence, they were infected with lentiviruses and selected with 2 µg/ml puromycin. Cells were then trypsinized and equal number of cells were plated. Confluent cells were differentiated in medium containing 850 nM insulin, 1 nM 3,3’,5-triiodo-L-thyronine, 0.5 µM dexamethasone, and 0.125 mM indomethacin for 48 hr, and then switched to medium containing 20 nM insulin and 1 nM 3,3’,5-triiodo-L-thyronine for another 4 days.
Oil red O staining
Cells were fixed with 3.7% (W/V) formaldehyde for 15 min at room temperature and then stained with 0.3% (W/V) Oil Red O solution in 60% isopropyl alcohol for 1 hr. The excess staining was removed by washing with water.
Brown fat removal
Surgery was performed in two-month old wild type and transgenic mice. Mice were anesthetized with isofluorane via a vaporizer. The fur in the interscapular area was clipped, and incisions were made around the brown fat depot, and brown fat was removed. Wound clips were used to close the skin incision. Animals were allowed to recover for 10 days before RNA isolation.
Statistical analysis
Two-tailed Student’s t test was used for statistical analysis. P<0.05 was considered significant. Data are presented as mean SEM. Sample size was determined based on power analysis and standard practice. No data were excluded. Randomization and blinding strategy was used whenever possible.
Supplementary Material
Acknowledgements
We thank the Transgenic Animal Modeling Core at University of Massachusetts Medical School for the generation of miR-378 transgenic mouse lines, Suzanne Wheeler and Nina Bishop at Animal Medicine for performing brown fat removal, and Dr. Michael Czech for helpful comments. This work was supported by NIH/NIDDK R01DK076118 and American Diabetes Association bridging grant 7-13-BS-179-BR (Y.-X.W.). Indirect calorimetry study was performed by the UMass Mouse Metabolic Phenotyping Center supported by NIH U24-DK093000 (J.K.K.).
Footnotes
Author contributions
D.P., C.M., B.Q., R.H.F, D.Y.J, J.K.K. and B.L. performed the experiments and analyzed the data. L.J.Z. performed the bioinformatics analyses. Y.-X.W. designed and supervised the project and wrote the manuscript.
Competing financial interests
The authors declare no conflicts of interest or competing financial interests.
REFERENCES
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Cousin B, et al. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci. 1992;103(Pt 4):931–942. doi: 10.1242/jcs.103.4.931. [DOI] [PubMed] [Google Scholar]
- 3.Guerra C, Koza RA, Yamashita H, Walsh K, Kozak LP. Emergence of brown adipocytes in white fat in mice is under genetic control. Effects on body weight and adiposity. The Journal of clinical investigation. 1998;102:412–420. doi: 10.1172/JCI3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrovic N, et al. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. The Journal of biological chemistry. 2010;285:7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. PPARgamma agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell metabolism. 2012;15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu J, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cypess AM, et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat Med. 2013;19:635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cypess AM, et al. Identification and importance of brown adipose tissue in adult humans. N Engl J Med. 2009;360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–E452. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 10.van Marken Lichtenbelt WD, et al. Cold-activated brown adipose tissue in healthy men. N Engl J Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 11.Virtanen KA, et al. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 12.Zingaretti MC, et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2009;23:3113–3120. doi: 10.1096/fj.09-133546. [DOI] [PubMed] [Google Scholar]
- 13.Saito M, et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes. 2009;58:1526–1531. doi: 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jespersen NZ, et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 2013;17:798–805. doi: 10.1016/j.cmet.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 15.Lidell ME, et al. Evidence for two types of brown adipose tissue in humans. Nat Med. 2013;19:631–634. doi: 10.1038/nm.3017. [DOI] [PubMed] [Google Scholar]
- 16.Sharp LZ, et al. Human BAT possesses molecular signatures that resemble beige/brite cells. PLoS One. 2012;7:e49452. doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: is beige the new brown? Genes Dev. 2013;27:234–250. doi: 10.1101/gad.211649.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 19.Kajimura S, Saito M. A New Era in Brown Adipose Tissue Biology: Molecular Control of Brown Fat Development and Energy Homeostasis. Annual review of physiology. 2013 doi: 10.1146/annurev-physiol-021113-170252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nedergaard J, Cannon B. The changed metabolic world with human brown adipose tissue: therapeutic visions. Cell metabolism. 2010;11:268–272. doi: 10.1016/j.cmet.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 23.Cristancho AG, Lazar MA. Forming functional fat: a growing understanding of adipocyte differentiation. Nat Rev Mol Cell Biol. 2011;12:722–734. doi: 10.1038/nrm3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARgamma. Annual review of biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 25.Francis SH, Blount MA, Corbin JD. Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions. Physiol Rev. 2011;91:651–690. doi: 10.1152/physrev.00030.2010. [DOI] [PubMed] [Google Scholar]
- 26.Rajakumari S, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kajimura S, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seale P, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tseng YH, et al. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jimenez-Preitner M, et al. Plac8 is an inducer of C/EBPbeta required for brown fat differentiation, thermoregulation, and control of body weight. Cell Metab. 2011;14:658–670. doi: 10.1016/j.cmet.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 31.Sellayah D, Bharaj P, Sikder D. Orexin is required for brown adipose tissue development, differentiation, and function. Cell Metab. 2011;14:478–490. doi: 10.1016/j.cmet.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 32.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013 doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villanueva CJ, et al. Adipose subtype-selective recruitment of TLE3 or Prdm16 by PPARgamma specifies lipid storage versus thermogenic gene programs. Cell metabolism. 2013;17:423–435. doi: 10.1016/j.cmet.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen Y, et al. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nature communications. 2013;4:1769. doi: 10.1038/ncomms2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol. 2012;14:1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- 40.Yin H, et al. MicroRNA-133 controls brown adipose determination in skeletal muscle satellite cells by targeting Prdm16. Cell Metab. 2013;17:210–224. doi: 10.1016/j.cmet.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu W, et al. miR-133a regulates adipocyte browning in vivo. PLoS Genet. 2013;9:e1003626. doi: 10.1371/journal.pgen.1003626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun L, et al. Mir193b-365 is essential for brown fat differentiation. Nat Cell Biol. 2011;13:958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol. 2009;218:444–449. doi: 10.1002/jcp.21621. [DOI] [PubMed] [Google Scholar]
- 44.Uldry M, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 45.Graves RA, Tontonoz P, Platt KA, Ross SR, Spiegelman BM. Identification of a fat cell enhancer: analysis of requirements for adipose tissue-specific gene expression. J Cell Biochem. 1992;49:219–224. doi: 10.1002/jcb.240490303. [DOI] [PubMed] [Google Scholar]
- 46.Schulz TJ, et al. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seale P, et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seale P, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindegaard B, et al. Interleukin-18 activates skeletal muscle AMPK and reduces weight gain and insulin resistance in mice. Diabetes. 2013;62:3064–3074. doi: 10.2337/db12-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen P, et al. Ablation of PRDM16 and Beige Adipose Causes Metabolic Dysfunction and a Subcutaneous to Visceral Fat Switch. Cell. 2014;156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 52.Butler AA, Kozak LP. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes. 2010;59:323–329. doi: 10.2337/db09-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haas B, et al. Protein kinase G controls brown fat cell differentiation and mitochondrial biogenesis. Sci Signal. 2009;2:ra78. doi: 10.1126/scisignal.2000511. [DOI] [PubMed] [Google Scholar]
- 54.Shin S, Heo J, Yeo J, Lee C, Chung E. Genetic association of phosphodiesterase 1B (PDE1B) with carcass traits in Korean cattle. Molecular biology reports. 2012;39:4869–4874. doi: 10.1007/s11033-011-1280-6. [DOI] [PubMed] [Google Scholar]
- 55.Stone RT, et al. Identification of genetic markers for fat deposition and meat tenderness on bovine chromosome 5: development of a low-density single nucleotide polymorphism map. J Anim Sci. 2005;83:2280–2288. doi: 10.2527/2005.83102280x. [DOI] [PubMed] [Google Scholar]
- 56.Knezevic I, et al. A novel cardiomyocyte-enriched microRNA, miR-378, targets insulin-like growth factor 1 receptor: implications in postnatal cardiac remodeling and cell survival. The Journal of biological chemistry. 2012;287:12913–12926. doi: 10.1074/jbc.M111.331751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carrer M, et al. Control of mitochondrial metabolism and systemic energy homeostasis by microRNAs 378 and 378*. Proc Natl Acad Sci U S A. 2012;109:15330–15335. doi: 10.1073/pnas.1207605109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eichner LJ, et al. miR-378(*) mediates metabolic shift in breast cancer cells via the PGC-1beta/ERRgamma transcriptional pathway. Cell Metab. 2010;12:352–361. doi: 10.1016/j.cmet.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 59.Trajkovski M, Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends in endocrinology and metabolism: TEM. 2013;24:442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cederberg A, et al. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106:563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 61.Gerin I, et al. Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab. 2010;299:E198–E206. doi: 10.1152/ajpendo.00179.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bronnikov GE, Zhang SJ, Cannon B, Nedergaard J. A dual component analysis explains the distinctive kinetics of cAMP accumulation in brown adipocytes. J Biol Chem. 1999;274:37770–37780. doi: 10.1074/jbc.274.53.37770. [DOI] [PubMed] [Google Scholar]
- 63.Ganesan J, et al. MiR-378 Controls Cardiac Hypertrophy by Combined Repression of MAP Kinase Pathway Factors. Circulation. 2013 doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 64.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopecky J, Clarke G, Enerback S, Spiegelman B, Kozak LP. Expression of the mitochondrial uncoupling protein gene from the aP2 gene promoter prevents genetic obesity. J Clin Invest. 1995;96:2914–2923. doi: 10.1172/JCI118363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ye L, et al. Fat cells directly sense temperature to activate thermogenesis. Proc Natl Acad Sci U S A. 2013;110:12480–12485. doi: 10.1073/pnas.1310261110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rothwell NJ, Stock MJ. Luxuskonsumption, diet-induced thermogenesis and brown fat: the case in favour. Clin Sci (Lond) 1983;64:19–23. doi: 10.1042/cs0640019. [DOI] [PubMed] [Google Scholar]
- 68.Pan D, Fujimoto M, Lopes A, Wang YX. Twist-1 is a PPARdelta-inducible, negative-feedback regulator of PGC-1alpha in brown fat metabolism. Cell. 2009;137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Campeau E, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fidock M, Miller M, Lanfear J. Isolation and differential tissue distribution of two human cDNAs encoding PDE1 splice variants. Cell Signal. 2002;14:53–60. doi: 10.1016/s0898-6568(01)00207-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.