Abstract
Phytoremediation makes use of plants and associated microorganisms to clean up soils and sediments contaminated with inorganic and organic pollutants. In this study, switchgrass (Panicum virgatum) was used to test for its efficiency in improving the removal of three specific polychlorinated biphenyl (PCB) congeners (PCB 52, 77 and 153) in soil microcosms. The congeners were chosen for their ubiquity, toxicity, and recalcitrance. After 24 weeks of incubation, loss of 39.9 ± 0.41% of total PCB molar mass was observed in switchgrass treated soil, significantly higher than in unplanted soil (29.5 ± 3.4%) (p<0.05). The improved PCB removal in switchgrass treated soils could be explained by phytoextraction processes and enhanced microbial activity in the rhizosphere. Bioaugmentation with Burkholderia xenovorans LB400 was performed to further enhance aerobic PCB degradation. The presence of LB400 was associated with improved degradation of PCB 52, but not PCB 77 or PCB 153. Increased abundances of bphA (a functional gene that codes for a subunit of PCB-degrading biphenyl dioxygenase in bacteria) and its transcript were observed after bioaugmentation. The highest total PCB removal was observed in switchgrass treated soil with LB400 bioaugmentation (47.3 ± 1.22 %), and the presence of switchgrass facilitated LB400 survival in the soil. Overall, our results suggest the combined use of phytoremediation and bioaugmentation could be an efficient and sustainable strategy to eliminate recalcitrant PCB congeners and remediate PCB-contaminated soil.
Keywords: PCBs, switchgrass, phytoextraction, Burkholderia xenovorans LB400, biphenyl dioxygenase gene
1. Introduction
Polychlorinated biphenyls (PCBs) are a group of synthetic chemicals that were widely used as heat transfer fluids during the last century. The production and commercial use of PCBs were prohibited by the EPA in 1979 because they were found to be toxic and carcinogenic (Longnecker et al., 1997; Ross, 2004). Nevertheless, because of their high stability and hydrophobicity, PCBs persist in the environment and accumulate through the food web, posing a potential risk to public health and ecosystems (Ross, 2004). An estimated 1.3 million tons of PCBs were produced worldwide between the 1930s and the mid-1980s, and the current total global PCB burden in background soils (i.e. soils not considered contaminated) is estimated to be 21,000 tons (Abraham et al., 2002; Meijer et al., 2003). Conventional remediation strategies such as soil excavation and the following incineration or landfilling are usually expensive and can be considered unsustainable (Amend and Lederman, 1992). Phytoremediation, the use of plants and associated microbes to remove and detoxify PCBs, represents a potentially cost effective and less disruptive approach to clean up PCB contaminated soils and sediments (Van Aken et al., 2010).
The presence of specific plant species is known to enhance the PCB removal efficiency in soils and sediments (Campanella et al., 2002; Chekol et al., 2004; Ionescu et al., 2009; Slater et al., 2011; Smith et al., 2007). Plants can take up PCBs from soil and convert lower chlorinated congeners into hydroxylated products (Ficko et al., 2010; Greenwood et al., 2011; Kučerová et al., 2000; Lee and Fletcher, 1992; Rezek et al., 2007; Zhai et al., 2010). Also, plants contribute to PCB biodegradation by providing a favorable environment for PCB-degrading microorganisms and can thus increase PCB-degrading populations in comparison to unplanted soil (de Cárcer et al., 2007; Leigh et al., 2006; Slater et al., 2011; Uhlík et al., 2009). Plants not only increase soil permeability and oxygen diffusion in the rhizosphere, but also release organic exudates as inducers, surfactants and microbial growth factors thereby stimulating and facilitating microbial PCB biodegradation in the root zone (Gilbert and Crowley, 1997; Hernandez et al., 1997; Leigh et al., 2002; Van Aken et al., 2010). For example, augmentation of soil with plant-derived compounds such as carvone and salicylic acid assisted aerobic PCB degraders and increased PCB removal (Singer et al., 2000; Singer et al., 2003).
Microorganisms are known to degrade PCBs via two general processes: aerobic oxidation with biphenyl ring cleavage and anaerobic reductive dechlorination (Adrian et al., 2009; Borja et al., 2005; Gibson and Parales, 2000; Pieper, 2005; Wiegel and Wu, 2000). Most aerobic PCB-degraders employ the upper biphenyl degradation pathway, with biphenyl 2,3-dioxygenase (BphA) catalyzing the initial step, transforming susceptible PCB congeners into the corresponding cis-dihydrodiol (Furukawa, 1994). Under anaerobic conditions, some members of the phylum Chloroflexi use certain PCB congeners as electron acceptors and transform them into less chlorinated congeners (Adrian et al., 2009; Fagervold et al., 2007; Fagervold et al., 2005; Wiegel and Wu, 2000). Some efficient microbial PCB-degrading isolates, such as Burkholderia xenovorans strain LB400, Arthrobacter sp. strain B1B, Ralstonia eutropha H850, Dehalocococoides chlorocoercia DF1 have been introduced to contaminated soils and sediments in efforts to improve PCB removal (Payne et al., 2013; Payne et al., 2011; Petrić et al., 2011; Singer et al., 2000).
In this study, we monitored the removal of three selected PCB congeners (PCB 52, PCB 77 and PCB 153) in switchgrass-planted soil microcosms and attempt to improve PCB degradation by introducing biphenyl-grown Burkholderia xenovorans strain LB400 into the rhizosphere of selected microcosms. The PCB congeners used here were chosen because they represent bottlenecks in environmental PCB degradation (Bedard et al., 1986). PCB 52 and 153 are both ortho-chlorinated, while PCB 77 has a dioxin-like structure, and is considered one of the most toxic congeners (Van den Berg et al., 2006). All three congeners are quite ubiquitous in the environment (Meggo et al., 2013). A combination of PCB congener analysis and molecular microbial analysis were employed to elucidate the mechanism of PCB removal in the soil reactors.
2. Materials and Methods
2.1. Soil reactor set up
The soil microcosm set-up procedure has been described previously (Meggo et al., 2013). Briefly, PCB-free soil from the village of Middle Amana in Iowa, USA, was passed through a 60 mesh sieve before experiments. The soil was spiked with a mixture of PCB 52, PCB 77 and PCB 153 (99% pure) (Accustandard Inc., New Haven, CT) at the concentration of 500 ng g−1 each. This level of PCB contamination has been reported in soils and sediments previously (Martinez and Hornbuckle, 2011; Van Metre and Mahler, 2005). In this study we investigated the possibility of using switchgrass to clean up PCB contamination that is below the dredging requirement (50 ppm) but still poses a potential risk to human and environmental health. PCB congeners were dissolved in hexane before adding to the soil. A quartering technique was used to homogenize soil. The soil was divided into four quadrants on a quartering canvas, and 20 diagonal trajectories were used to mix the soil components. The contaminated soil was aged for two months at 25°C in sealed tubs to allow PCBs sequestration into the soil matrix and thereby better represent field conditions.
Plastic containers (33.8 cm × 21.6 cm ×211.9 cm) and lids with aluminum foil covers were used for soil reactor set-up. Each reactor was filled with 2500 g sieved and homogenized soil. The spiked soil reactors were planted with switchgrass (Panicum virgatum) seeds (Adams-Briscoe Seed Co., Jackson, GA) (Figure S1). Switchgrass was grown at 25°C in a plant growth chamber under a 16 hour light/8 hour dark photo period (light intensity of 200 mmol m−2 s−1 and 60% humidity). Each treatment was performed to one PCB-spiked soil reactor. An unplanted reactor and a switchgrass planted reactor filled with clean soil were used as controls. A 5 ml sterile syringe with the narrow tip removed was used to take vertical core samples randomly from the reactors. The core samples were homogenized and subjected to PCB congener analysis and microbial analysis. Soil samples were taken at 12 week and 24 week for PCB congener analysis. Plant samples (roots and shoots) were collected at the end of experiment. PCB concentration measurements for soil and plant samples were performed in triplicate. For microbial analysis, soil samples were taken from each microcosm every four weeks starting at week 12.
2.2. Burkholderia xenovorans strain LB400 bioaugmentation
Bioaugmentation with Burkholderia xenovorans strain LB400 was conducted to one switchgrass-planted reactor and one unplanted reactor once every month. Prior to bioaugmentation strain LB400 was grown in K1 medium on solid biphenyl (1 g) as sole carbon and energy source until exponential phase (OD600 0.6~0.8) (Zaitsev and Karasevich, 1985). Cells were harvested by centrifugation (5000 × g, 15 min), washed once with sterile K1 medium, resuspended in K1 medium and inoculated into reactors (approximately 109 CFU per g soil). Autoclaved LB400 was added to a switchgrass planted reactor as a control.
The abundance of cultivable aerobic biphenyl-degrading bacteria was estimated by CFU enumeration one day after each bioaugmentation. Soil samples (1 g) were mixed with 9 ml of 0.9% sodium chloride solution and shaken for 1 h at 225 rpm on a platform shaker (New Brunswick Scientific, Pittsburgh, PA) at 25°C. Serially diluted supernatant was spread on K1 agar plates with biphenyl crystals provided on the lid of an inverted petriplate as sole carbon and energy source. CFUs were enumerated after 72 h of incubation at 30°C.
2.3. PCB extraction and quantification
The PCB extraction procedure has been previously described (Meggo et al., 2013). Briefly, for PCB extraction, 1:1 hexane/acetone mixture (3 ml/g) was added to homogenized soil or plant material (5 g) and sonicated for 1 h. Before sonication, surrogate standards including PCB 14 (3,5-dichlorobiphenyl), deuterated PCB 65 (2,3,5,6-tetrachlorobiphenyl) and PCB 166 (2,3,4,4′,5,6-hexachlorobiphenyl) (Cambridge Isotope Laboratories, Inc.) were added into the samples (50 ng of each surrogate) to account for any loss during the extraction. Surrogate recoveries were 103 ± 17.2% (PCB 14), 97.9 ± 15.2% (PCB 65) and 97.7 ± 17.7% (PCB 166).
The sonicated mixture was centrifuged at 1500 × g for 5 min, after which the supernatant was transferred to another sterile vial. The precipitates were subjected to a second extraction. The combined supernatants of the first and second extraction were evaporated to dryness using rotary evaporation and then dissolved in hexane. Double extraction with concentrated sulfuric acid and hexane was performed to remove of lipids and other polar substances. This hexane extract was concentrated to approximately 0.5 ml under a gentle nitrogen stream. The concentrate was eluted with 10 ml of hexane through a filter consisting of 0.1 g of silica (70–230 mesh, Fisher Scientific, Inc.), 0.1 g of anhydrous sodium sulfate and 0.9 g silica gel acidified with H2SO4 (silica:H2SO4 = 2:1).
The PCB congener analysis procedure was previously described (Hu et al., 2010). Briefly, the concentrated extracts were spiked with the internal standard containing 100 ng of PCB 204 (2,2′,3,4,4′,5,6,6′-octachlorobiphenyl). The samples were analyzed for PCB congeners using a gas chromatograph with mass selective detection (GC-MS/MS) modified from the EPA method 1668A (U.S.EPA 1999). The quantification of PCBs was performed by an Agilent 6890N gas chromatograph with an Agilent 7683 series auto sampler coupled to a Waters Micromass Quattro micro GC mass spectrometer (Milford, MA, USA) operating under electron impact (EI) positive mode at 70 eV and multiple reaction monitoring (MRM), and the trap current was 200 μA. The retention windows were defined by PCB parent/daughter ion pairs from mono- to deca- homologs which were 188/152, 222/152.10, 255.96/186, 291.92/222, 325.88/255.90, 359.84/289.90, 393.80/323.90, 427.76/357.80, 461.72/391.83, 497.68/427.70, respectively.
2.4. DNA and RNA extraction, reverse transcription, and quantitative PCR
Total RNA and DNA were extracted in replicate immediately after sampling from 2 g soil using the MoBio RNA PowerSoil Total RNA Isolation Kit (Mobio, Carlsbad, CA) and RNA PowerSoil DNA Elution Accessory Kit (Mobio, Carlsbad, CA) and stored at −80°C prior to further analysis. Before quantitative PCR (qPCR), RNA samples were subjected to contaminating DNA removal, RNA clean-up and reverse transcription. Contaminating DNA was removed by DNase I treatment according to the manufacturer’s instructions (Biolab, Ipswich, MA). Each 100 μl DNase I treatment reaction contained 30 μl RNA and 20U DNase I. RNA was then purified with the RNeasy Mini Kit (Qiagen, Germantown, MD) and reversed-transcribed to cDNA by SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. Each 80 μl reverse transcription reaction contained 40 μl purified RNA, 10mM dNTP mix, and 3 μg random primers.
To evaluate the RNA recovery efficiency during DNase I treatment, RNA purification and reverse transcription, 1 ng of luciferase control mRNA (Promega, Madison, WI) was added to RNA sample before DNase I treatment. After reverse transcription, luciferase cDNA was quantified by qPCR with the ref primer set (Table S1) as described previously (Johnson et al., 2005). The RNA recovery efficiency averaged 14.2% ± 11.5% among all RNA samples processed in this study.
The abundances of total bacteria, aerobic PCB-degrading bacteria, and Burkholderia xenovorans strain LB400 were estimated by qPCR using the bacterial 16S rRNA gene primer set 16SU f/r (Nadkarni et al., 2002), bphA primer set bphA 463f/674r (Petrić et al., 2011), and LB400 16S–23S rRNA internal transcribed spacers (ITS) primer set LB400 84f/278r (Norini et al., 2013), respectively (Table S1). Each 25 μl qPCR reaction contained 12.5 μl Power SYBR Green PCR Master Mix (Invitrogen, Carlsbad, CA), and variable amounts of primers and templates (Table S2). Bovine serum albumin (0.5 μg) was added to relieve possible PCR inhibition (Kreader, 1996). PCR conditions were as follows: 10 min at 95°C, followed by 40 cycles at 95°C (15 s) and 60°C (1 min) followed by a dissociation step. All qPCR was performed in triplicate with an ABI 7000 Sequence Detection System (Applied Biosystem, Grand Island, NY) and fluorescence data was analyzed by ABI 7000 System SDS Software (Applied Biosystems, Grand Island, NY). With each primer set, the target gene was not detected in no template (DI water) controls (Ct value > 35). Additional qPCR information, such as primer concentrations, template concentrations, qPCR linear range, qPCR efficiency range of the standard curves, and Y-intercepts are provided in Table S2, in accordance with MIQE guidelines (Bustin et al., 2009).
For luciferase mRNA qPCR, the standard DNA template was PCR products amplified from luciferase DNA with primer set ref ST (Table S1). For the bacterial 16S rRNA gene, the standard DNA template was prepared from PCR products amplified from Burkholderia xenovorans strain LB400 with primer set 8F/1492R (Klappenbach et al., 2000). For bphA, the standard DNA template was the LB400 bphA (amplified with the 463f/674r primer set) cloned into the pCR 2.1-TOPO vector. For LB400 16S–23S rRNA ITS gene, the standard DNA template was the LB400 16S–23S rRNA ITS gene (amplified with the LB400 84f/278r primer set) cloned into the pCR 2.1-TOPO vector.
2.5. Statistical analyses
PCB congener data was analyzed with an independent sample t test using R. Total bacterial 16S rRNA gene abundance, bphA (gene and transcript) abundance and LB400 16S–23S rRNA ITS abundance were analyzed with a two factor analysis of variance after log transformation using R (Rieu and Powers, 2009).
3. Results and Discussion
3.1. Enhanced degradation of specific PCB congeners in soil with switchgrass treatment
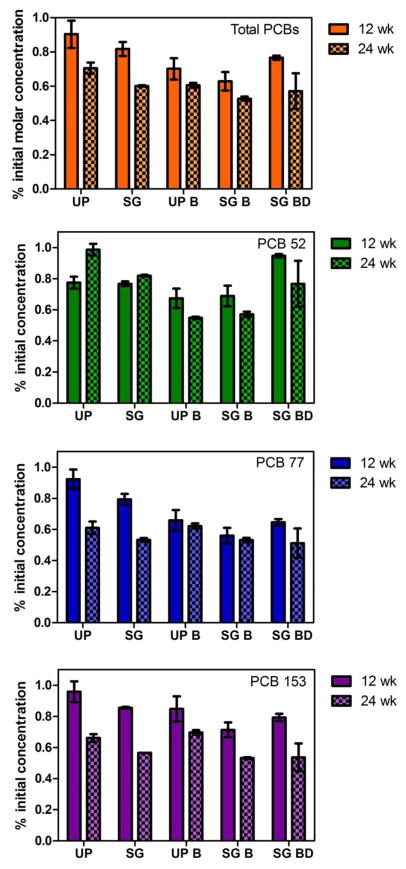
Significantly higher removal of PCB parent congeners (PCB 52, 77, 153) was achieved in the switchgrass-treated soil than in the unplanted soil after 24 weeks of incubation (p<0.05) (Figure 1). The presence of switchgrass enhanced PCB 52 removal more than the other two congeners (16.7%), while PCB 77 and PCB 153 removal was improved by 7.9% and 9.6%, respectively. Overall, 39.9 ± 0.4% of the total PCB molar mass loss was observed in switchgrass treated soil after 24 weeks of incubation, compared to 29.5 ± 3.4% in unplanted soil (Figure 1). Switchgrass has been observed previously to enhance PCB removal in soil and promote microbial activity in the rhizosphere (Chekol et al., 2004; Dzantor et al., 2000; Meggo et al., 2013). The increased PCB removal efficiency by switchgrass observed here is comparable to previous reports, although in those studies different plants were used and the soils were contaminated by PCB commercial mixtures rather than with specific congeners (Ding et al., 2009; Ionescu et al., 2009; Secher et al., 2013; Smith et al., 2007). In this study, addition of specific congeners allowed a focus on the variation in removals of congeners with differing chlorine positions.
Figure 1.
The percentage of initial molar concentrations for total PCB (orange), PCB 52 (green), PCB 77 (blue), and PCB 153(purple) after 12 weeks and 24 weeks incubation in unplanted soil (UP), switchgrass treated soil (SG), unplanted with bioaugmentaion soil (UP B), switchgrass treated with bioaugmentation soil (SG B), and switchgrass treated with bioaugmentation with dead LB400 soil (SG BD). Error bars indicate the standard deviation of three soil subsamples from the same reactor.
The enhanced PCB removal described above could be explained by rhizosphere biostimulation, rhizofiltration, phytoextraction, phytotransformation and phytovolatilization (Van Aken et al., 2010). We investigated a potential phytoextraction mechanism by quantifying total PCBs in switchgrass roots and shoots. At the end of the 24 week experiment, we found 4.3–7.3% of total PCBs in switchgrass roots, and 0.11–0.24% in shoots, suggesting that plant extraction processes were active as PCB removal mechanisms (Table 1). All three parent congeners and some transformation products were detected in switchgrass roots and shoots. Especially, the detection of PCB 153 in switchgrass shoots demonstrated its capability to take up and translocate highly chlorinated PCB congeners. Volatilization and deposition is excluded as a possible source of PCBs in the shoots because the shoot PCB concentration from the PCB-spiked reactor (9.30 ± 0.92 ng/g) is much higher than that from the blank reactor without PCB spiking (0.59 ± 0.21 ng/g) (Table S3). The uptake of PCBs by switchgrass and other plantshas been reported (Ficko et al., 2010; Greenwood et al., 2011; Liu et al., 2009), although in some studies the effect of phytoextraction in removing PCBs from soil was considered negligible because of the low percentage of total PCB mass observed in plant roots and shoots (<1% of total PCBs) (Di Gregorio et al., 2013; Huesemann et al., 2009). In this study, the fraction of total PCB mass in switchgrass was higher than previous studies, probably because of the high plant biomass for each switchgrass treated soil microcosm (560 ± 51 g root sand 360 ± 41 g shoots per soil microcosm). On the other hand, phytovolatilzation and phytotransformation were not considered to significantly affect PCB removal, given the relatively low PCB concentrations in switchgrass shoots. In a previous study it was also shown that hydroxylated metabolites were not detected in switchgrass hydroponically exposed to PCBs, suggesting that PCB transformation in switchgrass was not active(Liu et al., 2009).
Table 1.
Molar mass balance of PCBs in unplanted soil (UP), switchgrass treated soil (SG), unplanted with bioaugmentaion soil (UP B), switchgrass treated with bioaugmentation soil (SG B), and switchgrass treated with bioaugmentation with dead LB400 soil (SG BD) after 24 weeks of incubation. Results were the reported as mean ± standard deviation (n=3).
| PCB fraction in soils | PCB fraction in roots | PCB fraction in shoots | Total recovery | ||
|---|---|---|---|---|---|
| Overall | UP | 70.6 ± 4.1% | - | - | 70.6 ± 4.1% |
| SG | 60.1 ± 2.0% | 7.3 ± 0.3% | 0.11 ± 0.01% | 67.5 ± 2.3% | |
| UP B | 60.6 ± 2.5% | - | - | 60.6 ± 2.5% | |
| SG B | 52.7 ± 2.1% | 4.3 ± 0.2% | 0.24 ± 0.03% | 57.3 ± 2.3% | |
| SG BD | 57.1 ± 10.6% | - | - | 57.1 ± 10.6% | |
|
| |||||
| PCB 52 | UP | 98.6 ± 4.8% | - | - | 98.6 ± 4.8% |
| SG | 81.9 ± 2.4% | 9.4 ± 0.3% | 0.17 ± 0.01% | 91.4 ± 2.7% | |
| UP B | 54.8 ± 1.8% | - | - | 54.8 ± 1.8% | |
| SG B | 57.0 ± 2.4% | 4.7 ± 0.2% | 0.40 ± 0.05% | 62.2 ± 2.5% | |
| SG BD | 76.7 ± 14.9% | - | - | 76.7 ± 14.9% | |
|
| |||||
| PCB 77 | UP | 61.0 ± 4.9% | - | - | 61.0 ± 4.9% |
| SG | 53.2 ± 2.8% | 6.0 ± 0.3% | 0.06 ± 0.01% | 59.2 ± 3.0% | |
| UP B | 62.1 ± 3.4% | - | - | 62.1 ± 3.4% | |
| SG B | 53.1 ± 2.9% | 3.8 ± 0.3% | 0.16 ± 0.02% | 57.1 ± 3.1% | |
| SG BD | 51.1 ± 9.7% | - | - | 51.1 ± 9.7% | |
|
| |||||
| PCB 153 | UP | 66.1 ± 4.2% | - | - | 66.1 ± 4.2% |
| SG | 56.5 ± 2.9% | 8.2 ± 0.5% | 0.11 ± 0.02% | 64.8 ± 3.4% | |
| UP B | 69.7 ± 3.9% | - | - | 69.7 ± 3.9% | |
| SG B | 53.2 ± 2.8% | 5.0 ± 0.3% | 0.17 ± 0.02% | 58.4 ± 3.1% | |
| SG BD | 53.7 ± 9.4 % | - | - | 53.7 ± 9.4 % | |
Biostimulation of microbial PCB degradation is another mechanism potentially contributing to enhanced PCB removal in the rhizosphere soil. The occurrence of aerobic microbial PCB degradation in both unplanted and switchgrass treated soils was verified by the decrease in total PCB molar mass (Figure 1A). It is estimated that the enhanced microbial activity in the rhizosphere was responsible for the 7.1% lower molar mass recovery in the switchgrass-treated reactor than that in the unplanted reactor (p<0.05) (Table 1). However, the recoveries of PCB 77 and 153 in the switchgrass treated reactor are not significantly different from those in the unplanted reactor, suggesting microbial degradation of PCB 77 and 153 was not significantly improved with the presence of switchgrass (Table 1). PCB 52 appears to be the most susceptible to aerobic degradation by indigenous microorganisms. We propose that the dioxin-like structure of PCB 77 renders it relatively more toxic than other tetrachlorinated biphenyls. PCB 153 is the most recalcitrant probably because of its chlorine substituents at the 2,3,5,2′,3′,5′ - positions, which make the congener unsuitable for the oxidation at either the 2,3- or 3,4-position (Arnett et al., 2000; Mondello et al., 1997). The high chlorine content of PCB 153 also results in high hydrophobicity and reduces its accessibility to enzymes. The bulkiness of the chlorine atoms may also prevent access to the enzyme’s active site (Bedard et al., 1986).
3.2. Molecular biology analyses of unplanted and switchgrass planted soils
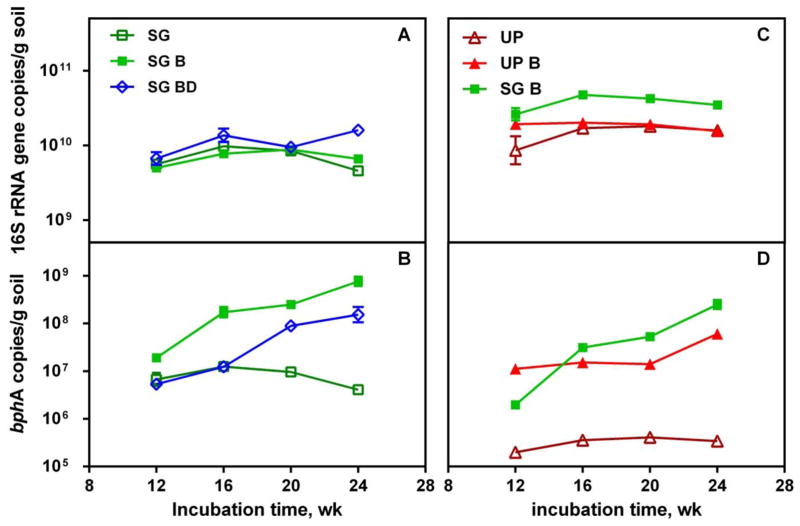
We estimated the abundance of total bacteria, biphenyl dioxygenase genes (bphA) and bphA transcripts as an independent means of tracking aerobic microbial PCB biodegradation during the experiment. Total bacterial abundance (as estimated by the abundance of bacterial 16S rRNA genes) was significantly higher in switchgrass reactors than in unplanted reactors (p<0.05, Figure 3A). We suspect that the switchgrass serves to improve oxygen diffusion and soil permeability (Singer et al., 2003). Switchgrass root exudates could also increase the microbial population in the rhizosphere by serving as a carbon and energy source (Kamath et al., 2004).
Figure 3.
qPCR analysis of bacterial 16S rRNA gene (A) and bphA (B) abundance with time in switchgrass treated soil (SG), switchgrass treated soil with bioaugmentation (SG B), switchgrass treated soil with autoclaved LB400 bioaugmention (SG BD); and bacterial 16S rRNA gene (C)and bphA (D) inunplanted soil (UP), unplanted soil with LB400 bioaugmentation (UP B), switchgrass treated soil with bioaugmentation (SG B). Error bars indicate the range of two soil subsamples from the same reactor.
Detecting bphA in both unplanted and switchgrass-treated soils indicates that the indigenous soil microbial community harbored the potential for aerobic PCB degradation. In the presence of switchgrass, bphA abundance was about 20 times higher (p<0.05, Figure 3B). However, bphA transcripts in both switchgrass planted and unplanted reactors were below our RT-qPCR quantification limit (40 copies/qPCR reaction), suggesting that bphA expression was not promoted by the presence of switchgrass. It is possible that the presence of other carbon sources in the rhizosphere, such as naturally-occurring organic carbon and organic plant exudates, made PCBs less favorable as substrates for indigenous microorganisms and subsequently inhibited bphA expression (Kamath et al., 2004; Parnell et al., 2010; Rentz et al., 2004). Soil extract may also inhibit bphA expression (Master and Mohn, 2001). Low levels of bphA expression would also manifest as low microbial PCB degradation in the switchgrass rhizosphere, which supports the PCB congener data presented above.
3.3. Enhanced microbial PCB degradation in soil with LB400 bioaugmentation
Thus, to further enhance aerobic degradation of PCB congeners in the rhizosphere, biphenyl-grown Burkholderia xenovorans strain LB400 was introduced to the soil (at a loading of approximately 109 CFU per g soil). LB400 is an effective aerobic PCB-degrader, capable of degrading a broad spectrum of PCB congeners including those used in this study (PCB 52, 77, 153) (Bedard et al., 1986; Bopp, 1986). With LB400 bioaugmentation, total PCB molar mass loss increased to 39.4 ± 1.4% and 47.3 ± 1.2 % in the unplanted and switchgrass-treated soil, respectively, after 24 weeks incubation, suggesting enhanced aerobic PCB degradation (Figure 1). Removal of PCB 52, 77 and 153 was not significantly improved in the control switchgrass reactor which was amended with autoclaved LB400 (Figure 1). This suggests that the bioaugmented LB400 biomass is not simply acting as a fertilizer for the plant.
Specifically, PCB 52 removal appeared to be most improved by LB400 bioaugmentation, achieving 43.7% and 24.9% more removal in unplanted and switchgrass-treated soils, respectively (p<0.05). PCB 153 removal in switchgrass-treated soil with bioaugmentation increased by 3.3% as compared to soil with only switchgrass treatment (p<0.05). In unplanted soil with bioaugmentation, PCB 77 and 153 removal was not improved (Figure 1). This observation is consistent with previous LB400 PCB biodegradation studies, where LB400 displayed a stronger capability to degrade PCB 52 than PCB 77 and 153 (Bedard et al., 1986; Bopp, 1986; Gibson et al., 1993; Rein et al., 2007). Compared with PCB 52, PCB 77 and 153 have higher Kow values and lower water solubilities (Van Noort et al., 2010). Although LB400 could degrade PCB 77 and 153 in resting cell assays and the biphenyl dioxygenase from LB400 is capable of oxidation at both the 2,3- and 3,4-position of the biphenyl ring, in the soil microcosms, the two congeners may be sorbed strongly to soil particles and thus are less bioavailable (Bedard et al., 1986; Haddock et al., 1995).
3.4. Molecular biology analyses of unplanted, switchgrass planted, and LB400 bioaugmented soils
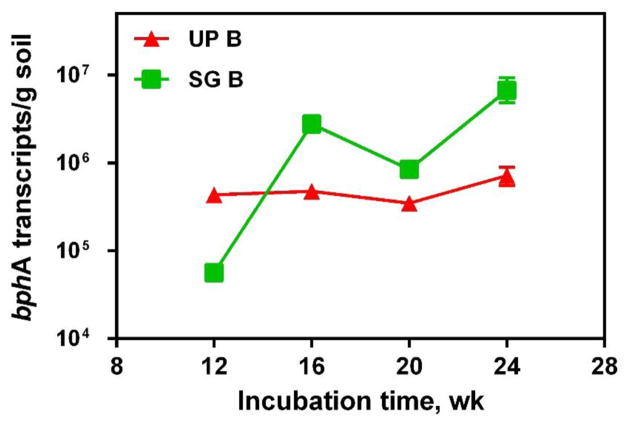
The enhanced PCB removal in bioaugmented soil was associated with elevated abundance of bphA and its transcripts, suggesting that aerobic PCB degradation was actively occurring (Figures 3 and 4). In bioaugmented soil, bphA abundance increased throughout incubation time, and was two orders of magnitude higher than in reactors without bioaugmentation (p<0.05, Figure 3B, 3D). The transcripts of bphA were detected (3.17 × 105 – 8.90 × 105 transcripts per g soil in unplanted soil with bioaugmentation, 4.73 × 104 – 4.14 × 106 transcripts per g soil in switchgrass treated soil with bioaugmentation), suggesting successful bphA expression (Figure 4).
Figure 4.
qPCR analysis of bphA transcripts with time in unplanted soil with bioaugmentation (UP B), and switchgrass treated soil with bioaugmention (SG B). Error bars indicate the range of two soil subsamples from the same reactor.
Bioaugmentation did not significantly increase the total microbial abundance (p>0.05, Figure 3A, 3C). Yet the reactor with autoclaved LB400 injection displayed greater total 16S rRNA gene abundance than the reactor with live LB400 bioaugmentation (p<0.05, Figure 3A), probably because that debris from the autoclaved LB400 cells was used as carbon and energy source by indigenous microorganisms.
The survival of bioaugmented bacteria is crucial to successful bioaugmentation and contaminant removal (Bouchez et al., 2000). To improve LB400 survival and keep a relatively stable LB400 population, we performed bioaugmentation once a month. The survival of LB400 after bioaugmentation was estimated by quantifying biphenyl-degrading bacteria with CFU enumeration. After bioaugmentation, the biphenyl-degrading bacterial abundance increased (4.7 × 108 –1.9 × 109 CFU per g soil for unplanted reactor, 2.1× 108 –2.3 × 109 CFU per g soil for switchgrass planted reactor). Yet this number decreased to the level of approximately 107 CFU per g soil after four weeks, prior to the next bioaugmentation event (Figure S2). The decrease may result from the competition with indigenous microbial community members and a possibly stressful environment caused by PCBs (Kim et al., 2001; Parnell et al., 2006; Van Veen et al., 1997). The LB400 16–23S internal transcribed spacer (ITS) region was quantified approximately two weeks after bioaugmentation, and ranged from 1.16 × 107 to 4.15 × 108 copies per g unplanted soil, and 9.61 × 107 to 7.04 × 108 copies per g switchgrass treated soil (Figure 5). An accumulation of ITSs was observed throughout the 24 week experiment, suggesting that after each bioaugmentation, a percentage of LB400 adapted to the soil environment and survived alongside indigenous microorganisms (Figure 5). The support of plants to bioaugmentation has been reported (Juhanson et al., 2009; Secher et al., 2013; Tam and Wong, 2008). Switchgrass facilitated LB400 survival possibly by improving aeration and providing root exudates as carbon and energy sources. The root system can help spread LB400 through the PCB contaminated soil. Furthermore, switchgrass may increase the bioavailability of PCB molecules by releasing organic compounds as surfactants (LeFevre et al., 2013).
Figure 5.
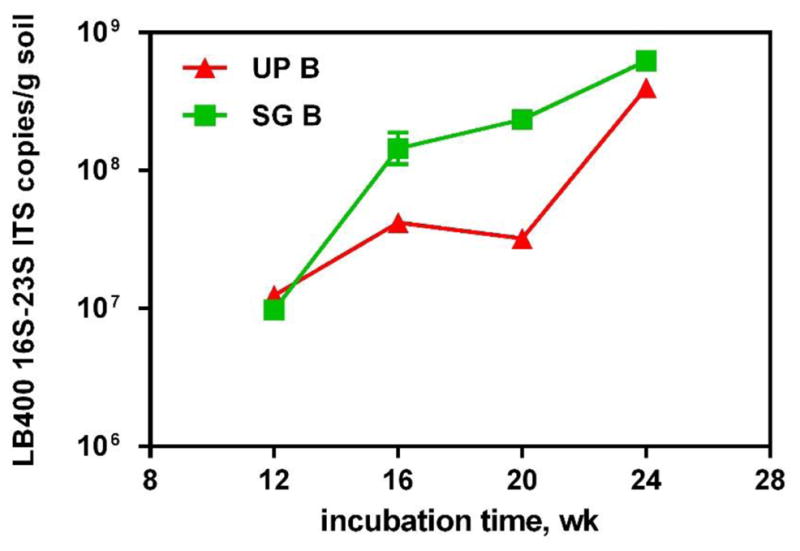
qPCR analysis of LB400 16S–23S ITS abundance with time in unplanted soil with bioaugmentation (UP B), and switchgrass treatedsoil with bioaugmention (SG B). Error bars indicate the range of two soil subsamples from the same reactor.
4. Conclusions
In this study, switchgrass improved the removal of a three congener mixture of PCB 52, 77 and 153, likely by phytoextraction and microbial biostimulation mechanisms. PCB 52 was most susceptible to microbial degradation, and its removal was improved greater in switchgrass-planted and LB400 bioaugmentated reactors than PCB 77 and 153. LB400 abundance was greater in the presence of switchgrass as revealed by the LB400 16–23S ITS gene quantification. Also, the most PCB removal and the highest bphA and transcript abundances were observed in bioaugmented soil with switchgrass treatment after 24 weeks, indicating switchgrass not only benefited LB400 survival, but also facilitated PCB removal by LB400. The combination of phytoremediation and bioaugmentation offers an efficient and environmental-friendly strategy to eliminate recalcitrant PCB congeners and remediate PCB contamination in the environment.
Supplementary Material
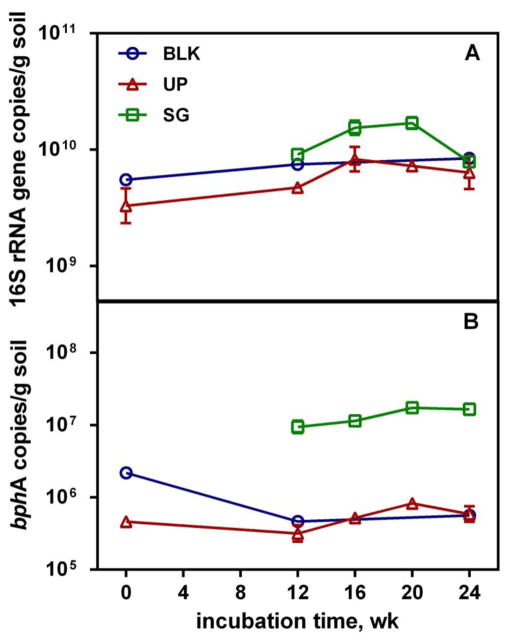
Figure 2.
qPCR analysis of (A) bacterial 16S rRNA genes and (B) bphA abundance with time in blank (BLK), PCB spiked and unplanted soil (UP), PCB spiked and switchgrass treated soil (SG). Error bars indicate the range of two soil subsamples from the same reactor.
Highlights.
Switchgrass improved PCB 52, 77 and 153 removal from soil by both plant and microbial activity.
PCB 52 removal in planted and LB400-augmented reactors was greater than PCB 77 and 153 removal.
Increased bphA abundance and expression was noted in LB400-augmented reactors.
Switchgrass presence appeared to improve LB400 survival and facilitated increased PCB removal.
Acknowledgments
This research was funded by the National Institute of Environmental Health Sciences (NIEHS) Superfund Research Program (SRP) Grant No. P42ES13661 and by a fellowship for YL from the Center for Biocatalysis and Bioprocessing at the University of Iowa. We thank Dr. Kai Wang of the University of Iowa for helping us with the statistical analyses. We thank Dr. Paige Novak of the University of Minnesota for graciously providing Burkholderia xenovorans LB400 cells. Finally, we thank Dr. Yang Oh Jin for developing the luciferase qPCR primer sets used in this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham WR, Nogales B, Golyshin PN, Pieper DH, Timmis KN. Polychlorinated biphenyl-degrading microbial communities in soils and sediments. Curr Opin Microbiol. 2002;5:246–253. doi: 10.1016/s1369-5274(02)00323-5. [DOI] [PubMed] [Google Scholar]
- Adrian L, Dudková V, Demnerová K, Bedard DL. “Dehalococcoides” sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol. 2009;75:4516–4524. doi: 10.1128/AEM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amend LJ, Lederman PB. Critical evaluation of PCB remediation technologies. Environ Prog. 1992;11:173–177. [Google Scholar]
- Arnett CM, Parales JV, Haddock JD. Influence of chlorine substituents on rates of oxidation of chlorinated biphenyls by the biphenyl dioxygenase of Burkholderia sp strain LB400. Appl Environ Microbiol. 2000;66:2928–2933. doi: 10.1128/aem.66.7.2928-2933.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard DL, Unterman R, Bopp LH, Brennan MJ, Haberl ML, Johnson C. Rapid assay for screening and characterizing microorganisms for the ability to degrade polychlorinated biphenyls. Appl Environ Microbiol. 1986;51:761–768. doi: 10.1128/aem.51.4.761-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp LH. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. Journal of Industrial Microbiology & Biotechnology. 1986;1:23–29. [Google Scholar]
- Borja J, Taleon DM, Auresenia J, Gallardo S. Polychlorinated biphenyls and their biodegradation. Process Biochem. 2005;40:1999–2013. [Google Scholar]
- Bouchez T, Patureau D, Dabert P, Juretschko S, Dore J, Delgenes P, Moletta R, Wagner M. Ecological study of a bioaugmentation failure. Environmental Microbiology. 2000;2:179–190. doi: 10.1046/j.1462-2920.2000.00091.x. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Campanella BF, Bock C, Schroder P. Phytoremediation to increase the degradation of PCBs and PCDD/Fs - potential and limitations. Environ Sci Pollut R. 2002;9:73–85. doi: 10.1007/BF02987318. [DOI] [PubMed] [Google Scholar]
- Chekol T, Vough LR, Chaney RL. Phytoremediation of polychlorinated biphenyl-contaminated soils: the rhizosphere effect. Environ Int. 2004;30:799–804. doi: 10.1016/j.envint.2004.01.008. [DOI] [PubMed] [Google Scholar]
- de Cárcer DA, Martín M, Karlson U, Rivilla R. Changes in bacterial populations and in biphenyl dioxygenase gene diversity in a polychlorinated biphenyl-polluted soil after introduction of willow trees for rhizoremediation. Appl Environ Microbiol. 2007;73:6224–6232. doi: 10.1128/AEM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio S, Azaizeh H, Lorenzi R. Biostimulation of the autochthonous microbial community for the depletion of polychlorinated biphenyls (PCBs) in contaminated sediments. Environ Sci Pollut R. 2013;20:3989–3999. doi: 10.1007/s11356-012-1350-x. [DOI] [PubMed] [Google Scholar]
- Ding N, Guo HC, Hayat T, Wu YP, Xu JM. Microbial community structure changes during Aroclor 1242 degradation in the rhizosphere of ryegrass (Lolium multifiorum L.) FEMS Microbiol Ecol. 2009;70:305–314. doi: 10.1111/j.1574-6941.2009.00742.x. [DOI] [PubMed] [Google Scholar]
- Dzantor EK, Chekol T, Vough LR. Feasibility of using forage grasses and legumes for phytoremediation of organic pollutants. Journal of Environmental Science and Health Part a-Toxic/Hazardous Substances & Environmental Engineering. 2000;35:1645–1661. [Google Scholar]
- Fagervold SK, May HD, Sowers KR. Microbial reductive dechlorination of Aroclor 1260 in Baltimore harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl Environ Microbiol. 2007;73:3009–3018. doi: 10.1128/AEM.02958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagervold SK, Watts JE, May HD, Sowers KR. Sequential reductive dechlorination of meta-chlorinated polychlorinated biphenyl congeners in sediment microcosms by two different Chloroflexi phylotypes. Appl Environ Microbiol. 2005;71:8085–8090. doi: 10.1128/AEM.71.12.8085-8090.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ficko SA, Rutter A, Zeeb BA. Potential for phytoextraction of PCBs from contaminated soils using weeds. Science of the Total Environment. 2010;408:3469–3476. doi: 10.1016/j.scitotenv.2010.04.036. [DOI] [PubMed] [Google Scholar]
- Furukawa K. Molecular genetics and evolutionary relationship of PCB-degrading bacteria. Biodegradation. 1994;5:289–300. doi: 10.1007/BF00696466. [DOI] [PubMed] [Google Scholar]
- Gibson DT, Cruden DL, Haddock JD, Zylstra GJ, Brand JM. Oxidation of polychlorinated-biphenyls by Pseudomonas sp strain LB400 and Pseudomonas Pseudoalcaligenes KF707. Journal of Bacteriology. 1993;175:6735–6735. doi: 10.1128/jb.175.14.4561-4564.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DT, Parales RE. Aromatic hydrocarbon dioxygenases in environmental biotechnology. Current Opinion in Biotechnology. 2000;11:236–243. doi: 10.1016/s0958-1669(00)00090-2. [DOI] [PubMed] [Google Scholar]
- Gilbert ES, Crowley DE. Plant compounds that induce polychlorinated biphenyl biodegradation by Arthrobacter sp. strain B1B. Appl Environ Microbiol. 1997;63:1933–1938. doi: 10.1128/aem.63.5.1933-1938.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood SJ, Rutter A, Zeeb BA. The absorption and translocation of polychlorinated biphenyl congeners by Cucurbita pepo ssp pepo. Environ Sci Technol. 2011;45:6511–6516. doi: 10.1021/es200598u. [DOI] [PubMed] [Google Scholar]
- Haddock JD, Horton JR, Gibson DT. Dihydroxylation and dechlorination of chlorinated bipenyls by purified biphenyl 2,3-dioxygenase from Pseudomonas sp. strain LB400. Journal of Bacteriology. 1995;177:20–26. doi: 10.1128/jb.177.1.20-26.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez BS, Koh SC, Chial M, Focht DD. Terpene-utilizing isolates and their relevance to enhanced biotransformation of polychlorinated biphenyls in soil. Biodegradation. 1997;8:153–158. [Google Scholar]
- Hu DF, Lehmler HJ, Martinez A, Wang K, Hornbuckle KC. Atmospheric PCB congeners across Chicago. Atmospheric Environment. 2010;44:1550–1557. doi: 10.1016/j.atmosenv.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesemann MH, Hausmann TS, Fortman TJ, Thom RM, Cullinan V. In situ phytoremediation of PAH- and PCB-contaminated marine sediments with eelgrass (Zostera marina) Ecological Engineering. 2009;35:1395–1404. [Google Scholar]
- Ionescu M, Beranova K, Dudkova V, Kochankova L, Demnerova K, Macek T, Mackova M. Isolation and characterization of different plant associated bacteria and their potential to degrade polychlorinated biphenyls. International Biodeterioration & Biodegradation. 2009;63:667–672. [Google Scholar]
- Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with a application to the tceA reductive dehalogenase gene. Appl Environ Microbiol. 2005;71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhanson J, Truu J, Heinaru E, Heinaru A. Survival and catabolic performance of introduced Pseudomonas strains during phytoremediation and bioaugmentation field experiment. FEMS Microbiol Ecol. 2009;70:446–455. doi: 10.1111/j.1574-6941.2009.00754.x. [DOI] [PubMed] [Google Scholar]
- Kamath R, Schnoor JL, Alvarez PJJ. Effect of root-derived substrates on the expression of nah-lux genes in Pseudomonas fluorescens HK44: implications for PAH biodegradation in the rhizosphere. Environ Sci Technol. 2004;38:1740–1745. doi: 10.1021/es0306258. [DOI] [PubMed] [Google Scholar]
- Kim IS, Lee H, Trevors JT. Effects of 2,2′,5,5′-tetrachlorobiphenyl and biphenyl on cell membranes of Ralstonia eutropha H850. FEMS Microbiol Lett. 2001;200:17–24. doi: 10.1111/j.1574-6968.2001.tb10686.x. [DOI] [PubMed] [Google Scholar]
- Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66:1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader CA. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kučerová P, Macková M, Chromá L, Burkhard J, Tríska J, Demnerová K, Macek T. Metabolism of polychlorinated biphenyls by Solanum nigrum hairy root clone SNC-9O and analysis of transformation products. Plant and Soil. 2000;225:109–115. [Google Scholar]
- Lee I, Fletcher JS. Involvement of mixed-function oxidase systems in polychlorinated biphenyl metabolism by plant-cells. Plant Cell Reports. 1992;11:97–100. doi: 10.1007/BF00235262. [DOI] [PubMed] [Google Scholar]
- LeFevre GH, Hozalski RM, Novak PJ. Root exudate enhanced contaminant desorption: an abiotic contribution to the rhizosphere effect. Environ Sci Technol. 2013;47:11545–11553. doi: 10.1021/es402446v. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Fletcher JS, Fu XO, Schmitz FJ. Root turnover: an important source of microbial substrates in rhizosphere remediation of recalcitrant contaminants. Environ Sci Technol. 2002;36:1579–1583. doi: 10.1021/es015702i. [DOI] [PubMed] [Google Scholar]
- Leigh MB, Prouzová P, Macková M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–2342. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JY, Hu DF, Jiang GB, Schnoor JL. In vivo biotransformation of 3,3′,4,4′-tetrachlorobiphenyl by whole plants - poplars and switchgrass. Environ Sci Technol. 2009;43:7503–7509. doi: 10.1021/es901244h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker MP, Rogan WJ, Lucier G. The human health effects of DDT (dichlorodiphenyltrichloroethane) and PCBs (polychorinated biphenyls) and an overview of organochlorines in public health 1. Annu rev publ health. 1997;18:211–244. doi: 10.1146/annurev.publhealth.18.1.211. [DOI] [PubMed] [Google Scholar]
- Martinez A, Hornbuckle KC. Record of PCB congeners, sorbents and potential toxicity in core samples in Indiana Harbor and Ship Canal. Chemosphere. 2011;85:542–547. doi: 10.1016/j.chemosphere.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master ER, Mohn WW. Induction of bphA, encoding biphenyl dioxygenase, in two polychlorinated biphenyl-degrading bacteria, psychrotolerant Pseudomonas strain Cam-1 and mesophilic Burkholderia strain LB400. Appl Environ Microbiol. 2001;67:2669–2676. doi: 10.1128/AEM.67.6.2669-2676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meggo RE, Schnoor JL, Hu D. Dechlorination of PCBs in the rhizosphere of switchgrass and poplar. Environ Pollut. 2013;178:312–321. doi: 10.1016/j.envpol.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer SN, Ockenden WA, Sweetman A, Breivik K, Grimalt JO, Jones KC. Global distribution and budget of PCBs and HCB in background surface soils: implications for sources and environmental processes. Environ Sci Technol. 2003;37:667–672. doi: 10.1021/es025809l. [DOI] [PubMed] [Google Scholar]
- Mondello FJ, Turcich MP, Lobos JH, Erickson BD. Identification and modification of biphenyl dioxygenase sequences that determine the specificity of polychlorinated biphenyl degradation. Appl Environ Microbiol. 1997;63:3096–3103. doi: 10.1128/aem.63.8.3096-3103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadkarni MA, Martin FE, Jacques NA, Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology-Sgm. 2002;148:257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- Norini MP, Secher C, Lollier M, Jezequel K, Cornu JY, Lebeau T. Quantification of the 16S–23S rRNA internal transcribed spacers of Burkholderia xenovorans strain LB400 using real-time PCR in soil samples. Lett Appl Microbiol. 2013;56:366–372. doi: 10.1111/lam.12057. [DOI] [PubMed] [Google Scholar]
- Parnell JJ, Denef VJ, Park J, Tsoi T, Tiedje JM. Environmentally relevant parameters affecting PCB degradation: carbon source- and growth phase-mitigated effects of the expression of the biphenyl pathway and associated genes in Burkholderia xenovorans LB400. Biodegradation. 2010;21:147–156. doi: 10.1007/s10532-009-9289-4. [DOI] [PubMed] [Google Scholar]
- Parnell JJ, Park J, Denef V, Tsoi T, Hashsham S, Quensen J, 3rd, Tiedje JM. Coping with polychlorinated biphenyl (PCB) toxicity: Physiological and genome-wide responses of Burkholderia xenovorans LB400 to PCB-mediated stress. Appl Environ Microbiol. 2006;72:6607–6614. doi: 10.1128/AEM.01129-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, Fagervold SK, May HD, Sowers KR. Remediation of polychlorinated biphenyl impacted sediment by concurrent bioaugmentation with anaerobic halorespiring and aerobic degrading bacteria. Environ Sci Technol. 2013;47:3807–3815. doi: 10.1021/es304372t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne RB, May HD, Sowers KR. Enhanced reductive dechlorination of polychlorinated biphenyl impacted sediment by bioaugmentation with a dehalorespiring bacterium. Environ Sci Technol. 2011;45:8772–8779. doi: 10.1021/es201553c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrić I, Hršak D, Fingler S, Udiković-Kolić N, Bru D, Martin-Laurent F. Insight in the PCB-degrading functional community in long-term contaminated soil under bioremediation. Journal of Soils and Sediments. 2011;11:290–300. [Google Scholar]
- Pieper DH. Aerobic degradation of polychlorinated biphenyls. Appl Microbiol Biotechnol. 2005;67:170–191. doi: 10.1007/s00253-004-1810-4. [DOI] [PubMed] [Google Scholar]
- Rein A, Fernqvist MM, Mayer P, Trapp S, Bittens M, Karlson UG. Degradation of PCB congeners by bacterial strains. Appl Microbiol Biotechnol. 2007;77:469–481. doi: 10.1007/s00253-007-1175-6. [DOI] [PubMed] [Google Scholar]
- Rentz JA, Alvarez PJJ, Schnoor JL. Repression of Pseudomonas putida phenanthrene-degrading activity by plant root extracts and exudates. Environmental Microbiology. 2004;6:574–583. doi: 10.1111/j.1462-2920.2004.00589.x. [DOI] [PubMed] [Google Scholar]
- Rezek J, Macek T, Mackova M, Triska J. Plant metabolites of polychlorinated biphenyls in hairy root culture of black nightshade Solanum nigrum SNC-90. Chemosphere. 2007;69:1221–1227. doi: 10.1016/j.chemosphere.2007.05.090. [DOI] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell. 2009;21:1031–1033. doi: 10.1105/tpc.109.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross G. The public health implications of polychlorinated biphenyls (PCBs) in the environment. Ecotoxicol Environ Saf. 2004;59:275–291. doi: 10.1016/j.ecoenv.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Secher C, Lollier M, Jezequel K, Cornu JY, Amalric L, Lebeau T. Decontamination of a polychlorinated biphenyls-contaminated soil by phytoremediation-assisted bioaugmentation. Biodegradation. 2013;24:549–562. doi: 10.1007/s10532-013-9625-6. [DOI] [PubMed] [Google Scholar]
- Singer AC, Gilbert ES, Luepromchai E, Crowley DE. Bioremediation of polychlorinated biphenyl-contaminated soil using carvone and surfactant-grown bacteria. Appl Microbiol Biotechnol. 2000;54:838–843. doi: 10.1007/s002530000472. [DOI] [PubMed] [Google Scholar]
- Singer AC, Smith D, Jury WA, Hathuc K, Crowley DE. Impact of the plant rhizosphere and augmentation on remediation of polychlorinated biphenyl contaminated soil. Environ Toxicol Chem. 2003;22:1998–2004. doi: 10.1897/02-471. [DOI] [PubMed] [Google Scholar]
- Slater H, Gouin T, Leigh MB. Assessing the potential for rhizoremediation of PCB contaminated soils in northern regions using native tree species. Chemosphere. 2011;84:199–206. doi: 10.1016/j.chemosphere.2011.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KE, Schwab AR, Banks MK. Phytoremediation of polychlorinated biphenyl (PCB)-contaminated sediment: a greenhouse feasibility study. J Environ Qual. 2007;36:239–244. doi: 10.2134/jeq2006.0089. [DOI] [PubMed] [Google Scholar]
- Tam NFY, Wong YS. Effectiveness of bacterial inoculum and mangrove plants on remediation of sediment contaminated with polycyclic aromatic hydrocarbons. Marine Pollution Bulletin. 2008;57:716–726. doi: 10.1016/j.marpolbul.2008.02.029. [DOI] [PubMed] [Google Scholar]
- Uhlík O, Jecna K, Macková M, Vlcek C, Hroudova M, Demnerová K, Paces V, Macek T. Biphenyl-metabolizing bacteria in the rhizosphere of Horseradish and bulk soil contaminated by polychlorinated biphenyls as revealed by stable isotope probing. Appl Environ Microbiol. 2009;75:6471–6477. doi: 10.1128/AEM.00466-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken B, Correa PA, Schnoor JL. Phytoremediation of polychlorinated biphenyls: new trends and promises. Environ Sci Technol. 2010;44:2767–2776. doi: 10.1021/es902514d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg M, Birnbaum LS, Denison M, De Vito M, Farland W, Feeley M, Fiedler H, Hakansson H, Hanberg A, Haws L, Rose M, Safe S, Schrenk D, Tohyama C, Tritscher A, Tuomisto J, Tysklind M, Walker N, Peterson RE. The 2005 World Health Organization reevaluation of human and mammalian toxic equivalency factors for dioxins and dioxin-like compounds. Toxicological Sciences. 2006;93:223–241. doi: 10.1093/toxsci/kfl055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Metre PC, Mahler BJ. Trends in hydrophobic organic contaminants in urban and reference lake sediments across the United States, 1970–2001. Environ Sci Technol. 2005;39:5567–5574. doi: 10.1021/es0503175. [DOI] [PubMed] [Google Scholar]
- Van Noort PCM, Haftka JJH, Parsons JR. Updated Abraham solvation parameters for polychlorinated biphenyls. Environ Sci Technol. 2010;44:7037–7042. doi: 10.1021/es102210g. [DOI] [PubMed] [Google Scholar]
- Van Veen JA, Van Overbeek LS, Van Elsas JD. Fate and activity of microorganisms introduced into soil. Microbiology and Molecular Biology Reviews. 1997;61:121. doi: 10.1128/mmbr.61.2.121-135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegel J, Wu Q. Microbial reductive dehalogenation of polychlorinated biphenyls. FEMS Microbiol Ecol. 2000;32:1–15. doi: 10.1111/j.1574-6941.2000.tb00693.x. [DOI] [PubMed] [Google Scholar]
- Zaitsev G, Karasevich Y. Primary steps in metabolism of 4-chlorobenzoate in Arthrobacter globiformis. Mikrobiologiya. 1985;50:423–428. [Google Scholar]
- Zhai GS, Lehmler FJ, Schnoor JL. Identification of hydroxylated metabolites of 3,3′,4,4′-tetrachlorobiphenyl and metabolic pathway in whole poplar plants. Chemosphere. 2010;81:523–528. doi: 10.1016/j.chemosphere.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.