Summary
Nfkb1 and Nfkb2 proteins p105 and p100 serve both as NF-κB precursors and inhibitors of NF-κB dimers. In a biochemical characterization of endogenous cytoplasmic and purified recombinant proteins, we found that p105 and p100 assemble into high-molecular-weight complexes that contribute to the regulation of all NF-κB isoforms. Unlike the classical inhibitors IκBα, -β, and -ε, high-molecular-weight complexes of p105 and p100 proteins bind NF-κB subunits in two modes: through direct dimerization of Rel homology domain-containing NF-κB polypeptides and through interactions of the p105 and p100 ankyrin repeats with preformed NF-κB dimers, thereby mediating the bona fide IκB activities, IκBγ and IκBδ. Our biochemical evidence suggests an assembly pathway in which kinetic mechanisms control NF-κB dimer formation via processing and assembly of large complexes that contain IκB activities.
Introduction
NF-κB (nuclear factor kappaB) denotes a group of structurally related dimeric transcription factors that share the ability to specifically bind DNA and to regulate the expression of many NF-κB-dependent genes. NF-κB-dependent gene expression is important for the differentiation of immune cells, for the development of lymphoid organs, and during immune activation (Hoffmann and Baltimore, 2006). In unstimulated cells, NF-κBs are bound by inhibitor proteins, IκBs (inhibitors of NF-kappaB), and are rendered inactive. Many stimuli, including the ligands of Toll-like receptors (TLR), such as bacterial lipopolysaccharide (LPS), induce phosphorylation and proteasomal degradation of IκBs, leading to the activation of NF-κB-responsive gene expression. The mammalian NF-κB signaling system consists of five NF-κB subunits—RelA, c-Rel, RelB, p50, and p52—and five proteins with inhibitory activity—IκBα, IκBβ, IκBε, p105, and p100. NF-κB subunits interact with each other in a combinatorial manner, assembling in homo- and heterodimers via the conserved dimerization interface in Rel homology domains (RHD). Dimerization is an essential property of NF-κB and is required for DNA binding (Hoffmann et al., 2006).
The Nfkb1 and Nfkb2 proteins, p105 and p100, have dual functions in the NF-κB signaling system. They are best known as precursors of p50 and p52. The rate of processing of the p105 and p100 regulates the availability of NF-κB dimers whose activity is regulated by IκB proteins. Maintaining a proper balance is physiologically important, as documented in engineered mouse strains in which the nfkb1 and nfkb2 genes produce only mature p50 or p52 proteins. The absence of p100 results in perinatal lethality (Ishikawa et al., 1997), and p105 deficiency results in an inflammatory phenotype and increased susceptibility to opportunistic infections (Ishikawa et al., 1998). Interestingly, these phenotypes are more severe than those of mice that lack expression of both p105 and p50 (Sha et al., 1995) or p100 and p52 (Caamano et al., 1998).
In addition to their role as precursors for p50 and p52, p105 and p100 can also inhibit preformed NF-κB dimers. In early studies, ectopic expression of these proteins was shown to inhibit NF-κB activity (Mercurio et al., 1992; Rice et al., 1992). More recently, p100 was shown to regulate preformed RelA:p50 dimer activity during LTβR signaling (Basak et al., 2007) and RSV-induced signaling (Liu et al., 2008), and stimulus-responsive expression of the nfkb2 gene can provide negative feedback onto RelA:p50 dimer during prolonged TLR-mediate IKK signaling (Shih et al., 2009). Further, in knockin mice, in which p105 degradation was impaired, preformed RelA:p50 dimers were inhibited, thereby causing T cell proliferation defects (Sriskantharajah et al., 2009). As signaling-responsive inhibitors of preformed NF-κB dimers, p105 and p100 satisfy the definition of IκB proteins. However, there is little understanding about the molecular architecture that allows p105 and p100 to function as IκBs.
Here, we report that p105 and p100 form high-molecular-weight (MW) complexes that incorporate other NF-κB subunits. These complexes not only provide “buffering” binding capacity for excess NF-κB subunits, but also contain the bona fide IκB activities IκBγ and IκBδ that mediate stimulus-responsive activation and postinduction repression of preformed NF-κB dimers. Based on biochemical evidence, we constructed a stoichiometric model for these complexes, which accounts for their high MW and compositional heterogeneity, and an assembly pathway that suggests that kinetic mechanisms controlled by an evolutionarily conserved helical dimerization domain in these precursor proteins determine the balance between NF-κB dimer generation and assembly into large complexes.
Results
Detection of p105 and p100 Complexes
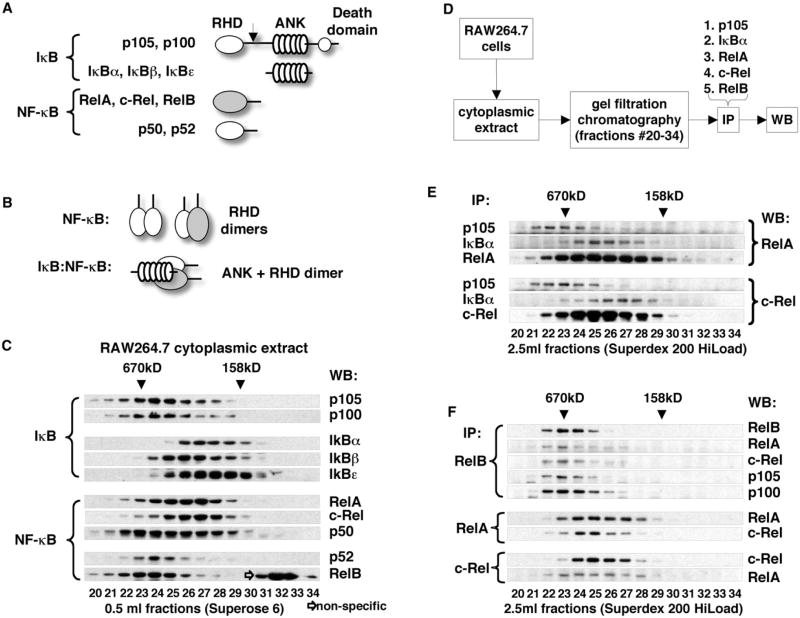
Nfkb1/p105 and Nfkb2/p100 share sequence similarity with both classical IκBs (IκBα, IκBβ, and IκBε) and NF-κBs (RelA, c-Rel, and RelB) because of the presence of the RHD and ANK domains (Figure 1A and Figure S1 available online). Thus, it is reasonable to suggest that p105 and p100 interact with other NF-κB subunits in both an “NF-κB”-like and “IκB”-like manner, utilizing their RHD and ANK domains, respectively (Figure 1B). To investigate these complex protein interactions, we fractionated cytoplasmic extracts of RAW264.7 macrophages by gel filtration chromatography and established the gel filtration profiles for all NF-κB subunits and IκB proteins using a panel of antibodies. We observed that p105 and p100 are detected in high-MW fractions distinct from lower-MW fractions containing IκBα, IκBβ, and IκBε. NF-κB subunits RelA, c-Rel, and p50 were distributed over a wide range of gel filtration fractions, overlapping with p105 and p100 in high-MW fractions and with IκBα, IκBβ, and IκBε in the lower-MW fractions. On the other hand, RelB and p52 preferentially partitioned to high-MW fractions, which also contained p105 and p100 (Figure 1C).
Figure 1. Detection of Endogenous High-MW p105 and p100 Complexes.
(A) Domain organization of IκB and NF-κB proteins. RHD, Rel homology domain; ANK, ankyrin repeat domain. Arrow indicates site of proteolysis in p105 and p100 that yields p50 and p52, respectively.
(B) Domain interactions between NF-κB and IκB proteins. Dimerization of RHD domains (top) and binding of ANK domain to RHD dimers (bottom).
(C) Cytoplasmic extract of RAW264.7 cells was fractionated by gel filtration chromatography (GF) on an analytical Superose 6 column. IκB and NF-κB proteins were detected by western blotting (WB).
(D) Flowchart illustrating immunoprecipitation (IP) experiments shown in (E) and (F).
(E) Cytoplasmic extract of RAW264.7 cells was fractionated by GF. p105, IκBα, RelA, and c-Rel were immunoprecipitated from GF fractions. RelA (top) and c-Rel (bottom) were detected by WB.
(F) RelB, RelA, and c-Rel were immunoprecipitated from GF fractions (as in E). RelB, RelA, c-Rel, p105, and p100 were detected by WB.
To evaluate the physical interaction of NF-κB subunits with IκB proteins and with each other, we fractionated RAW264.7 cytoplasmic extract using a preparative gel filtration column and immunoprecipitated p105, IκBα, RelA, c-Rel, and RelB from the resulting fractions. Immunoprecipitated proteins and their binding partners were detected by western blotting (see flowchart, Figure 1D). Immunoprecipitation of p105 and IκBα from gel filtration fractions showed that p105 specifically interacts with RelA and c-Rel in high-MW gel filtration fractions, whereas IκBα interacts with RelA and c-Rel in lower-MW fractions (Figure 1E). Direct immunoprecipitation of RelA and c-Rel, however, resulted in the detection of RelA and c-Rel in both the high- and low-MW fractions. We concluded from these observations that cytoplasmic RelA and c-Rel are indeed distributed between the larger, p105-containing complexes and the smaller complexes containing classical IκBs.
Immunoprecipitation of RelB (the NF-κB subunit that exclusively partitions to high-MW fractions, Figure 1C) resulted in co-precipitation of RelA and c-Rel. p105 and p100 were also detected in RelB immunoprecipitates (Figure 1F, top). The reciprocal immunoprecipitation of RelA and c-Rel showed that complexes containing these two NF-κB subunits were also present in lower-MW fractions (Figure 1F, bottom). Coimmunoprecipitation of RelA, c-Rel, p105, and p100 with RelB from high-MW fractions was interesting because it suggested that RelA and c-Rel interact with RelB as components of high-MW p105 and/or p100 complexes. This idea is also supported by our observation of specific interactions of p105 with RelA and c-Rel in high-MW fractions (Figure 1E) and by the sequential immunoprecipitation experiments of Mercurio and colleagues (Mercurio et al., 1993), the results of which also suggested that p105, p100, and RelA engage in higher-order complexes different from a simple heterodimer.
From the ability of p105 and p100 to compensate for the cytoplasmic retention of NF-κB in the absence of classical IκB proteins (Basak et al., 2007; Tergaonkar et al., 2005), we predicted that more RelA, c-Rel, and p50 will be associated with the high-MW complexes in IκB-deficient cells compared to the wild-type (WT) cells. To test this prediction, we fractionated the cytoplasmic extracts of ikba−/−ikbb−/−ikbe−/− triple-knockout mouse embryonic fibroblasts (MEF) and observed a shift in RelA, c-Rel, and p50 retention volumes toward higher-MW fractions (Figure S2A). The physical interaction of RelA with the high-MW p105 and/or p100 in ikba−/−ikbb−/−ikbe−/− MEF was confirmed by coimmunoprecipitation (Figure S2B). These results suggest that the large p105 and/or p100 complexes, detected by gel filtration, are not an artifact of fractionation experiments but represent the physiologically relevant high-MW protein assemblies that are distinct from complexes involving a single NF-κB dimer bound to DNA or a canonical IκB protein.
p105 and p100 Complexes Are Stimulus Responsive
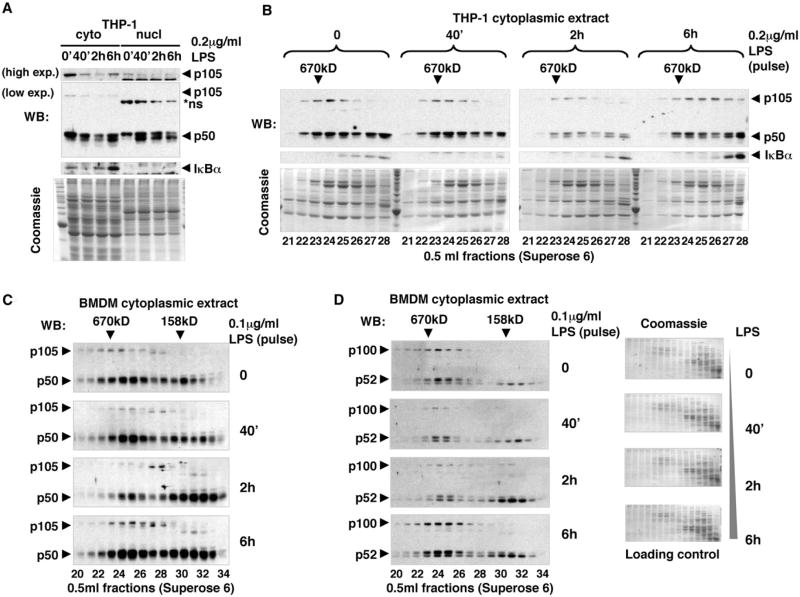
We next tested whether these high-MW complexes participate in NF-κB signaling. We treated THP-1 cells (differentiated by phorbol ester [PMA]) with 0.2 μg/ml LPS for 40 min and analyzed the amount and MW distribution of cytoplasmic p105, p50, and IκBα by gel filtration chromatography and western blotting at different time points after stimulation. We determined that the previously described (Salmeron et al., 2001) inducible degradation of p105 (Figure 2A, top) does indeed target high-MW p105 complexes (Figure 2B). The reduction in p105 levels 2 hr after LPS stimulation correlated with the decrease of p50 in high-MW fractions (Figure 2B), suggesting that the degradation of p105 releases p50 from high-MW complexes and, thus, contributes to LPS-induced NF-κB signaling in the nucleus (Figure 2A). However, 6 hr after LPS treatment, the amounts of p105 and p50 in high-MW fractions were partially restored, and no additional accumulation of p50 in the nucleus was detected, implying that newly synthesized p105 assembles with p50 into inhibited cytoplasmic complexes (Figure 2B). Similarly, we observed accumulation of p105 and p50 NF-κB subunits in high-MW cytoplasmic complexes 6 hr after LPS stimulation of bone marrow-derived macrophages (BMDM) (Figure 2C). The LPS-responsive disruption and formation of p100 high-MW complexes was also observed in BMDM (Figure 2D). At early time points, we detected a loss of high-MW p100, but by 6 hr, both p100 and p52 reaccumulated in high-MW fractions. We concluded, therefore, that de novo synthesized p100 complexes function to incorporate (and inhibit) p52 subunits, attenuating the effect of LPS on gene expression. Our results indicated that, collectively, high-MW complexes of p105 and p100 trap at least 50% of cytoplasmic p50 and p52 in macrophages post-LPS stimulation, thus participating in terminating NF-κB signaling.
Figure 2. High-MW Endogenous p105 and p100 Complexes Are Stimulus Responsive.
(A) THP-1 cells were stimulated with 0.2 μg/ml LPS for 40 min. Cytoplasmic (cyto) and nuclear (nucl) extracts were prepared at indicated time points and analyzed by western blotting (WB) to detect p105, p50, and IκBα. Exp., exposure.
(B) Cytoplasmic extracts of THP-1 cells (as in A) were further fractionated by gel filtration chromatography (GF) and analyzed by WB to detect p105, p50, and IκBα.
(C) BMDM were stimulated with 0.1 μg/ml LPS for 40 min. Cytoplasmic extracts were prepared at indicated time points and analyzed by GF followed by WB to detect p105 and p50 proteins.
(D) GF fractions of BMDM (as shown in C) were analyzed by WB to detect p100 and p52. Coomassie staining (in parallel with WB) provided controls for equivalent loading and for the efficiency of fractionation.
We noted that p50 and p52 appear as two isoforms in western blots. The slower-migrating (by SDS-PAGE) isoforms appeared to be overrepresented in p105 and p100 complexes (Figure 2). However, the relative amount of these isoforms might vary depending on cell type and culturing conditions (Figures 2 and S3), and their functional significance remains unclear.
Stoichiometry of p105:p50 Complexes
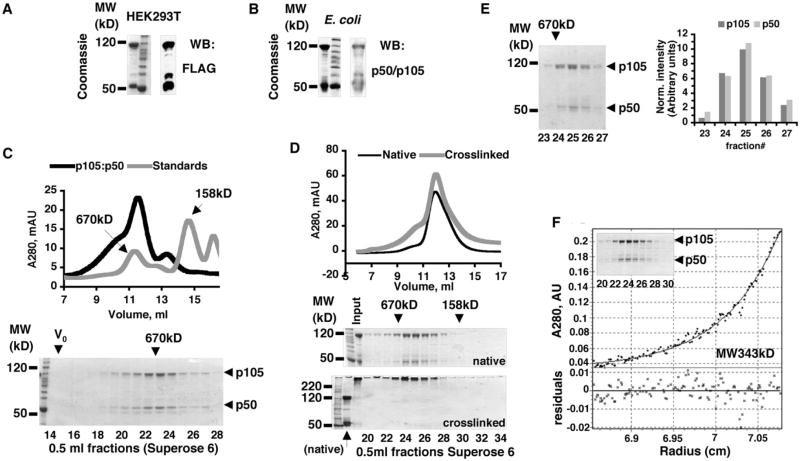
Based on the sequence conservation between p105 and p100 in the regions encompassing protein-protein interaction domains and on the comparable MW of endogenous p105 and p100 complexes, we hypothesized that the overall organization of p105 and p100 high-MW complexes is similar. To this end, we focused on p105:p50 complexes as a model system. We noted that overexpression of p105 in HEK293T cells or in E coli also yielded ∼50 kDa polypeptides that we copurified with p105 by affinity chromatography. These polypeptides were indeed proteolytic products of p105 because they were recognized by an antibody specific for N-terminal purification tag (Figure 3A) and by an antibody specific for p105/p50 (Figure 3B). Gel filtration chromatography of p105, purified from HEK293T cells, showed that p105 and its 50 kD N-terminal product (which we will refer to as p50) coelute, indicating that they interact by forming high-MW complexes (Figure 3C). Chemical crosslinking of the p105:p50 complexes (purified from E coli) followed by analytical gel filtration chromatography provided additional evidence that p105 and p50 interact by forming high-MW complexes (Figure 3D).
Figure 3. Stoichiometry of p105:p50 Complexes.
(A and B) p105 was purified by affinity chromatography from the mammalian (HEK293T, A) and bacterial (E. coli, B) expression systems. p105 and copurified 50 kD N-terminal fragment of p105 were detected by Coomassie staining and western blotting (WB).
(C) p105:p50 (purified from HEK293T) was analyzed by gel filtration chromatography (GF) and compared to MW standards. Absorbance at λ280 nm (A280) was plotted against retention volume (top). The resulting fractions were analyzed by SDS-PAGE and Coomassie staining (bottom).
(D) Chemically crosslinked p105:p50 complexes and native p105:p50 complexes were compared by GF on an analytical Superose 6 column. A280 was plotted against retention volume (top). The resulting fractions and input samples were analyzed by SDS-PAGE and Coomassie staining (bottom).
(E) 100 μg of p105:p50 complex was fractionated by GF followed by SDS-PAGE and Coomassie staining (left). The normalized values of the intensities of p105 and p50 bands were plotted against fraction number (right).
(F) p105:p50 complexes were purified from E. coli by Ni affinity chromatography followed by GF (insert, Coomassie-stained SDS-PAGE gel shows GF fractions) and analyzed by analytical ultracentrifugation (AUC). The representative result of the sedimentation equilibrium (SE) experiment performed at the rotor speed 8000 rpm is shown.
We noted that p105:p50 complexes contained approximately equal amounts of p105 and p50 polypeptides (Figures 3A–3C). To quantitatively test this observation, we analyzed 100 μg of recombinant p105:p50 complexes by analytical gel filtration followed by SDS-PAGE and Coomassie staining. The intensity of Coomassie staining was measured and normalized for the propensity of p105 and p50 polypeptides to bind Coomassie stain. The results showed that the p105:p50 complex indeed contains near equimolar amounts of both polypeptides (Figure 3E).
To determine the MW of p105:p50 complexes, we performed sedimentation equilibrium experiments with two p105:p50 protein preparations. Sedimentation equilibrium data from all three concentrations of each p105:p50 preparation analyzed at rotor speed 8000 rpm allowed for an acceptable fit of the experimental date to the single ideal species model, as indicated by low residuals (± 0.02 A280 units) distributed around the theoretical curve (Figures 3F and S6A).
Fitting the MW of p105 and p50 polypeptides in a 1:1 ratio into the experimentally determined MW (340–400 kD) of the p105:p50 complexes, we obtained a working stoichiometric model for the minimal core p105:p50 assemblies wherein two p105 polypeptides interact with two p50 molecules, (p105)2:(p50)2. The results of our analytical ultracentrifugation experiments demonstrated that the recombinant p105:p50 complex is larger than the simple p105:p50 heterodimer.
p105 and p100 Utilize Two Modes of Binding to NF-κB
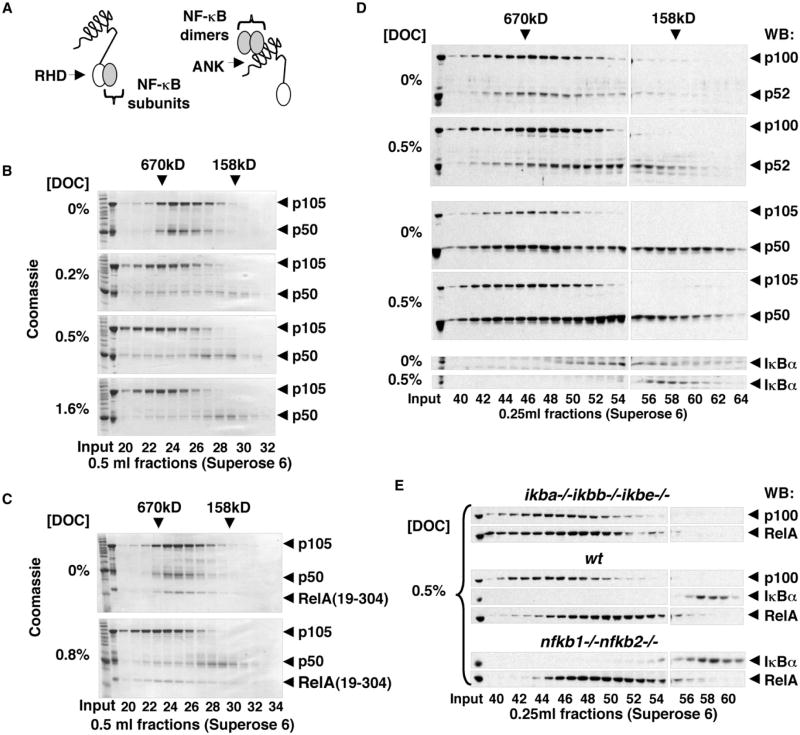
Because p105 and p100 proteins contain RHD and ANK domains, there are at least two types of interdomain interactions that can be simultaneously utilized in the assembly of the high-MW p105 and p100 complexes with other NF-κB subunits. The first type constitutes the individual NF-κB subunits binding to the N-terminal RHD domains of p105 and p100 (Figure 4A, left). This type of interaction was suggested in pioneering studies of the p105 protein (Rice et al., 1992). In addition, the preformed NF-κB dimers can bind to the ANK domains of p105 and p100 (Figure 4A, right), as proposed for the p100 protein (Basak et al., 2007). To obtain clear experimental evidence in support of these two binding modes, we combined in vitro sodium deoxycholate (DOC) treatment (Baeuerle and Baltimore, 1988) with gel filtration chromatography to discriminate between the preformed NF-κB dimers, which dissociate from ANK domains under these conditions, and the individual NF-κB subunits that remain bound to the RHD domains in the presence of DOC. Using this method, we analyzed the recombinant p105:p50 complexes (purified from E. coli) and found that approximately half of the p50 dissociated from p105 even at the lowest (0.2%) concentration of DOC, whereas some p50 remained bound to p105 at the highest (1.6%) concentration of DOC (Figure 4B). Based on these observations, we concluded that both modes of binding are involved in the assembly of the recombinant p105:p50 complexes.
Figure 4. Two Modes of Binding of NF-κB Subunits to p105 and p100.
(A) Individual NF-κB subunits interact with p105 and p100 via dimerization of RHD domains (left); preformed NF-κB dimers interact with p105 or p100 ANK domains (right).
(B) Recombinant p105:p50 complexes were analyzed by gel filtration chromatography (GF) in the presence of 0%-1.6% of sodium deoxycholate (DOC) followed by SDS-PAGE and Coomassie staining.
(C) Recombinant p105:p50:RelA(19–304) complexes were analyzed by GF in standard conditions and in the presence of 0.8% DOC followed by SDS-PAGE and Coomassie staining.
(D) Cytoplasmic extracts of THP-1 cells were analyzed by GF in standard conditions and in the presence of 0.5% DOC followed by western blotting (WB) to detect p100 and p52 (top), p105 and p50 (middle), or IκBα (bottom).
(E) Cytoplasmic extracts of ikba−/−ikb−/−ikbbe−/−, wild-type (WT), and nfkb1−/−nfkb2−/− MEF were analyzed by GF in the presence of 0.5% DOC. p100, IκBα, and RelA were detected by WB.
To characterize the interactions involved in the assembly of p105 with RelA, we tested the DOC sensitivity of the recombinant p105:p50:RelA(19–304) complexes. p105:p50:RelA(19–304) complexes were purified from E. coli after coexpression of p105 and RelA(19–304); the latter protein corresponds to the RHD of RelA. RelA(19–304) was stably incorporated into high-MW complexes with p105:p50 in a standard gel filtration conditions (Figure 4C, top). In 0.8% DOC-containing buffer, however, we observed that RelA(19–304) remained qualitatively associated with p105, whereas p50 partially dissociated from the complex (Figure 4C, bottom). Based on these results, we concluded that RelA could directly bind to RHD of p105.
To test how the endogenous p50 and p52 proteins interact with p105 and p100, we fractionated the cytoplasmic extract of THP-1 cells by gel filtration chromatography in the presence and absence of DOC and analyzed the resulting fractions by western blotting with antibodies specific for p52/p100 (Figure 4D, top), p50/p105 (Figure 4D, middle), and IκBα (Figure 4D, bottom). DOC treatment caused only partial dissociation of p52 from the high-MW complexes, which led us to conclude that p52 utilizes both binding modes to interact with p105 and/or p100 in high-MW complexes (Figure 4D, top). The dissociation of p50 from high-MW complexes was more difficult to establish because p50 is detected in the lower-MW fractions even before DOC treatment (Figure 4D, middle). The retention of p50 in high-MW gel filtration fractions (coincident with p105 and p100) in the presence of 0.5% DOC, however, was clearly observed, providing evidence of the direct interaction of endogenous p50 subunits with the RHD of p105 and/or p100. Complete dissociation of cytoplasmic IκBα complexes provided a positive control for this experiment (Figure 4D, bottom).
To test whether the endogenous RelA also utilizes the dimerization domain within the RHD to become incorporated into high-MW complexes, we fractionated the cytoplasmic extracts of ikba−/−ikbb−/−ikbe−/− MEF in the presence of 0.5% DOC and compared gel filtration profiles to WT MEF and to nfkb1−/− nfkb2−/− MEF deficient for both p105 and p100. More RelA was detected in high-MW fractions of cytoplasmic extracts from cells deficient for IκBα, IκBβ, and IκBε compared to fractionated cytoplasmic extracts of WT or p105/p100-deficient cells (Figure 4E). This result confirmed that endogenous p105 and p100 could interact with RelA by direct dimerization of their RHD domains.
In all, the results of our DOC experiments demonstrated that NF-κB subunits do indeed utilize two modes of binding to p105 and p100: (1) direct binding to p105 or p100 RHDs to RHDs of partner NF-κB polypeptides and (2) binding to p105 and p100 ANK domains to preformed NF-κB dimers.
C-Terminal Dimerization of p105
We suspected that the higher-order assembly of p105 and p100 complexes is facilitated by the C-terminal regions of these proteins, whereas the N-terminal RHDs form dimers. To identify the putative C-terminal homotypic interaction domain(s) in p105 and p100, we analyzed the conservation of p105 and p100 protein sequences (Figure S4) and mutagenized the conserved regions in p105.
First, we analyzed the recombinant p105 protein that lacks C-terminal death domain, p105(1–800). We found that p105(1–800) formed high-MW complexes similar to WT p105 as revealed by gel filtration chromatography (data not shown), suggesting that the death domain is not required for high-MW complex assembly. Of the remaining regions of p105, the protein sequences immediately preceding the ANK domain show the highest degree of conservation. We compared two recombinant C-terminal ANK domain-containing fragments of p105 that either included or lacked the conserved sequence immediately preceding ANK. A concentration-dependent shift in the retention volume of the ANK(491–800) fragment, containing the conserved sequence, indicated that this protein forms higher-order aggregates in a concentration-dependent manner in the conditions of our gel filtration chromatography experiment, whereas the p105 fragment lacking the conserved sequence, ANK(531–811), did not (Figure S5A). Both proteins had similar melting temperatures (Tm = 38.3°C and Tm = 36.3°C) (Figure S5B), comparable to the melting temperature of ANK domains from IκBα (Tm = 42°C) (Croy et al., 2004), and, thus, both proteins were folded at the conditions of our experiment (22°C). Interestingly, analytical ultracentrifugation experiments using similar p105 ANK domain-containing fragments of p105 previously suggested that the same p105 region contributes to homodimerization of p105 (Beinke et al., 2003).
To test whether the ANK(491–800) protein-containing C-terminal dimerization domain can interact with p105, we incubated purified p105:p50 complexes with ANK(491–800) prior to gel filtration chromatography. We observed a reduction of the MW of p105:p50 assembly and the apparent incorporation of ANK(491–800) into p105:p50 complexes (Figures S5C and S5D). As a control for the specificity of this interaction, we used the mixture of p105:p50 complexes with ANK(531–811) protein and observed that ANK(531–811) neither reduced the MW of nor incorporated into p105:p50 complex (Figures S5C and S5D). Thus, we concluded that ANK(491–800) interacted with p105 through the conserved helical domain preceding the ANK domain. The isolated complex of ANK(491–800) with p105:p50 was further purified and was stable (nondissociating) in our standard gel filtration conditions (Figure S5E).
The results of our in vitro experiments with recombinant proteins suggested that the evolutionarily conserved predicted α-helical dimerization domain of p105 (aa 503–530) and p100 (aa 453–480) plays a role in the higher-order assembly of p105 and p100 into high-MW complexes.
A Model for the High-MW p105 Complexes
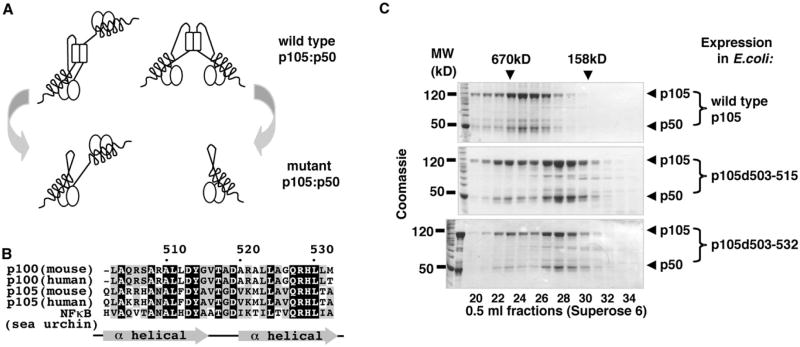
Endogenous p105 and p100 form high-MW complexes that contain other NF-κB subunits and are heterogeneous in composition. Our studies of recombinant p105:p50 complexes suggested that the core of these complexes might consist of two p105 polypeptides. We also determined that three types of protein interactions are involved in the formation of the high-MW p105 complexes: dimerization of RHD domains, binding of preformed RHD dimers to ANK domains of p105, and dimerization of the evolutionarily conserved α-helical domain in the C-terminal half of p105. Based on these observations, we predicted that p105 complexes are a pool of complexes of two core architectures. The first is the RHD dimer of p105, in which one ANK domain folds back onto the dimeric RHD “platform” of p105, and the other is available to bind a p50:p50 dimer in trans. The complexes of the second type are built around p105 dimer mediated by the C-terminal helical dimerization domain. In the latter type of complexes, the p105 RHDs dimerize with RHDs of p50 subunits (Figure 5A, top). To test our model, we utilized a deletion mutagenesis approach aimed to remove the helical dimerization interface (Figure 5B) in p105 protein. We predicted that such a mutation in p105 would disrupt the second type of complexes, whereas the first type would not be affected (Figure 5A, bottom).
Figure 5. Molecular Architecture of p105:p50 Complexes.
(A) The predicted architectures of the minimal (core) wild-type p105:p50 complexes (top) and complexes formed by the C-terminal helical dimerization domain mutants of p105 (bottom).
(B) A multiple sequence alignment of the predicted α-helical dimerization domain of human and mouse p105, p100, and their ortholog (NFkB) from sea urchin.
(C) Wild-type and mutant p105 proteins, p105d503–515 and p105d503–532, were expressed in E.coli and purified by the Ni affinity chromatography (via N-terminal His tag) followed by gel filtration chromatography (GF). GF fractions were analyzed by SDS-PAGE and Coomassie staining.
We expressed and purified two p105 mutant protein complexes that lack one or two predicted helixes in this region of p105, p105d503–515, and p105d503–532. These mutations were designed based on the sequence alignments of p105 and p100 proteins and their evolutionarily distant ortholog from sea urchin (Figure 5B). The chromatographic properties of mutant p105 complexes were consistent with our model. Specifically, we observed two populations, the higher MW and the lower MW, of the mutant p105 complexes (Figure 5C). The MW of the major (low-MW) population of the p105 mutant complexes was determined by analytical ultracentrifugation under equilibrium condition and was consistent with the MW of p105:p50 heterodimer predicted by our model (Figure S6 and data not shown).
Biogenesis of High-MW p105:p50 Complexes
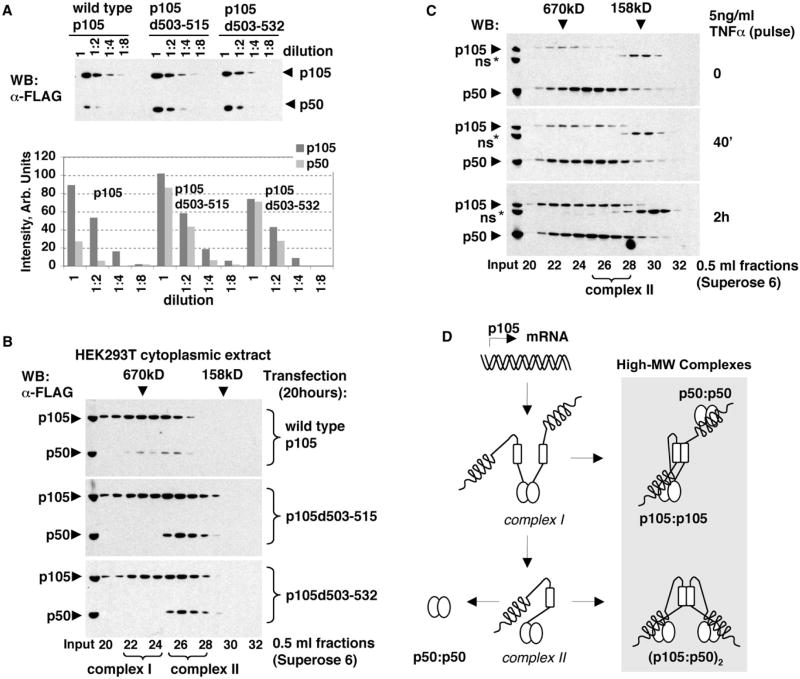
To examine the function of higher-order assembly of p105 complexes, we ectopically expressed the wild-type p105 and the α-helical dimerization domain-deficient p105 mutants in HEK293T cells for 20 hr and analyzed the state of p105 complexes under the condition of continuous protein expression. First, we noted that more p50 was generated in cells expressing mutant p105 proteins compared to the cells expressing wild-type p105 (Figure 6A), suggesting that the higher-order assembly of p105 via the helical dimerization domain inhibits p105 processing. We suspected that the deficiency in the helical dimerization domain led to the accumulation of complex assembly intermediates susceptible to constitutive processing. Indeed, gel filtration chromatography of cytoplasmic extracts of HEK293T cells, expressing p105 mutants, showed that a greater portion of the unprocessed p105 is present in lower-MW fractions (complex II) than when the wild-type p105 was expressed (Figure 6B). Interestingly, when we treated HEK293T cells with TNFα to induce the synthesis of the endogenous p105, the formation of the low-MW p105 complexes (complex II) was also observed (Figure 6C).
Figure 6. p105 Processing and the Assembly of p105 High-MW Complexes.
(A) FLAG-tagged wild-type (WT) and mutant p105 proteins, p105d503–515 and p105d503–532, were transiently expressed in HEK293T cells. Cytoplasmic extracts were prepared 20 hr after transfection, and their serial dilutions were analyzed by western blotting (WB, top). The intensities of WT and mutant p105 and p50 bands were quantitated and plotted against dilution factor (bottom).
(B) Cytoplasmic extracts (as in A)were analyzed by gel filtration chromatography (GF) followed by WB. Complex I and complex II denote p105 complexes distinct in their chromatographic properties.
(C) HEK293T cells were stimulated with 5 ng/ml TNFα for 40 min. Cytoplasmic extracts were prepared at indicated time points and analyzed by GF followed by WB to detect endogenous p105 and p50 proteins. Complex II denotes the distinct lower-MW p105 complexes detected after stimulation.
(D) A model for the biogenesis of p50 homodimers and the high-MW p105:p50 complexes.
These results led us to speculate that, during the assembly of the high-MW p105:p50 complexes, the lower-MW simple p105:p50 heterodimers are transiently present as an assembly intermediates. Such p105:p50 heterodimers are likely derived from p105 homodimers (complex I) and may be further processed to p50:p50 or may dimerize with another p105:p50 via the C-terminal helical dimerization domain. The higher-order assembly of the p105:p50 heterodimers results in the “mature” high-MW p105:p50 complexes in which p50 is bound to p105 via the RHD dimerization domain. We suspected that p105 homodimer (complex I) is indeed the first intermediate (in the molecular pathway that may give rise to complexes between p50 homodimers and p105) because it was detected at 20 hr of overexpression of p105d503–531 in HEK293T cells (Figure 6B) but largely disappeared after 48 hr, when the accumulation of complex II was more evident in this expression system (Figure S7). Based on the results of the DOC sensitivity experiments, we suggest that, during the assembly of p105 complexes with p50, complex I may incorporate preformed p50:p50 homodimers via the p105 ANK domain interactions (Liou et al., 1992) (Figure 6D). Attempts to perturb the endogenous high-MW assemblies by expressing the helical dimerization domain were not successful, perhaps due to lack of sufficient cellular expression of the short peptide (data not shown).
Compositional Heterogeneity of the High-MW p105 Complexes
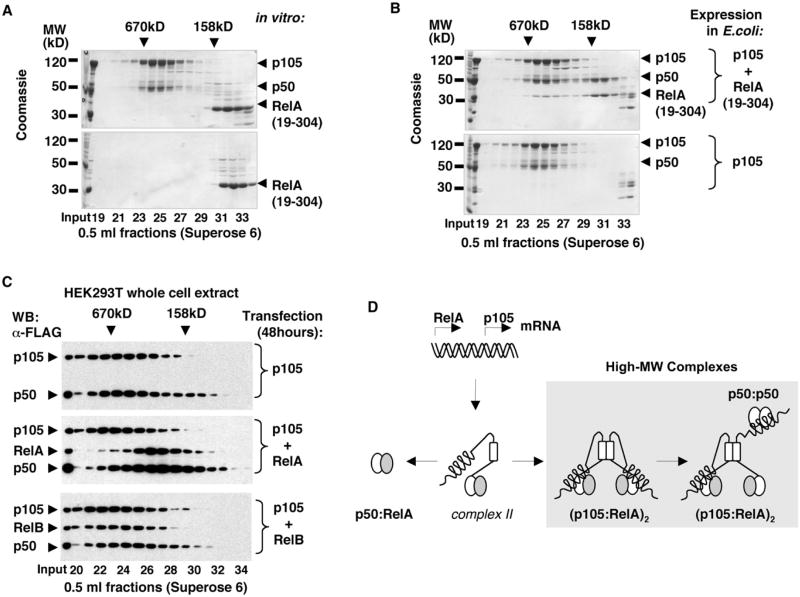
Our immunoprecipitation experiments showed that endogenous p105 interacts with RelA and c-Rel in high-MW complexes (Figure 1E). To test how RelA assembles with p105, we incubated purified p105:p50 complexes with the recombinant RelA RHD homodimers, RelA(19–304), in vitro. The results of gel filtration chromatography of preincubated p105:p50 and RelA:RelA complexes showed that RelA subunits do not incorporate into p105:p50 complexes in the conditions of our experiment (Figure 7A). Coexpression of RelA with p105, however, resulted in the incorporation of RelA(19–304) into p105:p50 complexes. In addition, we detected a separate pool of p50:RelA(19–304) complexes (Figure 7B). The coexpression of RelA, but not RelB, with p105 in HEK293T cells also resulted in the formation of p50:RelA complexes, as detected by gel filtration chromatography of whole-cell lysates (Figure 7C). These results led us to propose that biogenesis of p50:RelA heterodimer requires cosynthesis of both proteins. (Figure 7D). Because we have previously determined that the assembly of high-MW p105 complexes via helical dimerization domain inhibits p105 processing, we propose that the rate of p105 higher-order assembly might regulate the availability of the p50:RelA heterodimers. According to our model, posttranslational dimerization of p105 and RelA RHD domains, followed by the assembly of p105 high-MW complexes, may result in the immediate inhibition of the de novo synthesized RelA, as opposed to the inhibition of RelA:p50 heterodimers by classical IκBs.
Figure 7. Biogenesis of p50:RelA Heterodimers and RelA Complexes with p105.
(A) Recombinant p105:p50 complexes were incubated with RelA RHD homodimers, RelA(19–304), and the resulting complexes were analyzed by gel filtration chromatography (GF) followed by SDS-PAGE and Coomassie staining (top) and compared to the similarly treated isolated RelA(19–304) (bottom).
(B) His-tagged p105 was coexpressed with untagged RelA(19–304) in E. coli, and the resulting complexes were isolated by Ni affinity chromatography followed by GF (top) and compared to the recombinant p105:p50 complexes (bottom).
(C) FLAG-tagged p105 was expressed in HEK293T cells alone or coexpressed with FLAG-tagged RelA or RelB. At 48 hr after transfection, the whole-cell extracts were analyzed by GF followed by western blotting (WB).
(D) A model for the biogenesis of p50:RelA heterodimers and the high-MW p105:p50:RelA complexes.
Discussion
In this study, we applied biochemical methods to provide insights into the mechanism of how p105 and p100 inhibit NF-κB. Our studies reveal a relationship between their function as precursors of p100 and p105, which control NF-κB dimer formation and their functions as bona fide IκB inhibitors that inhibit latent NF-κB dimers and release them in response to specific cellular stimulation. We show that both p105 and p100 assemble into large complexes governed by three distinct interaction domains and that the resulting large complexes are heterogeneous in composition.
We discovered an intricate and functionally significant “life cycle” of p105 and p100 molecules, from synthesis to degradation: newly synthesized p105 and p100 proceed through assembly intermediates before maturing into stable high-MW complexes observed in the steady state. Several competing interactions and events are embedded within the assembly or maturation pathway; these determine the functional role of individual domains of p105 and p100 polypeptides.
When p105 or p100 is synthesized, the first protein-protein interaction that affects their ultimate function is the RHD-mediated dimerization with another NF-κB polypeptide. NF-κB is an obligate dimer, and RHD dimerization is kinetically fast and only slowly reversible. The abundance of binding partners is determined by their rate of synthesis. Thus, the formation of heterodimers involving one of the activator proteins RelA, c-Rel, or RelB versus the formation of p105:p105 or p100:p100 homodimers is determined by their relative synthesis rates and binding affinities. Which dimer is formed will determine the most likely pathway that follows.
If p105 or p100 associate with RelA, c-Rel, or RelB, they may be processed to give rise to NF-κB heterodimers containing p50 or p52, as has been previously shown for p50:c-Rel and p50:RelA (Rice et al., 1992). The processing may be constitutive or stimulus inducible, but the partner NF-κB subunit is likely to influence its efficiency (Figure 7C). When ineffective, p105 or p100 escape processing, and the heterodimers with RelA, c-Rel, or RelB constitute a building block for the formation of high-MW complexes via the C-terminal helical dimerization domain.
If p100 or p105 proteins self-associate upon synthesis, they may either be completely processed to give rise to p50 or p52 homodimers, or they may form stable high-MW assemblies via two intermediates, complex I and complex II. We suspect that complex I is a p105 or p100 homodimer, in which one ANK domain may interact with the RHD dimeric interface (in cis). However, the second ANK domain of the resulting self-inhibited dimer remains unoccupied, allowing it to bind other preformed RHD dimers in trans. But even if this second ANK domain is processed to form complex II, higher-order assembly may continue when the first ANK domain detaches and binds other NF-κB dimers or the helical dimerization domain drives complex formation.
It is plausible that p105 and p100 can form even higher-order complexes via combinatorial interactions between the three domains: the dimerization domains in the RHD shared by all NF-κB proteins and the helical and ANK domains present in p100 and p105. Gel filtration chromatography of endogenous cytoplasmic p105 and p100 complexes (Figure 1) shows the presence of higher-MW complexes and suggests that such higher-order, conformationally heterogeneous complexes do indeed form. In the case of the purified recombinant p105 complexes, the accumulation of the stable “very high” MW species was also detected (Figure 3). We suspect that these very high-MW endogenous species do not readily function as substrates to form NF-κB dimers via processing, but they may have buffering functions to terminally remove excess NF-κB polypeptides from the pool susceptible to regulation.
Our model suggests that the kinetics of ANK-RHD interactions is an important parameter controlling the ability of high-MW complex formation in which ANK domains bind preformed NF-κB dimers in trans. As trans inhibition is a defining criterion for bona fide IκB function, this kinetic competition determines the abundance of IκB activities residing in the resulting high-MW complexes; these have been termed IκBδ for p100 and IκBγ for p105. In the case of p100, IκBδ activity was shown to be stimulus responsive, as LTβR stimulation leads to degradation of the ANK domain, releasing bound RelA:p50 and RelB:p50 dimers to the nucleus and leaving p52:p100 cis-inhibited dimers (Basak et al., 2007).
The kinetics of both the ANK-RHD interactions and the helical dimerization domain interactions control the rate of processing of p105and p100. Deletion of the dimerization domain disrupts multi-protein assembly, resulting in excessive processing (Figure 6). In contrast, coexpression of RHD-binding partners that result in high-affinity ANK-RHD interactions may inhibit processing, and “matured” endogenous p105 isolated by immunoprecipitation from unstimulated cells was found to be resistant to further processing in vitro (O.V.S., unpublished data). The consequence of this step is highly significant. Because all cellular p50 observed within NF-κB dimers (such as p50:RelA) derives from the p105-processing event, duration of the intermediate p105 assembly steps and the regulation of p105 oligomerization are critical determinants of the abundance of cellular NF-κB dimers. The rate of p105 synthesis, along with the rates of RelA, c-Rel, and IκB synthesis, all will have an influence on the duration of maturation of p105 high-MW complexes. In essence, the assembly steps could differ between basal and stimulated states in which both synthesis of p105 and proteolysis (controlled by IKK2 activity) are elevated.
In conclusion, our study provides a framework for understanding the role of p105 and p100 as inhibitors of NF-κB and the mechanisms of inhibition. We showed that NF-κB p105 and p100 form large heterogeneous assemblies, which include various NF-κB subunits and the stimulus-responsive IκBγ and IκBδ activities. In elucidating the assembly pathway, we suggest that the kinetics of interactions both in trans and in cis, along with the rates of de novo synthesis of its components and the relevant proteolytic activities, determine the rate of new NF-κB dimer formation. Thus, the high-MW assembly pathway plays a key role in proper control of the NF-κB-signaling system, which is critical to physiological regulation and human health.
Experimental Procedures
Cytoplasmic and Nuclear Fractionation
Cytoplasmic/nuclear fractionation was performed as described (Werner et al., 2005) with some modifications. In brief, cells were lysed in at least two cell pellet volumes of CE buffer: 10 mM HEPES-KOH (pH 7.9), 60 mM KCl, 1 mM EDTA, 0.5% NP-40, and 1 mM DTT supplemented with Protease Inhibitor Cocktail. After collecting cytoplasmic fractions (by centrifugation at 500 × g), nuclei were washed with five pellet volumes of CE buffer and lysed in three pellet volumes of NE buffer: 250 mM Tris-HCl (pH 7.5), 60 mM KCl, 1 mM EDTA, and 1 mM DTT supplemented with Protease Inhibitor Cocktail.
Gel Filtration Chromatography
Cytoplasmic extracts for analytical gel filtration were prepared from 350 cm2 of 90% confluent adherent cell cultures (equivalent to two 150 mm plates). From each sample, 200 μl, filtered and normalized for protein concentration, was injected onto Superose 6 10/300GL column and resolved in 25 mM Tris-HCl (pH 7.5), 140 mM NaCl, and1 mM DTT at 0.5 ml/min flow rate. DOC was added to the gel filtration buffer in the indicated experiments. Either 250 or 500 μl fractions were collected, and 15–30 μl from each fraction shown was analyzed by western blotting and Coomassie staining. For preparative gel filtration, we scaled up sample preparation and injected 3 ml of cytoplasmic extract (24 mg of total protein) into a HiLoad 16/60 column packed with Superdex 200 gel filtration resin. The column was developed with 25 mM Tris-HCl (pH 7.5), 140 mM NaCl, 1 mM DTT, 1 mM EDTA, and 1 mM of phenylmethylsulphonyl fluoride (PMSF) at a flow rate of 1 ml/min. Fractions (2.5 ml) were collected, and 210 μl from each fraction shown was analyzed by immunoprecipitation.
For recombinant protein analysis, 250–500 μg of protein was injected in 200 μl volume into 10/300GL columns prepacked with Superose 6 or Superdex 75 gel filtration resins, except where noted. Columns were developed with 25 mM Tris-HCl (pH 7.5), 140 mM NaCl, and 1 mM DTT at 0.5 ml/min. DOC was added to the gel filtration buffer in the indicated experiments. Absorbance at λ280 nm was monitored. Gel filtration profiles were recorded and analyzed with Unicorn software (Pharmacia).
DOC-Gel Filtration Chromatography Assay
The DOC treatment (Baeuerle and Baltimore, 1988) was used in combination with gel filtration chromatography. To determine the conditions for our assay, we first tested DOC sensitivity of the endogenous IkBa complexes. Second, we tested the DOC sensitivity of two in vitro reconstituted protein complexes. The first complex, p50:ANK(491–800), recapitulated the interactions of p50 homodimers with the C-terminal ANK domain of p105. The second complex, p52:p100CTD, represented the interactions of p52 homodimers with C-terminal domain (CTD) of p100 (aa 406–899) that included the ANK and death domains. Lastly, we determined that p50:ANK(368–800) complex obtained by coexpression of two proteins in HEK293T cells is also sensitive to DOC treatment (Figure S8). The biochemical conclusions about the modes of protein interactions were drawn from a comparison of the gel filtration profiles of the identical samples in the absence and presence of DOC, except for cytoplasmic extracts of MEF of different genetic backgrounds, which we compared with each other in the presence of 0.5% DOC. Recombinant proteins were detected by Coomassie staining of PAGE gels, and endogenous proteins were detected by western blotting.
Chemical Crosslinking
Purified p105:p50 complexes diluted to 0.25 mg/ml were treated with 0.1% glutaraldehyde at room temperature for 15 min in the standard gel filtration buffer and stopped by the addition of sodium borohydride. The extent of crosslinking was determined by SDS-PAGE (6%) followed by Coomassie staining. Crosslinked p105:p50 was concentrated to 1.6 mg/ml, and homogeneity of sample was assessed by gel filtration chromatography.
In-Gel Protein Quantitation
Analytical amounts of protein were resolved by SDS- PAGE and stained with Coomassie G250. Resulting gels were scanned at 300 dpi resolution and converted into TIFF files. The intensity of protein bands was quantified using NIH ImageJ software and normalized for the propensity of corresponding polypeptides to bind Coomassie stain, calculated based on the primary amino acid sequence according to Compton and Jones (1985).
Differential Scanning Calorimetry
Differential scanning calorimetry experiments were carried out using an NDSC II calorimeter (CSC) at a scanning rate of 1 K/min under 3.0 atm of pressure. Protein samples were purified by gel filtration chromatography in PBS (10 mM sodium phosphate [pH 7.4] and 140 mM NaCl) and analyzed at 0.5–1.0 mg/ml.
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed using ProteomeLab XL-I ultracentrifuge equipped with scanning UV/Visible optics and An-50 Ti eight-hole rotor. Absorbance at λ280 nm was monitored. Protein samples in standard gel filtration buffer were analyzed at two to three different concentrations in the range 0.1–1.1 mg/ml at three rotor speeds: 5,000, 8,000, and 12,000 rpm. The data were modeled as a single ideal species using the Hetero-analysis software (by J.L. Cole and J.W. Lary, University of Connecticut).Molecular mass was calculated using values of vbar = 0.7300 and density = 1.00300.
Supplementary Material
Acknowledgments
We thank Soumen Basak, Erika Mathes, and Alexei Savinov for valuable discussions and reagents; Bruce Worcester for his critical reading of this manuscript; Anu Moorthy and Vivien Wang for molecular cloning; Andrey Bobkov for conducting AUC and DSC experiments at the BIMR protein biophysical facility; and Nancy Rice and Mary Ernst for antibody reagents. This research was supported by NIH grants to G.G. (A1064326) and A.H. (GM071862) and by an American Heart Association predoctoral fellowship to O.V.S. (0715041Y).
Footnotes
Supplemental Data: Supplemental Data include Supplemental Experimental Procedures and eight figures and can be found with this article online at http://www.cell.com/molecular-cell/supplemental/S1097-2765(09)00316-.
References
- Baeuerle PA, Baltimore D. Activation of DNA-binding activity in an apparently cytoplasmic precursor of the NF-kappa B transcription factor. Cell. 1988;53:211–217. doi: 10.1016/0092-8674(88)90382-0. [DOI] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beinke S, Deka J, Lang V, Belich MP, Walker PA, Howell S, Smerdon SJ, Gamblin SJ, Ley SC. NF-kappaB1 p105 negatively regulates TPL-2 MEK kinase activity. Mol Cell Biol. 2003;23:4739–4752. doi: 10.1128/MCB.23.14.4739-4752.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamano JH, Rizzo CA, Durham SK, Barton DS, Raventos-Suarez C, Snapper CM, Bravo R. Nuclear factor (NF)-kappa B2 (p100/p52) is required for normal splenic microarchitecture and B cell-mediated immune responses. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton SJ, Jones CG. Mechanism of dye response and interference in the Bradford protein assay. Anal Biochem. 1985;151:369–374. doi: 10.1016/0003-2697(85)90190-3. [DOI] [PubMed] [Google Scholar]
- Croy CH, Bergqvist S, Huxford T, Ghosh G, Komives EA. Biophysical characterization of the free IkappaBalpha ankyrin repeat domain in solution. Protein Sci. 2004;13:1767–1777. doi: 10.1110/ps.04731004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Baltimore D. Circuitry of nuclear factor kappaB signaling. Immunol Rev. 2006;210:171–186. doi: 10.1111/j.0105-2896.2006.00375.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R. Gastric hyperplasia and increased proliferative responses of lymphocytes in mice lacking the COOH-terminal ankyrin domain of NF-kappaB2. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Claudio E, Dambach D, Raventos-Suarez C, Ryan C, Bravo R. Chronic inflammation and susceptibility to bacterial infections in mice lacking the polypeptide (p)105 precursor (NF-kappaB1) but expressing p50. J Exp Med. 1998;187:985–996. doi: 10.1084/jem.187.7.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC, Nolan GP, Ghosh S, Fujita T, Baltimore D. The NF-kappa B p50 precursor, p105, contains an internal I kappa B-like inhibitor that preferentially inhibits p50. EMBO J. 1992;11:3003–3009. doi: 10.1002/j.1460-2075.1992.tb05370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Li K, Garofalo RP, Brasier AR. Respiratory syncytial virus induces RelA release from cytoplasmic 100-kDa NF-kappa B2 complexes via a novel retinoic acid-inducible gene-I-NF-kappa B-inducing kinase signaling pathway. J Biol Chem. 2008;283:23169–23178. doi: 10.1074/jbc.M802729200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercurio F, Didonato J, Rosette C, Karin M. Molecular cloning and characterization of a novel Rel/NF-kappa B family member displaying structural and functional homology to NF-kappa B p50/p105. DNA Cell Biol. 1992;11:523–537. doi: 10.1089/dna.1992.11.523. [DOI] [PubMed] [Google Scholar]
- Mercurio F, DiDonato JA, Rosette C, Karin M. p105 and p98 precursor proteins play an active role in NF-kappa B-mediated signal transduction. Genes Dev. 1993;7:705–718. doi: 10.1101/gad.7.4.705. [DOI] [PubMed] [Google Scholar]
- Rice NR, MacKichan ML, Israel A. The precursor of NF-kappa B p50 has I kappa B-like functions. Cell. 1992;71:243–253. doi: 10.1016/0092-8674(92)90353-e. [DOI] [PubMed] [Google Scholar]
- Salmeron A, Janzen J, Soneji Y, Bump N, Kamens J, Allen H, Ley SC. Direct phosphorylation of NF-kappaB1 p105 by the IkappaB kinase complex on serine 927 is essential for signal-induced p105 proteolysis. J Biol Chem. 2001;276:22215–22222. doi: 10.1074/jbc.M101754200. [DOI] [PubMed] [Google Scholar]
- Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- Shih VFS, Kearns JD, Basak S, Savinova OV, Ghosh G, Hoffmann A. Kinetic control of negative feedback regulators of NF-kB determines their pathogen- and cytokine-receptor signaling specificity. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.812367106. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskantharajah S, Belich MP, Papoutsopoulou S, Janzen J, Tybulewicz V, Seddon B, Ley SC. Proteolysis of NF-kappaB1 p105 is essential for T cell antigen receptor-induced proliferation. Nat Immunol. 2009;10:38–47. doi: 10.1038/ni.1685. [DOI] [PubMed] [Google Scholar]
- Tergaonkar V, Correa RG, Ikawa M, Verma IM. Distinct roles of IkappaB proteins in regulating constitutive NF-kappaB activity. Nat Cell Biol. 2005;7:921–923. doi: 10.1038/ncb1296. [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.