SUMMARY
Nuclear factor κB (NF-κB) regulates gene expression by binding to specific DNA elements, known collectively as κB sites, that are contained within the promoters/enhancers of target genes. We found that the identity of the central base pair (bp) of κB sites profoundly affects the transcriptional activity of NF-κB dimers. RelA dimers prefer an A/T bp at this position for optimal transcriptional activation (A/T-centric) and discriminate against G/C-centric κB sites. The p52 homodimer, in contrast, activates transcription from G/C-centric κB sites in complex with Bcl3 but represses transcription from the A/T-centric sites. The p52:Bcl3 complex binds to these two classes of κB sites in distinct modes, permitting the recruitment of coactivator, corepressor, or both coactivator and corepressor complexes in promoters that contain G/C-, A/T-, or both G/C- and A/T-centric sites. Therefore, through sensing of bp differences within κB sites, NF-κB dimers modulate biological programs by activating, repressing, and altering the expression of effector genes.
INTRODUCTION
The nuclear factor κB (NF-κB) family of transcription factors regulates the transcription of a vast collection of inducible effector genes whose coordinated function results in signal-dependent cellular responses to diverse stimuli (Hoffmann et al., 2006). Despite >25 years of intensive study, it is still not clear how, or even whether, the precise DNA sequences targeted by various NF-κB dimers direct gene expression levels in response to signal transduction in vivo.
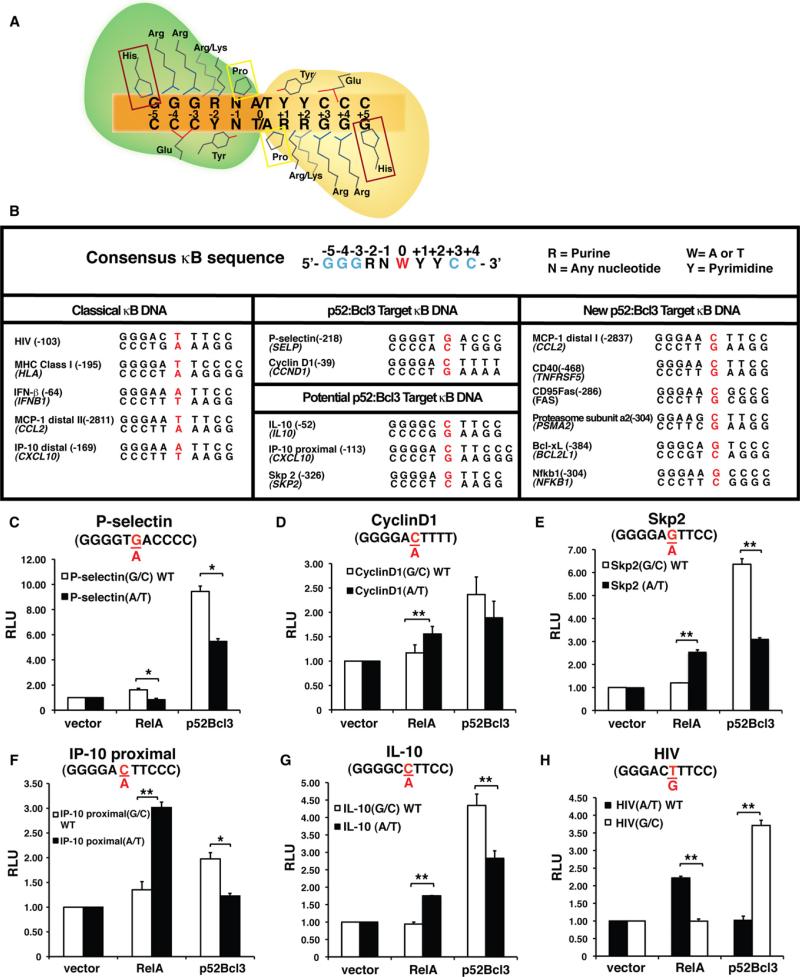
Five NF-κB family member subunits (p50, p52, RelA/p65, cRel, and RelB) assemble combinatorially into functioning homo- and heterodimers that bind a set of specific DNA elements, known as κB sites, in the enhancers/promoters of target genes. DNA binding activity is confined to a highly conserved ~300-residue region, referred to as the rel homology region (RHR), that is present near the N termini of NF-κB subunits. Initial discovery and characterization of several physiological κB sites established the pseudo-symmetric consensus κB site as 5’-GGGRNW YYCC-3’ (R = purines, N = any nucleotide, W = adenine or thymine, and Y = pyrimidine). X-ray crystal structures of several distinct NF-κB homo- and heterodimers in complex with different κB sites revealed that diverse NF-κB dimers recognize κB sites through a relatively conserved mode in which the RHR of each monomer mediates base-specific contacts through the DNA major groove to one half site wherein the flanking (G)GG/ (C)CC sequences are contacted by a set of invariant residues. The inner, more variable base pairs (bps) participate in less conserved interactions with the NF-κB subunits (Figure 1A).
Figure 1. NF-κB Dimers Recognize κB Sites Using a Conserved Mode, and the p52:Bcl3 Complex Activates Reporters with G/C-Centric κB Sites.
(A) Cartoon representation of conserved base-specific contacts between an NF-κB dimer and a κB DNA. The RHR regions of the two NF-κB monomers are marked in green and yellow. Invariant Pro residue (boxed in yellow) makes van der Waals contacts. Invariant Tyr residue makes both van der Waals and hydrogen-bonding contacts. His residues in the box are present only in p50 and p52, replaced by an Ala in RelA, cRel, and RelB. Arg/Lys denotes the presence of Lys in p50 and p52, and Arg in RelA, cRel, and RelB at equivalent positions.
(B) Sequences of κB DNAs. Top: Consensus of known κB sites (the two half-sites are marked by ± 1 to ± 4/5, and the central position is marked by 0; left: well-known A/T-centric κB sites; middle: G/C-centric κB sites; right: newly identified G/C-centric κB sites.
(C–H) Luciferase reporter activity driven by a promoter containing a single WT G/C- or mutant A/T-centric P-selectin κB site (C), cyclin D1 κB site (D), Skp2 κB site (E), IP-10 proximal κB site (F), IL-10 κB site (G), and WT A/T- or mutant G/C-centric HIV κB site (H) cotransfected with vectors expressing p52 and Bcl3 or RelA. RLU, relative luciferase unit. *p < 0.05, **p < 0.01. Error bars represent SD.
Expanded genomic analyses have led to the identification of a large number of κB sites present within promoter/enhancer (cis-regulatory) regions of several hundred genes. In vitro binding experiments have been carried out to classify these κB sites according to their binding specificity for different NF-κB dimers. These analyses led to the observation that the binding affinity displayed by the various NF-κB dimers for distinct κB sites does not necessarily correlate with what occurs during regulation of gene expression by NF-κB in vivo. For example, the p50/RelA heterodimer binds tightly to most κB sites, whereas RelA and cRel homodimers bind to many of the same sites with relatively low affinity. However, ablation of nfκb1 p105/p50 or nfκb2 p100/ p52 genes in mice does not greatly affect the expression of NF-κB target genes, suggesting that RelA and cRel homo- and heterodimers alone are sufficient for the transcriptional activation. Other detailed genetic experiments have shown that some genes are activated only in the presence of one or a subset of NF-κB subunits (Hoffmann et al., 2003; Natoli et al., 2005; Ogawa et al., 2005). Structural and biochemical analyses of DNA binding by NF-κB dimers have also revealed the existence of a large number of κB sites that display relatively similar affinities compared with consensus κB sites even though they lack one consensus half site entirely (Ghosh et al., 2012; Siggers et al., 2012).
It was demonstrated that a single bp difference between the κB sites within IFN-gamma-inducible protein 10(IP-10) and monocyte chemoattractant protein 1 (MCP-1) promoters alters the genes’ responsiveness to different NF-κB dimers (Leung et al., 2004). Whereas both p50/RelA heterodimer and RelA homo-dimer can activate MCP-1 gene expression, only the heterodimer can activate IP-10 gene expression under specific conditions. Interestingly, both RelA homodimer and p50/RelA heterodimer bind both of these κB sites with similar affinities in vitro. It was shown that coactivator recruitment differs between the two promoters (Leung et al., 2004). A later report supported these observations by showing promoter-specific activity of RelA (Ogawa et al., 2005).
In addition to dimer-specific gene activation, NF-κB also represses transcription. For example, RelA represses expression of the nrp1 gene, which is essential for osteoclast differentiation (Hayashi et al., 2012). Interestingly, the sequence of the RelA-repressive κB site displays significant deviation from the consensus in one of its half sites. The p50 homodimer was shown to repress interferon-stimulated response element (ISRE) sites (Cheng et al., 2011). These sites also display only half-site similarity to the κB consensus. Taken together, these reports strongly suggest that κB sites may dictate NF-κB transcriptional outcomes. However, no systematic analysis has been carried out to define the link among the κB site sequence, NF-κB dimer binding specificity, and regulation of target gene expression.
Using in vitro binding, cell-based gene expression, and bioinformatic experiments, we show the following: (1) κB sites can be classified into two distinct classes based on the bp identity of the central position. (2) The transcriptional responses of RelA dimers and p52 homodimer complexed to Bcl3 (henceforth referred to as the p52:Bcl3 complex) are different for two classes of κB sites. RelA dimers bind G/C-centric sites poorly in vitro and fail to activate transcription from these sites in vivo; instead, RelA dimers activate transcription by binding to A/T-centric sites. However, the p52:Bcl3 complex activates transcription by binding to G/C-centric sites. Strikingly, the same complex represses transcription by binding to A/T-centric sites. (3) The p52:Bcl3 complex accomplishes this dual feat by recruiting coactivator and corepressor complexes to G/C- and A/T-centric sites, respectively. The same NF-κB complex thus can simultaneously activate and repress distinct κB sites located in the same promoter where the strength of activation and repression dictates the overall outcome. (4) In vitro experiments reveal that the p52:Bcl3 complex binds to these κB sites in distinct modes, presumably because of the distinct structural states imposed by the sequence difference. Structural studies revealed that the central bp of a κB DNA does not make base-specific contacts with NF-κB dimers, which suggests that the conformational state of κB sites is an essential component of gene regulation specificity. Overall, this study provides the underpinnings of a model that integrates κB site sequence, NF-κB dimer selectivity, and regulation of target gene expression.
RESULTS
The Central bp of κB DNA Sequences Plays a Key Role in Providing the Transcriptional Activity of NF-κB Dimers
To investigate whether NF-κB dimers select specific κB sites for transcriptional activity, we searched for promoters that are stringently regulated by specific dimers. It was previously shown that the promoters of P-selectin and CyclinD1 are specifically bound by the p52 homodimer (Guttridge et al., 1999; Pan and McEver, 1995). Along with p52, these promoters also require Bcl3, which is consistent with the notion that p52 alone, lacking an activation domain, cannot activate transcription as a homodimer. p52 and Bcl3 are also involved in the transcriptional regulation of S-phase kinase-associated protein 2 (Skp2), IP-10, and inter-leukin 10 (IL-10) genes (Barré and Perkins, 2007; Leung et al., 2004; Mühlbauer et al., 2008). Two κB sites are known to regulate IP-10 gene expression, but neither of them is specifically linked to the p52:Bcl3 complex. To date, no κB sites that drive NF-κB-dependent activation of IL-10 and Skp2 genes are known. We identified κB sites within IL-10 and Skp2 promoters by searching sequences from –10 κB to +1 κB of the transcription start site of these genes. Alignment of the κB sites of these five promoters (P-selectin, CyclinD1, IP-10, IL-10, and Skp2) revealed that all five contain at least one G/C-centric κB site (Figure 1B). Because nearly all known κB sites contain A/T bp at the central position, we set to investigate whether the central bp is a critical determinant of transcriptional specificity of the p52:Bcl3 complex. Luciferase reporter constructs with either natural (G/C-centric) or mutant (A/T-centric) bp for all five of these κB sites were generated. In all cases, the G/C-centric κB sites induced luciferase expression in the presence of both p52 and Bcl3 (Figures 1C–1G). Deletion of the N and/or C terminus of Bcl3 nearly abolished luciferase activity, further signifying the critical role of Bcl3 (Figures S1A–S1C). Conversion of the central G/C to A/T bp reduced luciferase expression, although the extent of reduction varied. All natural G/C-centric κB sites were discriminated by RelA. In some cases, alteration of G/C to A/T bp of these sites enhanced luciferase expression in response to RelA. These observations support the notion that, at least in the in vitro system, the central bp in κB sites plays an important role in imposing NF-κB dimer function. Several other NF-κB dimers were also tested for their transcriptional activation specificity. Again, we observed that only the p52:Bcl3 complex was able to drive luciferase expression from this site (see Figures S1E–S1G; Extended Results).
We further tested two well-characterized κB sites: the HIV-κB site and the major histocompatibility complex (MHC)-κB site. As shown in Figure 1H, RelA activates the wild-type (WT) A/T-centric HIV-κB site but not the mutant G/C-centric HIV-κB site. However, the p52:Bcl3 complex showed the opposite activity. Similar effects were observed for the MHC-κB site (Figure S2A). Together, these observations suggest a dominant role of the central bp in NF-κB dimer selectivity. RelA dimers select A/T-centric sites for transcriptional activation, whereas the p52: Bcl3 complex selects G/C-centric sites. However, it is noticeable that the activation potential of RelA dimers from classical A/T-centric sites is far greater than that of the p52:Bcl3 complex from G/C-centric sites.
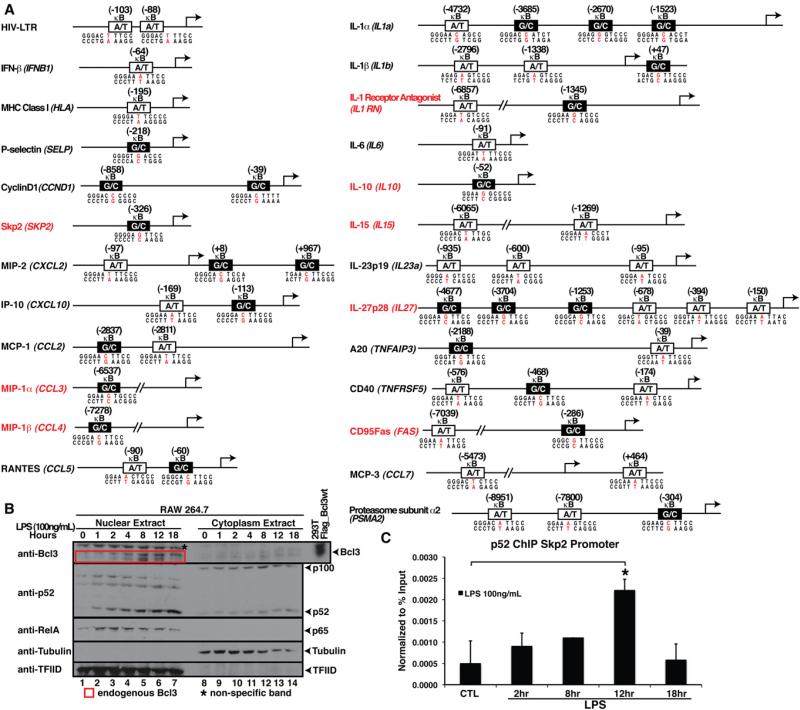
Architecture of the κB Sites in NF-κBB Target Gene Promoters
We next investigated the promoters of NF-κB target genes from published literature and a database (http://bioinfo.lifl.fr/NF-KB/) that has compiled promoters for over 100 mouse/human NF-κB target genes. We also examined microarray data obtained with the use of Kdo2-Lipid A (a synthetic lipid that has endotoxin activity equal to that of LPS and activates macrophages via TLR-4) and generated by the LIPID MAPS consortium (http://www.lipidmaps.org/data/index.html). We focused on known NF-κB target genes and inspected the sequences for the presence of κB sites from –10 kB to +1 kB of the transcription start site (a subset of these sites are listed in Figure 2A). The κB sites of a large number of these genes have not been annotated previously. We targeted several of these genes for further investigation (Figure 2A). Some of these promoters contain only classical A/T-centric κB sites, and some contain only G/C-centric κB sites, but most contain both A/T- and G/C-centric sites. From this list, we tested three G/C-centric sites located in the promoters of CD95Fas(FAS), CD40(TNFRSF5), and proteasome subunit α2 (PSMA2) by reporter assay (Figures S2B–S2D). Our analysis confirmed a strong correlation between G/C-centric sites and their activation by the p52:Bcl3 complex. In addition, RelA activated the mutant A/T-centric sites. However, alteration of G/C to A/T at the central position did not significantly reduce the p52:Bcl3 complex transcriptional activity in these three cases, suggesting that other bps in κB sites also contribute to gene regulation. That is, mutation in the context of naturally occurring κB sites could not overpower the auxiliary effect of other bps in the sequence. Further studies are required to assess the role of the other bps in dimer specificity.
Figure 2. Promoter Architecture and LPS-Mediated NF-κB Activation.
(A) Identification of κB sites in the promoter/enhancer region of known NF-κB target genes. Genes with newly identified κB sites are indicated in red. A/T and G/C-centric κB sites are indicated by white and black boxes, respectively.
(B) Western blot analysis of nuclear and cytoplasmic extracts of RAW 264.7 cells after LPS stimulation for different times. RelA accumulation in the nucleus peaked between 1 to 2 hr of LPS stimulation; Bcl3 and p52 accumulated in the nucleus at later time points; peaked at 8–12 hr of LPS stimulation.
(C) ChIP assay showing p52 loading onto the Skp2 promoter in RAW 264.7 cells treated with LPS for the indicated times. *p < 0.05. Error bars represent SD. See also Figure S2 and Table S1.
We used a lentiviral system that integrates NF-κB-driven promoters with destabilized green fluorescent protein (dGFP) expression as a reporter into the genome of monocyte THP-1 cells to further validate the transfection-based luciferase activity. Using this lentivirus system, we generated stable THP-1 cell lines with promoters containing WT HIV, P-selectin, and Skp2-κB sites or their mutational variants, and measured dGFP expression in LPS-stimulated cells (Figure S2F; Extended Results). Cells with G/C-centric P-selectin and Skp2-κB sites showed low dGFP expression at 8 hr and higher expression at 24 hr; however, the corresponding A/T mutants behaved in a manner opposite to that of their WT counterparts. In sum, the lentiviral and transfection-based reporter results are consistent. These observations allowed us to speculate that the early induction was likely to be mediated by RelA dimers, whereas the late expression was mediated by the p52:Bcl3 complex.
To test whether the promoters are similarly regulated in the endogenous context as depicted in the artificial reporter systems, we used the RAW 264.7 macrophage cell line as a model system. This cell line has been extensively investigated for its LPS-responsive gene regulation program by NF-κB (Lee et al., 2006; Ogawa et al., 2005; Wessells et al., 2004). Two events control the gene activation profiles: the kinetics of activation of specific NF-κB subunits, and their selectivity toward κB sites. Therefore, the activation kinetics of p52 and Bcl3 are expected to converge in order for them to function together. Indeed, we found nuclear accumulation of Bcl3 and p52 at 8–12 hr after LPS induction (Figures 2B and 2C). In contrast, nuclear RelA peaked between 1 and 2 hr, and slowly diminished afterward. These observations suggest that early-induced genes may be activated by RelA through binding to A/T-centric κB sites, whereas late-activated genes may be induced by the p52:Bcl3 complex through binding to the G/C-centric κB sites. Promoters that contain both types of κB sites would be persistently activated, at the early stage by RelA and at the late stage by the p52:Bcl3 complex.
The p52:Bcl3 Complex Is Recruited to Both G/C- and A/T-centric κB Sites, with Opposite Transcriptional Outcomes
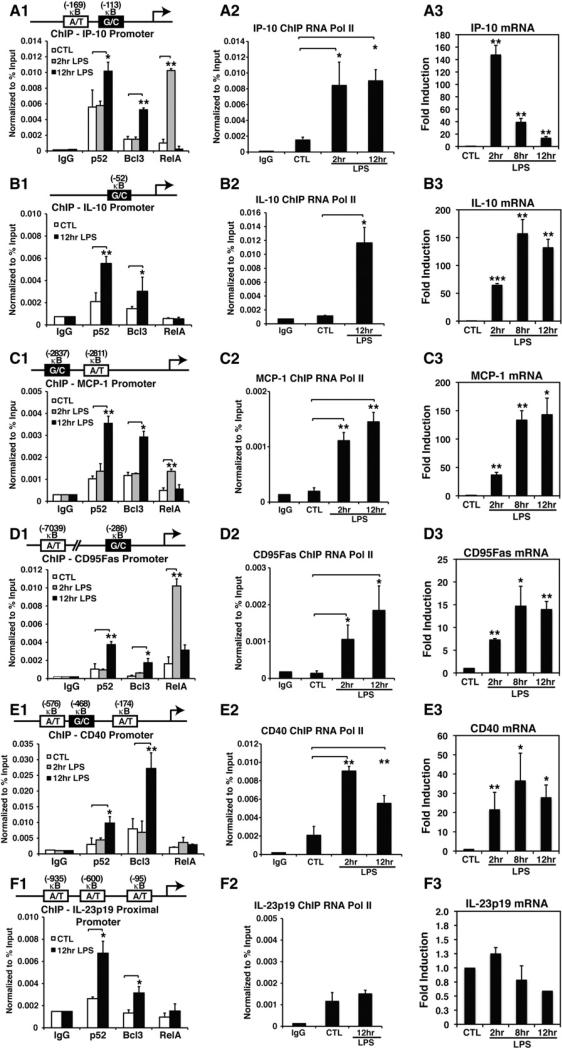
To validate our hypothesis regarding κB-site selectivity in cells, we carried out chromatin immunoprecipitation (ChIP) assays on several promoters that contain at least one G/C-centric κB site. Both p52 and Bcl3 were bound to IL-10, MIP-1α, MIP-1β, IP-10, MCP-1, CD95Fas, CD40, A20, and IL-1 receptor antagonist (IL-1RN) promoters at 12 hr, but were significantly less bound at 2 hr of LPS induction (Figures 3A1–3E1 and S3B). We also tested a subset of these promoters for RelA binding. With the exception of CD40, RelA bound to these promoters at 2 hr but not at 12 hr. We also found that RNA Pol II is recruited to these promoters, suggesting that these promoters are transcriptionally active (Figures 3A2–3F2). The pattern of DNA binding by the NF-κB dimers is consistent with the promoter architecture. These results led us to propose that A/T-centric κB sites primarily recruit RelA dimers at early times after LPS stimulation, whereas G/C-centric κB sites recruit the p52:Bcl3 complex at late stages. Two promoters, IL-23p19 and IL-6, which do not contain any G/C-centric κB sites, were also tested. A ChIP assay confirmed that both p52 and Bcl3 occupied the IL-23p19 promoter at 12 hr of LPS induction (Figure 3F). However, RNA Pol II was not recruited to this promoter, suggesting transcriptional silencing. This result also suggests that the recruitment of p52 and Bcl3 may lead to promoter silencing. Surprisingly, the IL-6 promoter, which contains an A/T-centric κB site, did not recruit either p52 or Bcl3 (Figure S3B). IL-6 is known to be activated by the p50:IκBζ complex in macrophages (Yamamoto et al., 2004). Therefore, at least a subset of A/T-centric sites are regulated by NF-κB dimers other than RelA and the p52:Bcl3 complex.
Figure 3. p52 and Bcl3 Binding to Endogenous Promoters, and Gene Activation upon LPS Stimulation.
(A1–F1) ChIP assay showing recruitment of p52, Bcl3, and RelA to indicated promoters at different times in RAW 264.7 cells untreated or treated with LPS for the indicated times.
(A2–F2) ChIP assay showing recruitment of RNA Pol II in RAW 264.7 cells treated with LPS at the indicated times. *p < 0.05, **p < 0.01.
(A3–F3) mRNA levels of the same set of genes monitored by qRT-PCR in RAW 264.7. **p < 0.01, ***p < 0.001 versus untreated controls. Error bars represent SD.
See also Figure S3 and Tables S1 and S2.
We next tested whether the kinetics of gene expression correlates with the recruitment of the NF-κB complexes (Figures 3A3–3F3 and S3A; Extended Results). In agreement with the microarray data obtained with Kso2-Lipid A and generated by the LIPID MAPS consortium, quantitative RT-PCR (qRT-PCR) revealed that all of the tested genes, with the exception of IL-23p19, were expressed in macrophages by LPS stimulation. However, the amplitudes and kinetics of expression varied. The IP-10 messenger RNA (mRNA) level showed prolonged induction from 2 to 12 hr, with a peak at 2 hr, suggesting a continuous transcription from the early to late phases of induction. The early massive IP-10 induction is probably due to RelA recruitment, whereas the late induction results from p52:Bcl3 recruitment. In other test cases, the expression pattern was different. The mRNA levels were lower at early times than at late times. CD40 was activated at early times of LPS induction; however, RelA was not recruited to this promoter, suggesting that other NF-κB subunits, such as cRel, may have compensated for RelA's function at early times. Consistent with the fact that IL-23p19 contains three A/T-centric κB sites, we found that the expression of IL-23p19 was completely blocked even though both p52 and Bcl3 were recruited to the promoter upon LPS induction. Altogether, these observations prompted us to propose an intriguing hypothesis: RelA activates genes from the A/T-centric κB sites at early times; however, the co-occupancy of p52 and Bcl3 in A/T-centric κB sites results in repression of transcription, whereas it activates transcription by binding to G/C-centric κB sites at late times.
The observation that the p52/RelB heterodimer is the key dimer of noncanonical NF-κB signaling pathways raises the question as to whether the p52/RelB heterodimer might also be responsible for the late gene activation. We measured RelB mRNA in both RAW 264.7 and bone-marrow-derived macrophages (BMDM), and found that RelB was induced early by LPS and its levels were significantly lower at late times (Figure S3C). These observations are consistent with the results shown in the LIPID MAPS, which showed that RelB levels were only 1.47- and 1.28-fold higher at 8 and 12 hr after stimulation, suggesting that RelB dimers were not significantly activated by LPS in macrophages during the timeframe investigated in this study. Altogether, these observations suggest that p52:Bcl3 is a key NF-κB complex that is responsible for regulating κB sites at late times. Moreover, unlike stimuli such as LTβ, which stringently activates the noncanonical NF-κB pathway, LPS activates both the canonical and noncanonical pathways. Therefore, dimers that are activated and regulated by LPS are expected to be different from those activated only through the noncanonical pathway.
Bcl3 Is Essential for p52-Mediated Transcriptional Regulation
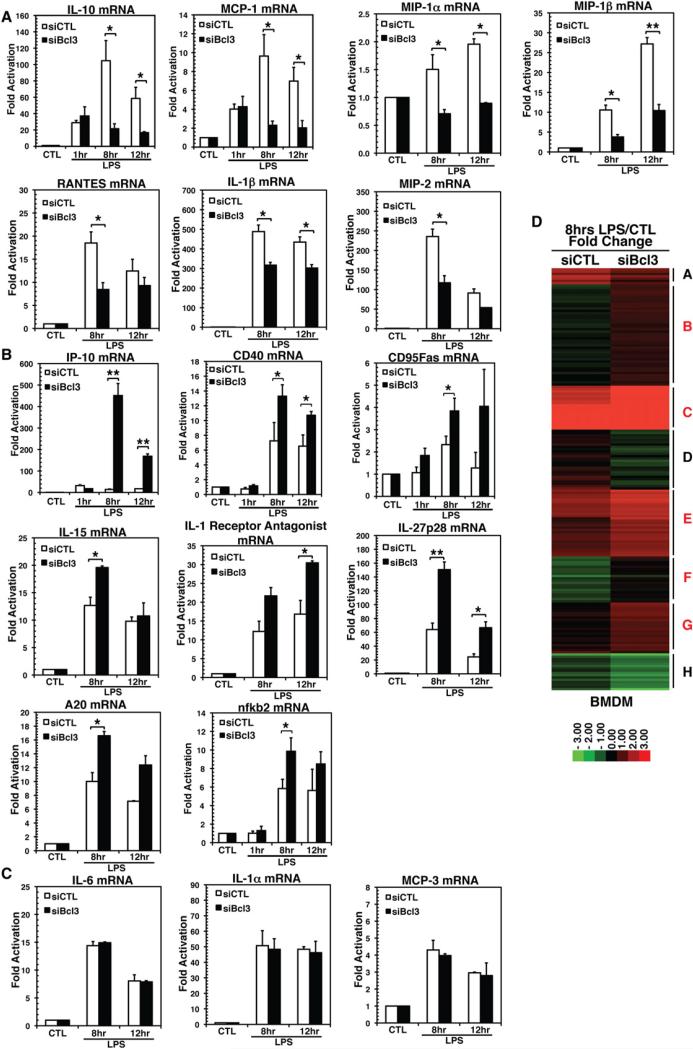
To further confirm the role of the p52:Bcl3 complex in gene regulation, we carried out small interfering (siRNA)-mediated Bcl3 knockdown in BMDM and tested how it affects target gene expression. We found that NF-κB target genes that were expressed in RAW 264.7 cells were also expressed in BMDM (Figure S4B), although the amplitude and kinetics of expression varied. For instance, Bcl3 levels were highest at 8 hr, and most target genes showed the highest levels at 8 hr in BMDM, compared with 12 hr in RAW 264.7 cells. We were able to obtain ~70% siBcl3 knockdown compared with control scramble siRNA (Figure S4A). We performed qRT-PCR to test 18 NF-κB target genes for their Bcl3 responsiveness. In LPS-stimulated cells, the mRNA levels of some genes were repressed (IL-10, MCP-1, MIP-1α, MIP-1β, MIP-2, RANTES, and IL-1β; Figure 4A), some were enhanced (IP-10, CD40, CD95Fas, nfκb2, IL-1RN, IL-15, IL-27p28, and A20; Figure 4B), and three remained unchanged (IL-6, IL-1α, and MCP-3; Figure 4C). These observations further support the notion that the p52:Bcl3 complex not only activates transcription but also represses gene transcription. Because IL-10, MIP-1α, and MIP-1β contain only a single G/C-centric κB site, repression of these genes in siBcl3 cells clearly suggests a critical transcriptional activation role of the p52:Bcl3 complex. The expression profiles of other genes in this category (containing both A/T- and G/C-centric sites) that are repressed in siBcl3 cells suggested that the activation function of the p52:Bcl3 complex is dominant over its repression function. The second class of genes was activated upon siBcl3, suggesting that the repression function of the p52:Bcl3 complex from the A/T-centric sites is dominant over its activation function from the G/C-centric sites. The third class of genes was not sensitive to siBcl3, which suggests that other NF-κB dimers could play a role in their activation. For example, IL-6 was shown to be activated by the p50:IκBζ complex. Further investigation is required to understand the responsiveness of each κB DNA to different NF-κB dimers.
Figure 4. Bcl3 Is Essential for Target Gene Expressions.
(A–C) mRNA levels showing genes that are (A) downregulated, (B) upregulated, or (C) unaffected upon siBcl3 knockdown in BMDM by qRT-PCR analysis. *p < 0.05, **p < 0.01, ***p < 0.001 versus nontreated control or as indicated. Error bars represent SD.
(D) Microarray mRNA expression data from mouse scramble siRNA (siCTL) and siBcl3 BMDM cells stimulated with LPS (100 ng/ml) for 8 hr were analyzed by K-means clustering. Red represents stimulus-responsive gene induction, whereas green represents repression. Cluster identifiers are indicated on the right, with the red symbol indicating clusters with increased expression in siBcl3 cells compared with siCTL cells.
To further test our hypothesis on an unbiased, genome-wide scale, we profiled gene induction and repression by LPS stimulation (0 or 8 hr) in siBcl3 BMDM compared with that in siCTL cells by microarray analysis. LPS-stimulated samples at 8 hr were used because Bcl3 levels were highest at this time of induction in WT BMDM. Genes with ~1.5-fold enhanced activation or repression upon siBcl3 were selected. Genes that showed a change in basal-level activation or repression but no change post-LPS stimulation were excluded from analysis. K-means clustering was performed with selected 184 genes (Figure 4D), which resulted in eight clusters that were further categorized based on the extent of activation or repression. This resulted in four main categories: genes that were activated in siCTL samples that were further activated (Figure 4D, clusters C, E, and G) or repressed (clusters A and D) in siBcl3 cells, and genes that showed no change in siCTL samples but were activated (Figure 4D, clusters B and F) or repressed (cluster H) in siBcl3 cells. All of the target genes tested by qRT-PCR (Figures 4A–4C) showed similar expression patterns in microarray analysis. The biological functions of more than two-thirds of these 184 genes are known. Collectively, these gene products are known for their activities in inflammation, growth and proliferation, vascular function, hemoglobin metabolism, and redox regulation. Proinflammatory genes (Nos2, Ptgs2, Ptges, CD69, and Vcam1), antiproliferation genes (Slfn1, Dnajb6, and Jdp2), and genes that regulate vascular activity (Itga5, Vegfa, Cxcl4, Tpbg, collagen, and Mmp13/14) are activated in siBcl3 cells listed in clusters B, C, and E. Growth- and proliferation-promoting genes (Egr1, Dek, CD74, Ccnd1, Areg, Dusp6, Gdf15, Ptpre, and Lmna) are downregulated, as are the genes encoding redox and pH-sensing enzymes (Hmox1, Sepp1, Srxn1, and Hvcn1) listed in clusters A, D, and H. These observations are consistent with earlier reports that the p52:Bcl3 complex promotes cell proliferation (Brenne et al., 2009; Massoumi et al., 2006; Ong et al., 1998; Rocha et al., 2003; Westerheide et al., 2001); however, the significance of NF-κB/Bcl3 in redox and pH sensing remains to be established. In addition, the Samhd1 gene, an HIV-1 restriction factor that encodes deoxynucleoside triphosphatase, was upregulated in siBcl3, suggesting an inhibitory role of Bcl3 in HIV infection (Figure S4C).
Although most of the siBcl3-affected genes are functionally connected to NF-κB, it is not clear whether their expression is directly regulated by NF-κB. We randomly picked 28 genes that showed significant up- or downregulation in the microarray NF-κB binding-site analysis, and found that they all contain κB sites and most of them contain multiple sites (Figure S4E). The expression profiles of some of these genes are shown in Figures S4C–S4D. Genes that are upregulated contain predominantly or exclusively A/T-centric sites, whereas genes that are downregulated contain both A/T- and G/C-centric sites. Without a functional analysis, one cannot be certain that these genes are regulated by the p52:Bcl3 complex; however, in general, the genome-wide analysis further strengthens our hypothesis that the G/C-centric sites are activated by the p52:Bcl3 complex, whereas A/T-centric sites are repressed by the same complex. As mentioned above, the activity of the p52:Bcl3 complex on promoters containing both types of κB sites is expected to be determined by the combinatorial effect of the relative strengths of each site.
Bcl3 Alters the Mode of Interaction between the p52 Homodimer and Two Classes of κB Sites In Vitro
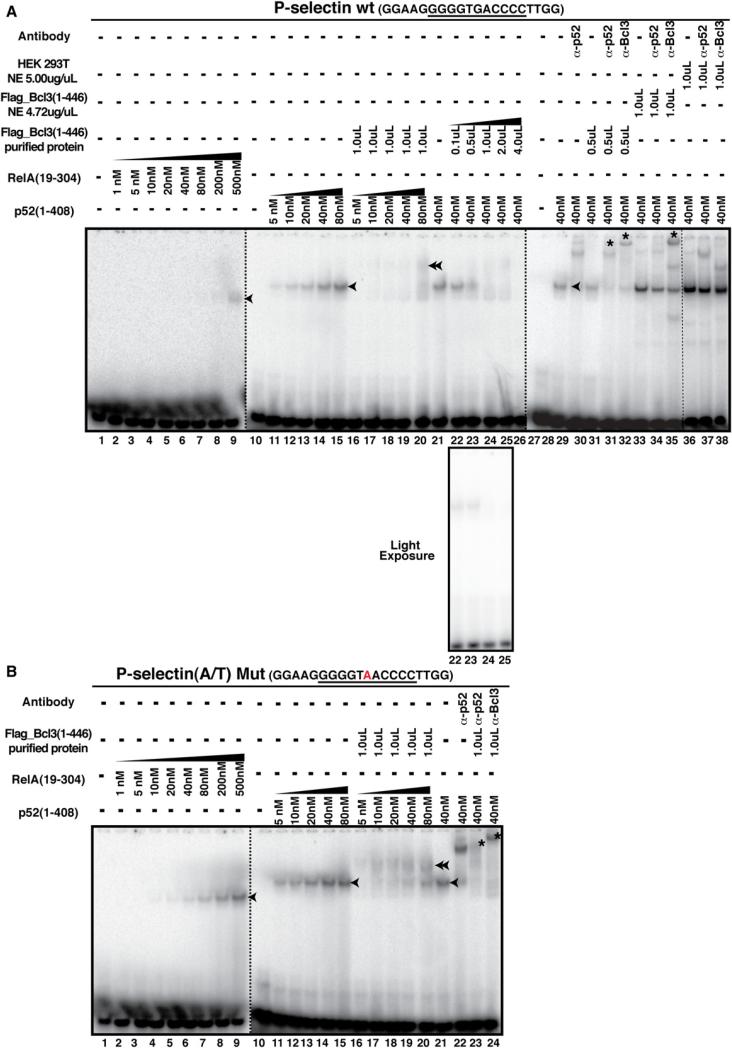
To further assess whether the p52:Bcl3 complex senses A/T-and G/C-centric κB sites differently, we carried out in vitro DNA-binding experiments using the electrophoretic mobility shift assay (EMSA). Surprisingly, we found that the p52 homo-dimer bound to the A/T-centric κB sites with higher affinity than the G/C-centric sites. To test whether Bcl3 reverses this effect, we used both Bcl3-transfected nuclear extracts (NE) and affinity-purified Flag-tagged Bcl3 from HEK293T cells, and tested the binding of the p52:Bcl3 complex to five WT G/C-and corresponding mutant A/T-centric κB sites (P-selectin, Skp2, IP-10, CyclinD1, and IL-10; Figures 5 and S5A–S5D). We also tested WT A/T- and corresponding mutant G/C-centric κB sites of HIV, IL-23p19, and MHC (Figures S5E–S5G). Contrary to our expectation, we found that with increasing concentrations of Bcl3, the p52:G/C-centric P-selectin κB DNA complex disappeared with no visible ternary complex in the gel (Figure 5A, lanes 16–20). This behavior is reminiscent of IκBα-mediated inhibition of DNA binding by RelA or p50/RelA heterodimer. IκBα actively dissociates the RelA:DNA complex, forming the IκBα: RelA complex and liberating free DNA. In this case, however, no enhancement of free P-selectin κB probe with increasing amounts of Bcl3 was observed, suggesting that the p52:DNA complex does not dissociate (Figure 5A, light exposure, lanes 22–25). Antibody supershift identified distinct slow-mobility complexes in the presence of antibodies against both anti-Bcl3 and anti-p52 (Figure 5A). This suggested that although no ternary complex was visible, both proteins were bound to the probe simultaneously. In contrast, association between A/T-centric κB DNA with p52 and Bcl3 was detected clearly as a distinct slow-mobility complex (Figure 5B, lanes 16–20). The presence of both proteins in this complex was also confirmed by antibody supershift analysis. All other probes with G/C- and A/T-centric κB sites showed similar trends (Figure S5). One possible reason for the apparent absence of the ternary complex between the G/C-centric κB site and the p52:Bcl3 complex is its kinetic instability. Transient dissociation and reassociation results in the lack of a discrete complex band in the gel. Antibody binding alters the nature of the complex, resulting in a clear supershifted band. We also noted that the CyclinD1 κB site was the only exception in which both G/C- and A/T-centric probes revealed nearly similar behaviors with no visible ternary complex in the gel (Figure S5C). However, in both cases, antibody supershift confirmed the presence of both proteins. CyclinD1-κB site differs significantly from most other κB sites.
Figure 5. EMSA of p52, p52:Bcl3, and RelA on WT G/C-Centric or Mutant A/T-Centric P-Selectin κB DNAs.
(A) Recombinant p52, RelA, and Flag-tagged full-length Bcl3 protein or NE binding to WT (G/C)-centric P-selectin κB DNA. Single arrowheads denote the NF-κB:κB DNA binary complex, the double arrowhead denotes the p52:Bcl3:κB DNA ternary complex, and * indicates the supershifted complex by specific antibody.
(B) Recombinant p52, RelA, Flag-tagged Bcl3 protein or NE binding to mutant (A/T)-centric P-selectin κB DNA.
The presence of one T-rich half site may influence the stability of the (p52:Bcl3):CyclinD1-κB DNA complex. This may explain why both WT G/C- and mutant A/T-centric CyclinD1 luciferase reporters were transcriptionally active, as shown earlier (Figure 1D). On the other hand, we found that both WT A/T- and mutant G/C-centric MHC-κB sites formed stable ternary complexes, although the complex with the mutant G/C-centric κB site was less prominent (Figure S5G). Consistent with these observations, we found that both WT and mutant MHC luciferase reporters were somewhat defective in the p52:Bcl3-driven activation (Figure S2A). We suggest that whereas the identity of the central bp is a crucial determinant of the transcriptional activity of the p52:Bcl3 complex, this bp works in conjunction with other bps within the κB sites. Possibly, every bp in κB sites plays a role in the determination of NF-κB dimer specificity. Taken together, these experiments suggest that the p52:Bcl3 complex binds to two distinct types of κB sites with distinct kinetic stabilities and/or conformations. We refer to this binding difference as distinct binding modes, and suggest that the different transcriptional outcomes are related to the binding modes.
The p52:Bcl3 Complex Recruits Coactivators to G/CCentric Sites, and Corepressors to A/T-Centric κB Sites
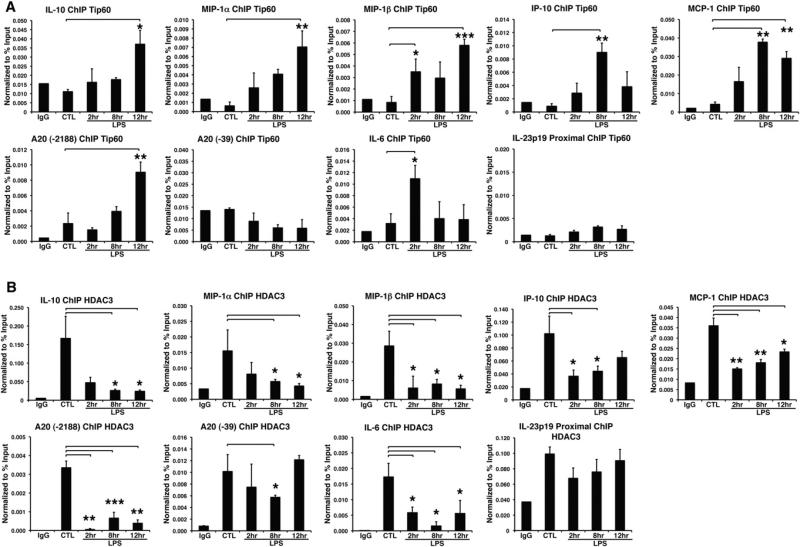
We hypothesized that the distinct binding modes of the p52:Bcl3 complex allow it to interact with specific cofactors, such as core-pressors to A/T-centric sites and coactivators to G/C-centric sites. Earlier reports showed that Bcl3 recruits the Tip60 histone acetyl transferase (HAT) complex to activate transcription (Baek et al., 2002; Dechend et al., 1999). ChIP experiments showed that indeed Tip60 was recruited to the IL-10, MIP-1α, MIP-1β, IP-10, and MCP-1 promoters with kinetics of recruitment that coincided with p52 and Bcl3 recruitment (Figure 6A). Although this experiment did not specify whether Tip60 was recruited to the G/C-centric sites in IP-10 and MCP-1 promoters, because these promoters also contain A/T-centric κB sites; however, the recruitment of Tip60 to the IL-10, MIP-1α, and MIP-1β κB sites clearly suggested that Tip60 was recruited to the G/C-centric sites. To further assess whether Tip60 was recruited to the G/C-centric site of a promoter containing both A/T- and G/C-centric sites, we chose to test A20 (TNFAIP3). In this promoter, two κB sites are >2 κB apart from each other. Because fragmentation generates on average ~500 bp DNA fragments, one can in principle show that Tip60 is specifically recruited to G/C-centric sites (–2,188) but not to A/T-centric sites (–39). Our results thus clearly demonstrate the specificity of G/C-centric κB sites for Tip60 recruitment. Consistent with our model, no Tip60 was found to be recruited to the IL-23p19 promoter, a pure A/T-centric κB promoter, even though the p52:Bcl3 complex was recruited. The lack of Tip60 recruitment to the IL-6 promoter further supports the report that p50:IκBζ is primarily responsible for activation of the IL-6 gene (Yamamoto et al., 2004).
Figure 6. Selective Recruitment of Cofactors to the Target Gene Promoters.
(A) ChIP assay showing recruitment of Tip60 to IL-10, MIP-1α, MIP-1β, IP-10, MCP-1, A20 (–2188), and IL-6 promoters, but not A20 (–39) and IL-23p19 promoters in RAW 264.7.
(B) ChIP assay showing the recruitment of HDAC3 to the IP-10, MCP-1, A20 (–39) and IL-23p19 promoters in RAW 264.7. *p < 0.05, **p < 0.01, ***p < 0.001 versus nontreated control or as indicated. Error bars represent SD.
See also Table S1.
To determine whether the converse is true (i.e., whether the p52:Bcl3 complex recruits a corepressor complex to the A/T-centric κB sites), we investigated the recruitment of repressors to the same set of five promoters. Previous studies showed that Bcl3 interacts with CtBP, HDAC3, and TBLR1, which are part of the NCoR/SMRT corepressor complex (Choi et al., 2010; Keutgens et al., 2010). Here we found the presence of HDAC3, a histone deacetylase, in the IL-23p19 promoter before induction, and it remained throughout the time of induction. In contrast, HDAC3 repressed the IL-10, MIP-1α, and MIP-1β promoters preinduction but disappeared during the course of LPS induction (Figure 6B). We next tested for the presence of HDAC3 in composite κB-site promoters such as IP-10, MCP-1, and A20. We found a different pattern of HDAC3 recruitment: postinduction recruitment was partial with respect to preinduction in the IP-10 and MCP-1 promoters. Because the A/T- and G/C-centric sites are close to each other in both cases, we proposed that both p52:Bcl3:Tip60 and p52:Bcl3:HDAC3 complexes might be present in these promoters at late times of induction. The A20 promoter clarified the κB-site specificity for HDAC3. We found that HDAC3 was present at both sites in the basal state, was absent at 2–8 hr, and then returned at 12 hr postinduction. HDAC3 recruitment at 12 hr coincided with the recruitment of the p52:Bcl3 complex at 12 hr to the A/T-centric site (–39). HDAC3 was not recruited to the G/C- centric site (–2,188) postinduction. These observations allow us to suggest that the transcriptional activation complex is present on the G/C-centric site, whereas the repression complex is present on the A/T-centric site. Altogether, our results suggest that the p52:Bcl3 complex “senses” the DNA sequence difference and binds two classes of κB sites in distinct modes, and thus recruits either coactivator or corepressor complexes, leading to opposite transcriptional outcomes.
DISCUSSION
Despite the wealth of basic biochemical knowledge that has been gained, deciphering the underlying principle behind NF-κB dimer-specific target gene activation has remained elusive, for several reasons. The cis-regulatory regions of most NF-κB target genes contain multiple nonidentical κB sites that function noncooperatively (Giorgetti et al., 2010). Moreover, the majority of NF-κB-inducing stimuli activate several NF-κB dimers. These facts make the task of determining which dimer binds to which κB sequence within a promoter/enhancer a challenge. Furthermore, no obvious correlation exists between a κB site and an NF-κB dimer for selective binding in vitro. Through a detailed examination of NF-κB target gene promoter sequences, loading of specific NF-κB subunits onto the κB sites, and genome-wide gene expression patterns (Figures 2A and S4), we show that the identity of the bp at the central position of κB sites strongly affects NF-κB dimer-specific target gene expression. NF-κB dimers bind to different κB sequences with different binding affinities and binding modes. In this study, we show that the p52:Bcl3 complex binds to both A/T- and G/C-centric κB sites; however, the binding affinities and binding modes are different, leading to different transcriptional outcomes. The complex recruits HDAC3 to the A/T-centric sites to repress transcription and recruits Tip60 to the G/C-centric site to activate transcription. The simultaneous recruitment of transcriptional coactivators and corepressors to a composite promoter/enhancer is a compelling finding. Here, we discuss how sequence variations affect the affinity and conformation of NF-κB:DNA complexes, as well as the functional consequences of κB sequence variations.
DNA Sequence Variations Affect NF-κB:DNA Complex Affinity and Conformation
Previous structural and binding studies of NF-κB:κB DNA complexes have provided some mechanistic insights into how κB sequences affect affinity and conformation. Whereas the flanking GC-rich sequences are mostly responsible for affinity and κB selectivity, NF-κB dimers “read” the central bp indirectly. This indirect-readout mechanism has been shown to be crucial for many protein-DNA recognition events (Chen-Park et al., 2002; Meijsing et al., 2009). For instance, κB sites with AT-rich sequences (A-2A-1A0T+1T+2 or A-2A-1T0T+1T+2) in the middle segment undergo bending that is apparently essential for RelA and cRel binding (Chen et al., 2000). If the AT richness is broken with a G/C bp at the center, the bendability of the DNA is expected to be different and hence the protein:DNA complex conformation would differ. It is important to note that A-2T-1A/T0A+1T+2 sequences are not acceptable for RelA even though they are AT rich. T at −1 and A at +1 positions not only alter DNA bendability but also put incorrect function groups for RelA to interact. Thus, variations with the inner positions of κB sequences would significantly affect RelA and possibly cRel binding. At the same time, variability in DNA bending would also affect the conformations of NF-κB:DNA complexes. This explains why RelA homodimer binds poorly to the P-selectin κB site (with T at −1 and A at +1) but relatively better to other G/C-centric sites.
Both p50 and p52 are more tolerant to sequence variability within the inner positions. Remarkably, however, even p50 and p52 read the central bp differently, as revealed by the structures of p50 and p52 homodimers bound to two nearly identical κB sites, with the only difference being at the central position (A/T bp versus A/A mismatch; Cramer et al., 1997; Müller et al., 1995). These variations allow us to make some speculations (Figure S6): Conserved residues from the p50 and p52 subunits bind identical DNA sites differently because the DNA-noncontacting protein residues of these two subunits differ. Therefore, an argument can be made that the central bp transduces specific contact networks, resulting in different protein conformations at a distal surface. NF-κB subunits are sensitive to small variations in the κB sites because the RHR region of NF-κB, which is responsible for both DNA binding and protein dimerization, is bimodular (including the N-terminal domain and C-terminal dimerization domain) and connected through a small linker peptide. The relative flexibility of the modules enables the dimer to scan far greater DNA conformational spaces to find correct functional groups for direct contact compared with a single-module DNA-binding domain as in other DNA-binding proteins. This greater scanning ability helps NF-κB to bind diverse κB sites with similar binding affinities (Ghosh et al., 2012). Therefore, NF-κB:DNA complex conformation is distinct for distinct DNA sites even when the bp difference between these sites is as small as one.
The protein-DNA recognition process cannot be fully captured by static structures derived from crystallographic studies. A microsecond molecular simulation study of an A/T-centric κB site embedded within a 20-bp-long DNA showed unusual DNA structural transitions at the long timescales of the simulations (Mura and McCammon, 2008). This study revealed that one of the bases of the central bp flipped out of the DNA axis. It is likely that the dynamic behavior of target DNA affects the binding modes of a transcription factor, and that such differential binding modes control the assembly of the larger transcription complex. It appears that binding of the p52:Bcl3 complex to the G/C- and A/T-centric κB sites is differentially regulated, resulting in kinetic instability of complexes involving G/C-centric sites but not those involving A/T-centric sites (Figures 5 and S5). Whether the binding behaviors are also different in vivo is not known. However, the fact that the two types of κB sites recruit different cofactors by binding to the same p52:Bcl3 complex in vivo strongly suggests a link between modes of NF-κB binding to the DNA sequence.
Transcriptional regulation through DNA allostery has been observed in other transcription factors, such as glucocorticoid receptor (GR), nuclear receptors, and Drosophila Hox family transcription factors (Joshi et al., 2010; Meijsing et al., 2009; Ogawa et al., 2005). Yamamoto and colleagues (2004) extensively studied the transcriptional activity of GR for different target genes. They found that the binding affinity between GR and its response element did not correlate with transcriptional activity. Instead, small variations in DNA sequence affected the conformation of a loop within the DNA-binding domain. Mutations at various positions within the loop had diverse effects on the expression of different genes containing different GR-response elements. Although the functional consequence is unclear, Kitayner et al. (2010) found that a protein noncontacting bp in the interior of the DNA response element forms Hoogsteen bp when bound to transcription factor p53. They showed that Hoogsteen bp alters the DNA shape, electrostatic potential, and configuration of neighboring sequences, resulting in differential protein-DNA interactions as compared with sequences with non-Hoogsteen bp. Furthermore, a recent NMR study showed that CA and TA base steps in DNA duplexes show transient formation of Hoogsteen bp (Nikolova et al., 2011). Such sequence-specific continuous fluctuations in bp mode may allow a transcription factor to bind two sequences differently, resulting in distinct functional outcomes, as we observed in the case of the p52:Bcl3 complex.
Functional Consequences of Sequence Variations in κB Sites
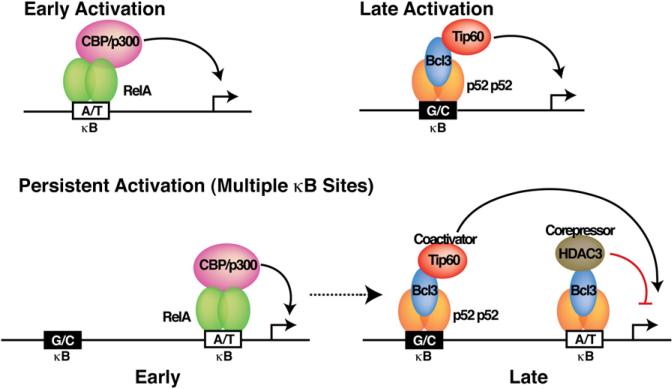
This study provides insights into the mechanism of temporal gene expression by NF-κB dimers. During early induction, preexisting RelA dimers (presumably the p50/RelA heterodimer) are activated through degradation of prototypical IκB inhibitors, which in turn activate early response genes by binding to the A/T-centric κB sites (Figure 7). Along with many early response genes, RelA activates several NF-κB and IκB genes, such as nfκb2 p100/p52, bcl3, and IkBz (Brasier et al., 2001), that contain A/T- centric sites. At the late phase of induction, the newly formed p52:Bcl3 complex activates genes with G/C-centric κB sites such as IL-10. Promoters that contain multiple κB sites, both A/T- and G/C-centric, such as IP-10, MCP-1 and A20, would persistently be activated by sequential engagement of different NF-κB dimers/complexes, such as early activation by RelA dimers and late activation by the p52:Bcl3 complex. However, the extent of persistent activation is dictated by the strength of each κB site. Our results are consistent with the report that κB sites in a promoter/enhance act independently (Giorgetti et al., 2010), and in line with another, earlier report showing that different NF-κB dimers bind to the same promoter at different kinetics (Saccani et al., 2003). Transcriptional repression by the p52:Bcl3 complex through its binding to the A/T-centric κB sites prevents uncontrolled, prolonged activation. RelA is not the only NF-κB dimer that activates target gene at early times. Genes such as CD40, IL-6, and MCP-3 are not regulated by RelA in macrophages. The p50:IkκBζ complex has been shown to activate IL-6. Sequences of the κB sites are different outside of the central A/T bp. These variations may be sufficient for the p50:IκBζ complex to outcompete the p52:Bcl3 complex in cells. It remains to be determined which NF-κB dimer represses the A/T-centric IL-23p19 promoter. Future studies will reveal how bps in other positions affect NF-κB recruitment and transcriptional activity.
Figure 7. Model Depicting Temporal Control of Target Genes by Different NF-κB Dimers and Coactivator and Corepressor Complexes.
Promoters with A/T-centric κB site(s) are activated by RelA supported by CBP/p300 at early times after induction. Composite promoters containing A/T- and G/C-centric κB sites are simultaneously regulated by corepressor and coactivator complexes nucleated by the p52:Bcl3 complex.
The κB sequence-specific gene regulatory dynamics of NF-κB dictates the setting of biological programs. For instance, our results clarify how the p52:Bcl3 complex participates in coordinating biological programs, such as the control of inflammation and proliferation. It controls inflammation by activating antiinflammatory cytokines such as IL-10 through binding to their G/C-centric κB sites. At the same time, it can repress proinflammatory cytokines such as IL-23, IL-6, and IL-8, and other inflammatory factors such as NOS2, COX2, and prostaglandin E synthase by binding to their A/T-centric κB sites. Our observation is consistent with the report that p52 and Bcl3 jointly dampen inflammation of effector genes that would otherwise undergo uncontrolled expression, as seen in nfκb2−/− bcl3−/− double knockout mice (Zhang et al., 2007). Through highly selective regulation of the G/C-centric κB sites within the promoters of Skp2 and CyclinD1, the p52:Bcl3 complex controls cell proliferation. The transcriptional activation of CyclinD1 from a specific κB site is well documented (Guttridge et al., 1999), and the CyclinD1 κB site has G/C bp at the central position. Because CyclinD1 and Skp2 promoters contain only G/C-centric κB sites, the absolute requirement of p52 and Bcl3 for activation and in turn cell-cycle regulation can now be properly explained (Barré and Perkins, 2007). In addition to these two, our microarray analysis identified new growth- and proliferation-promoting genes: Egr1, Dek, CD74, Areg, Dusp6, Gdf15, Ptpre, and Lmna. These genes are expressed at reduced levels in the absence of Bcl3. They contain at least one G/C-centric κB site in their promoters (Figure S4E), which further supports the paradigm that the coordination of a biological program is controlled at the level of the κB DNA response element of the effector genes. Interestingly, several genes that inhibit cell-cycle progression, such as Slfn1, Dnajb6, and Jdp2, are activated in siBcl3. All of these genes contain A/T-centric κB sites in their promoters. Thus, the p52:Bcl3 complex regulates cell proliferation by activating genes that promote proliferation and repressing genes that block proliferation by sensing the κB sites in their promoters differently.
Our study also clarifies a long-standing confusion about the function of Bcl3. Some earlier reports showed that, like the classical IκBs, Bcl3 could dissociate its preferred p50 or p52 homodimer from κB DNAs and thus function as a repressor of transcription (Nolan et al., 1993). Other studies also showed that, under some circumstances, Bcl3 could associate with p50 or p52 homodimer, forming an activation complex on the promoter (IκB:NF-κB:κB DNA), and therefore function to coactivate NF-κB transcription (Bours et al., 1993; Dechend et al., 1999; Fujita et al., 1993; Massoumi et al., 2006; Rocha et al., 2003). Here we have shown how Bcl3 could accomplish this dual role. Its oncogenic function may be linked to both transcriptional activation and repression: through its continuous presence, Bcl3 induces both uncontrolled proliferation and inflammation.
In sum, we have shown how sequence variations in κB sites diversify gene regulatory programs by imposing conformational restrictions on bound NF-κBs. This work presents a description of rules that govern the in vivo transcriptional specificity of NF-κB dimers, and describes how simultaneous activation and repression by an NF-κB dimer are dictated by κB DNA response elements. Our study sets the stage for genome-wide analysis of the binding of p52:Bcl3 and other NF-κB dimers in correlation with functions to fully elucidate the scope of response-element-specific conformational variations in the pattern of gene regulation.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
RelA (sc-372), tubulin (sc-5286), RNA Pol II (sc-900), HDAC3 (sc-11417), normal rabbit IgG, and all secondary HRP-conjugated antibodies were purchased from Santa Cruz Biotechnology. HDAC3 (ab7030) antibody was obtained from Abcam. Flag (M2) (F3165) antibody and LPS (L6529) were obtained from Sigma. Tip60 (07-038) was obtained from Millipore. p100/p52 (#1494 for ChIP assays and #1495 for western blot and EMSA) and Bcl3 (#1348) rabbit antisera were a gift from Dr. Nancy Rice (National Institutes of Health, Bethesda, MD).
Mammalian Cell Culture
BMDMs were obtained from 5- to 7-week-old C57BL/6 mice as previously described (Sawka-Verhelle et al., 2004). In brief, macrophages were cultured in RPMI 1640 medium (Invitrogen) supplemented with 25% fetal bovine serum (FBS) and 25% L-cell-conditioned medium as a source of macrophage colony-stimulating factor. RAW 264.7, HeLa, and HEK293T cells were grown in Dulbecco's modified Eagle's medium (CellGro) supplemented with 10% FBS, 2 mM glutamine, and antibiotics.
For RNA interference experiments, scramble control or smart-pool siRNAs (Dharmacon) against Bcl3 were transfected into BMDM using Lipofectamine 2000 (Invitrogen) as previously described (Ghisletti et al., 2009). Cells were used for experiments after 48 hr of incubation, and target gene knockdown efficiency was validated by qPCR.
Protein Expressions and Purifications
Recombinant mouse RelA(19-304) and human p52(1-408) were expressed and purified from Escherichia coli BL21(DE3) cells. Flag-Bcl3(1-446) was expressed in HEK293T cells using Lipofectamine 2000 (Invitrogen) for 48 hr according to the manufacturer's protocol, and purified with Anti-Flag M2 Affinity resin (A2220; Sigma).
Luciferase Reporter Assays
HeLa cells were transiently transfected with Flag-RelA(1-551), or Flag-p52(1-415) together with Flag-Bcl3(1-446) expression vectors, and the lucif-erase reporter DNA with specific κB DNA promoters. Luciferase activity assays were performed with the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. Details regarding the protocol are available in the Extended Experimental Procedures.
ChIP Assays
ChIP assays were performed in RAW 264.7 cells as previously described (Ghisletti et al., 2009). The sequences of the promoter-specific primers are listed in Table S1.
RNA Isolation and qRT-PCR
Total RNA (isolated by RNeasy kit; QIAGEN) was prepared from RAW 264.7 cells treated with LPS (100 ng/ml) as indicated; 0.1 μg of total RNA was used for complementary DNA (cDNA) synthesis (SuperScriptIII First-Strand Synthesis System; Invitrogen), and 1 μl of cDNA was used for qRT-PCR analysis. Values were normalized to the glyceraldehyde 3-phosphate dehydrogenase (GAPDH) content (Livak and Schmittgen, 2001). Data are represented as the mean ± standard deviation (SD) of three independent experiments, each performed in duplicate. Primer sequences are listed in Table S2.
Microarray Analysis
RNA was extracted from BMDM cells with the QIAGEN RNeasy kit and hybridized to Illumina Mouse RefSeq Sentrix-8 V2 BeadChip arrays at the University of California-San Diego Biogem Facility. Probes with an ~1.5-fold phenotype in siBcl3 with siCTRL upon 8 hr of LPS (100 ng/ml) stimulation were selected (biological duplicated samples). K-means clustering was performed with selected probes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Susan Taylor for providing the fluorescence plate reader, Dr. Leor Weinberger and Brandon Razooky for helping with the lentivirus experiments, and Kristyn E. Feldman for help and valuable discussion regarding the micro-array analysis. This work was supported by National Institutes of Health grants GM085490 and AI064326.
Footnotes
EMSA
DNA-binding assays were performed using EMSA as previously described (Moorthy et al., 2007). Sequences of the κB DNAs used are listed in Table S3.
ACCESSION NUMBERS
The National Center for Biotechnology Information GEO accession number for the microarray data reported in this work is GSE39922.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Results, Extended Experimental Procedures, six figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2012.08.042.
REFERENCES
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- Barré B, Perkins ND. A cell cycle regulatory network controlling NF-kappaB subunit activity and function. EMBO J. 2007;26:4841–4855. doi: 10.1038/sj.emboj.7601899. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. The oncoprotein Bcl-3 directly transactivates through kappa B motifs via association with DNA-binding p50B homodimers. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- Brasier AR, Lu M, Hai T, Lu Y, Boldogh I. NF-kappa B-inducible BCL-3 expression is an autoregulatory loop controlling nuclear p50/NF-kappa B1 residence. J. Biol. Chem. 2001;276:32080–32093. doi: 10.1074/jbc.M102949200. [DOI] [PubMed] [Google Scholar]
- Brenne AT, Fagerli UM, Shaughnessy JD, Jr., Våtsveen TK, Rø TB, Hella H, Zhan F, Barlogie B, Sundan A, Børset M, Waage A. High expression of BCL3 in human myeloma cells is associated with increased proliferation and inferior prognosis. Eur. J. Haematol. 2009;82:354–363. doi: 10.1111/j.1600-0609.2009.01225.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YQ, Sengchanthalangsy LL, Hackett A, Ghosh G. NF-kappaB p65 (RelA) homodimer uses distinct mechanisms to recognize DNA targets. Structure. 2000;8:419–428. doi: 10.1016/s0969-2126(00)00123-4. [DOI] [PubMed] [Google Scholar]
- Chen-Park FE, Huang DB, Noro B, Thanos D, Ghosh G. The kappa B DNA sequence from the HIV long terminal repeat functions as an allosteric regulator of HIV transcription. J. Biol. Chem. 2002;277:24701–24708. doi: 10.1074/jbc.M200007200. [DOI] [PubMed] [Google Scholar]
- Cheng CS, Feldman KE, Lee J, Verma S, Huang DB, Huynh K, Chang M, Ponomarenko JV, Sun SC, Benedict CA, et al. The specificity of innate immune responses is enforced by repression of interferon response elements by NF-κB p50. Sci. Signal. 2011;4:ra11. doi: 10.1126/scisignal.2001501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Lee JM, Kim H, Nam HJ, Shin HJ, Kim D, Ko E, Noh DY, Kim KI, Kim JH, Baek SH. Bcl3-dependent stabilization of CtBP1 is crucial for the inhibition of apoptosis and tumor progression in breast cancer. Biochem. Biophys. Res. Commun. 2010;400:396–402. doi: 10.1016/j.bbrc.2010.08.084. [DOI] [PubMed] [Google Scholar]
- Cramer P, Larson CJ, Verdine GL, Müller CW. Structure of the human NF-kappaB p52 homodimer-DNA complex at 2.1 A resolution. EMBO J. 1997;16:7078–7090. doi: 10.1093/emboj/16.23.7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechend R, Hirano F, Lehmann K, Heissmeyer V, Ansieau S, Wulczyn FG, Scheidereit C, Leutz A. The Bcl-3 oncoprotein acts as a bridging factor between NF-kappaB/Rel and nuclear co-regulators. Onco-gene. 1999;18:3316–3323. doi: 10.1038/sj.onc.1202717. [DOI] [PubMed] [Google Scholar]
- Fujita T, Nolan GP, Liou HC, Scott ML, Baltimore D. The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-kappa B p50 homodimers. Genes Dev. 1993;7(7B):1354–1363. doi: 10.1101/gad.7.7b.1354. [DOI] [PubMed] [Google Scholar]
- Ghisletti S, Huang W, Jepsen K, Benner C, Hardiman G, Rosenfeld MG, Glass CK. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G, Wang VY, Huang DB, Fusco A. NF-κB regulation: lessons from structures. Immunol. Rev. 2012;246:36–58. doi: 10.1111/j.1600-065X.2012.01097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Siggers T, Tiana G, Caprara G, Notarbartolo S, Corona T, Pasparakis M, Milani P, Bulyk ML, Natoli G. Noncooperative interactions between transcription factors and clustered DNA binding sites enable graded transcriptional responses to environmental inputs. Mol. Cell. 2010;37:418–428. doi: 10.1016/j.molcel.2010.01.016. [DOI] [PubMed] [Google Scholar]
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS., Jr. NF-kappaB controls cell growth and differentiation through transcriptional regulation of cyclin D1. Mol. Cell. Biol. 1999;19:5785–5799. doi: 10.1128/mcb.19.8.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Nakashima T, Taniguchi M, Kodama T, Kumanogoh A, Takayanagi H. Osteoprotection by semaphorin 3A. Nature. 2012;485:69–74. doi: 10.1038/nature11000. [DOI] [PubMed] [Google Scholar]
- Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- Joshi R, Sun L, Mann R. Dissecting the functional specificities of two Hox proteins. Genes Dev. 2010;24:1533–1545. doi: 10.1101/gad.1936910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keutgens A, Shostak K, Close P, Zhang X, Hennuy B, Aussems M, Chapelle JP, Viatour P, Gothot A, Fillet M, Chariot A. The repressing function of the oncoprotein BCL-3 requires CtBP, while its polyubiquitination and degradation involve the E3 ligase TBLR1. Mol. Cell. Biol. 2010;30:4006–4021. doi: 10.1128/MCB.01600-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, Shakked Z. Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 2010;17:423–429. doi: 10.1038/nsmb.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KY, Ito K, Hayashi R, Jazrawi EP, Barnes PJ, Adcock IM. NF-kappaB and activator protein 1 response elements and the role of histone modifications in IL-1beta-induced TGF-beta1 gene transcription. J. Immunol. 2006;176:603–615. doi: 10.4049/jimmunol.176.1.603. [DOI] [PubMed] [Google Scholar]
- Leung TH, Hoffmann A, Baltimore D. One nucleotide in a kappaB site can determine cofactor specificity for NF-kappaB dimers. Cell. 2004;118:453–464. doi: 10.1016/j.cell.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fässler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy AK, Huang DB, Wang VY, Vu D, Ghosh G. X-ray structure of a NF-kappaB p50/RelB/DNA complex reveals assembly of multiple dimers on tandem kappaB sites. J. Mol. Biol. 2007;373:723–734. doi: 10.1016/j.jmb.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlbauer M, Chilton PM, Mitchell TC, Jobin C. Impaired Bcl3 up-regulation leads to enhanced lipopolysaccharide-induced interleukin (IL)-23P19 gene expression in IL-10(−/−) mice. J. Biol. Chem. 2008;283:14182–14189. doi: 10.1074/jbc.M709029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- Mura C, McCammon JA. Molecular dynamics of a kappaB DNA element: base flipping via cross-strand intercalative stacking in a microsecond-scale simulation. Nucleic Acids Res. 2008;36:4941–4955. doi: 10.1093/nar/gkn473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natoli G, Saccani S, Bosisio D, Marazzi I. Interactions of NF-kappaB with chromatin: the art of being at the right place at the right time. Nat. Immunol. 2005;6:439–445. doi: 10.1038/ni1196. [DOI] [PubMed] [Google Scholar]
- Nikolova EN, Kim E, Wise AA, O'Brien PJ, Andricioaei I, Al-Hashimi HM. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan GP, Fujita T, Bhatia K, Huppi C, Liou HC, Scott ML, Baltimore D. The bcl-3 proto-oncogene encodes a nuclear I kappa B-like molecule that preferentially interacts with NF-kappa B p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol. 1993;13:3557–3566. doi: 10.1128/mcb.13.6.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, Glass CK. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell. 2005;122:707–721. doi: 10.1016/j.cell.2005.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong ST, Hackbarth ML, Degenstein LC, Baunoch DA, Anastasi J, McKeithan TW. Lymphadenopathy, splenomegaly, and altered immunoglobulin production in BCL3 transgenic mice. Oncogene. 1998;16:2333–2343. doi: 10.1038/sj.onc.1201771. [DOI] [PubMed] [Google Scholar]
- Pan J, McEver RP. Regulation of the human P-selectin promoter by Bcl-3 and specific homodimeric members of the NF-kappa B/Rel family. J. Biol. Chem. 1995;270:23077–23083. doi: 10.1074/jbc.270.39.23077. [DOI] [PubMed] [Google Scholar]
- Rocha S, Martin AM, Meek DW, Perkins ND. p53 represses cyclin D1 transcription through down regulation of Bcl-3 and inducing increased association of the p52 NF-kappaB subunit with histone deacetylase 1. Mol. Cell. Biol. 2003;23:4713–4727. doi: 10.1128/MCB.23.13.4713-4727.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Mol. Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Sawka-Verhelle D, Escoubet-Lozach L, Fong AL, Hester KD, Herzig S, Lebrun P, Glass CK. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J. Biol. Chem. 2004;279:17772–17784. doi: 10.1074/jbc.M311991200. [DOI] [PubMed] [Google Scholar]
- Siggers T, Chang AB, Teixeira A, Wong D, Williams KJ, Ahmed B, Ragoussis J, Udalova IA, Smale ST, Bulyk ML. Principles of dimer-specific gene regulation revealed by a comprehensive characterization of NF-κB family DNA binding. Nat. Immunol. 2012;13:95–102. doi: 10.1038/ni.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells J, Baer M, Young HA, Claudio E, Brown K, Siebenlist U, Johnson PF. BCL-3 and NF-kappaB p50 attenuate lipopolysaccha-ride-induced inflammatory responses in macrophages. J. Biol. Chem. 2004;279:49995–50003. doi: 10.1074/jbc.M404246200. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Mayo MW, Anest V, Hanson JL, Baldwin AS., Jr. The putative oncoprotein Bcl-3 induces cyclin D1 to stimulate G(1) transition. Mol. Cell. Biol. 2001;21:8428–8436. doi: 10.1128/MCB.21.24.8428-8436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Yamazaki S, Uematsu S, Sato S, Hemmi H, Hoshino K, Kaisho T, Kuwata H, Takeuchi O, Takeshige K, et al. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 2004;430:218–222. doi: 10.1038/nature02738. [DOI] [PubMed] [Google Scholar]
- Zhang X, Wang H, Claudio E, Brown K, Siebenlist U. A role for the IkappaB family member Bcl-3 in the control of central immunologic tolerance. Immunity. 2007;27:438–452. doi: 10.1016/j.immuni.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.