Abstract
The actin cytoskeleton, which regulates cell polarity, adhesion, and migration, can influence cancer progression, including initial acquisition of malignant properties by normal cells, invasion of adjacent tissues, and metastasis to distant sites. Actin-dependent molecular motors, myosins, play key roles in regulating tumor progression and metastasis. In this review, we examine how non-muscle myosins regulate neoplastic transformation and cancer cell migration and invasion. Members of the myosin superfamily can act as either enhancers or suppressors of tumor progression. This review summarizes the current state of knowledge on how mutations or epigenetic changes in myosin genes and changes in myosin expression may affect tumor progression and patient outcomes and discusses the proposed mechanisms linking myosin inactivation or upregulation to malignant phenotype, cancer cell migration, and metastasis.
Keywords: cancer, metastasis, myosin, actin
Introduction
Myosin superfamily
Myosins are actin-dependent molecular motors that utilize the energy of ATP hydrolysis to generate force. The many functions of myosins include cell contractility, cell signaling, endocytosis, vesicle trafficking and protein/RNA localization [Hartman and Spudich, 2012; Krendel and Mooseker, 2005; Woolner and Bement, 2009]. All myosins share certain structural and functional features, particularly the presence of an actin-binding head domain, which is also responsible for myosin ATPase activity. Actin-dependent ATPase activity is used by myosin motors to generate force and to translocate cargo along actin filaments. Myosins also contain a neck domain, which can bind light chains, and a tail, which is variable in sequence and length and is likely to determine myosin intracellular localization and cargo binding specificity.
The first myosin to be studied biochemically was skeletal muscle myosin, also known as myosin II [reviewed in Szent-Gyorgyi, 2004]. Muscle myosin forms heterohexamers consisting of a dimer of heavy chains associated with four light chains, which can be further assembled into thick filaments that promote the contraction of muscle fibers. Following the discovery of non-muscle myosins, myosin family was subdivided into two groups: class I (small, mostly monomeric motors) and class II (dimeric myosins similar to skeletal muscle myosin; this class includes both muscle and non-muscle myosins). Class II myosins are also known as conventional myosins. Subsequent identification of a multitude of other non-muscle myosins, along with the progress of the human genome sequencing, dramatically changed our understanding of myosin diversity [Berg et al., 2001]. Currently, myosin superfamily of motor proteins is divided into multiple classes; this classification is based primarily on the phylogenetic analysis of the motor domain [Berg et al., 2001; Foth et al., 2006; Hodge and Cope, 2000; Odronitz and Kollmar, 2007; Richards and Cavalier-Smith, 2005]. Of these, 12 classes (I, II, III, V, VI, VII, IX, X, XV, XVI, XVIII, XIX) are found in vertebrates, including humans. All myosin classes distinct from myosin II are known as unconventional myosins and typically function as either monomers or dimers of heavy chains associated with calmodulin-like light chains, which bind to the IQ motifs in the neck region. The light chains stabilize the neck region and amplify the conformational changes in the motor domain to produce movement along actin filaments. Myosin velocity and step size are proportional to the length of the neck region (the lever arm) [Tyska and Warshaw, 2002].
Functional diversity of myosin motors
Myosin isoforms exhibit variations in their mechanochemical properties, such as the duty ratio (fraction of the ATPase cycle spent in the strong actin-bound state) and processivity (the ability to move continuously along an actin filament without diffusing away) [O'Connell et al., 2007]. Low duty ratio myosins may function in rapid contractile events or serve as dynamic, short-lived tethers between actin filaments and myosin cargo while high duty ratio myosins are better adapted to generating sustained tension. Processive myosins are best suited for the long-range transport of organelles and macromolecules. The diversity of mechanochemical properties and cargo binding domains allows non-muscle myosins to contribute to many processes important for cancer progression, including cell migration and invasion through the extracellular matrix, regulation of protein and organelle localization, cell shape changes, and cell signaling.
Non-muscle myosins are associated with a wide variety of actin-containing cellular structures. These include microvilli, stereocilia, and filopodia (membrane protrusions supported by the linear, unipolar bundles of actin filaments), stress fibers (linear structures composed of actin filaments of mixed polarity that are reminiscent of muscle sarcomeres in their periodic organization and are associated with cell-substrate contacts), adhesion belts (parallel bundles of actin filaments running along the apical portion of epithelial cells and associated with cell-cell junctions), and other types of actin assemblies (Figure 1). Actin polymerization in cells relies on the activity of actin nucleators, protein complexes that promote formation of short nuclei composed of 2–3 actin monomers, to initiate filament assembly. Distinct actin structures are formed through the activity of different nucleators (including Arp2/3 complex, which promotes formation of branched actin networks, formins, which nucleate parallel bundles, and other nucleators) and are associated with distinct subsets of actin-binding and crosslinking proteins [Arjonen et al., 2011; Campellone and Welch, 2010; Michael and Yap, 2013; Vindin and Gunning, 2013; Yang and Svitkina, 2011].
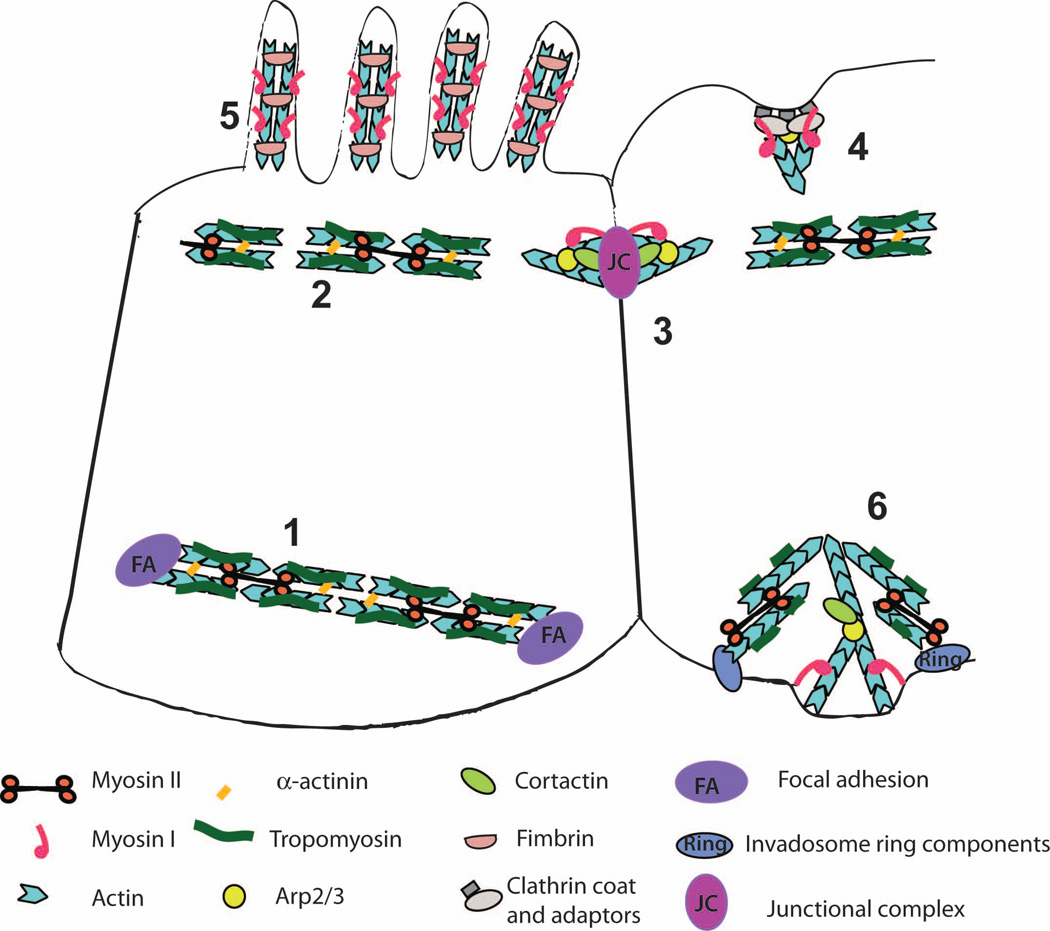
Figure 1.
Organization of the actin-based cytoskeletal structures associated with myosin motors. Stress fibers (1) and apical adhesion belts (2) are composed of linear actin bundles cross-linked by alpha-actinin and associated with tropomyosin and myosin II; assembly of these structures requires the activity of formins. These are highly contractile structures that can promote elongation and maturation of focal adhesions (stress fibers) or apical constriction of epithelial cells during morphogenesis (adhesion belts). Nascent cell-cell adhesions (3) are formed following the engagement of transmembrane adhesion receptors such as cadherins on the surfaces of adjacent cells, leading to assembly of junctional complexes. Junctional complexes initiate formation of branched, Arp2/3-nucleated actin assemblies associated with myosin I and cortactin but devoid of tropomyosin. The transition from nascent cell-cell adhesions to apical adhesion belts during junctional maturation is not well understood. Actin networks assembled at clathrin-coated endocytic invaginations (4) consist of branched, Arp2/3 nucleated actin and are associated with myosin I. Microvilli (5) contain parallel bundles of actin filaments cross-linked by fimbrin that exclude tropomyosin and myosin II but are enriched in myosin I. Invadosomes (6) are specialized adhesion complexes involved in ECM degradation. Invadosome structure is especially complicated since they consist of an actin core made up of a branched actin meshwork and a ring of adhesion and adaptor proteins that surrounds the core and is associated with a more loosely organized actin cloud. This structural complexity is reflected in the diverse assortment of cytoskeletal proteins associated with invadosomes, including both Arp2/3 and formins, class I and II myosins, and tropomyosin. We have presented a hypothetical model of invadosome organization, where the core consists of branched actin networks associated with cortactin and myosin I while the ring and cables contain tropomyosin and myosin II; whether this accurately reflects invadosome organization remains to be determined.
Some myosin classes display preferences for specific actin structures. For example, yeast and metazoan myosins II and yeast myosin V have been shown to preferentially associate with parallel actin bundles nucleated by formins and containing the actin-binding protein tropomyosin, while class I myosins are excluded from tropomyosin-containing actin assemblies and preferentially bind to the Arp2/3-nucleated branched actin networks [Ostap, 2008; Pollard and Lord, 2014; Pruyne, 2008; Stark et al., 2010; Tojkander et al., 2011; Wang and Coluccio, 2010]. In mammalian cells, tropomyosin is present in stress fibers, cytokinetic contractile rings, and apical adhesion belts; these structures are highly contractile due to myosin II activity. Members of myosin class I, on the other hand, bind to actin structures that do not contain tropomyosin, such as branched actin networks surrounding clathrin-coated vesicles, actin filaments associated with nascent (immature) junctional complexes, and fimbrin-crosslinked actin bundles in the microvilli (Figure 1). Another example of a preferential association between a specific actin structure and a particular myosin class is found in filopodia, which contain parallel bundles of actin filaments cross-linked by fascin and enriched in Myo10; the precise mechanism that governs Myo10 localization to filopodia has not been determined [Kerber and Cheney, 2011]. Myosin specificity toward particular cytoskeletal structures provides an additional mode of controlling myosin activity; for example, changes in tropomyosin or fascin expression that are often observed in cancer cells [Arjonen et al., 2011; Wang and Coluccio, 2010] could lead to changes in myosin-mediated cell motility and tension.
Myosin activity and the hallmarks of cancer
This review will provide an overview of the roles that non-muscle myosins play in cancer progression and metastasis. We will discuss the known associations between myosin mutations and cancer, correlation between myosin expression levels and tumor progression, and the mechanisms (either experimentally confirmed or hypothetical) that may link myosin activity to tumor invasion and metastasis. Many of the findings on the role of myosins in cancer progression are quite recent and limited to a relatively small number of patient cases; however, as research continues to uncover the functions of myosins as tumor suppressors or promoters, additional large-scale studies will undoubtedly follow.
The discussion of the contributions of the myosin superfamily to neoplastic transformation and metastasis can be framed in terms of the more general pathways that drive tumor progression. It has been proposed that most changes that lead to tumor formation and metastasis can be described as associated with several hallmarks of cancer cells, which represent the key properties that are necessary for cancer cell survival and tumor spreading [Hanahan and Weinberg, 2011]. These hallmarks include uncontrolled proliferation and the ability to prevent growth suppression, disruption of the programmed cell death mechanisms, induction of angiogenesis, maintenance of replicative immortality, evasion of immune surveillance, and the ability to invade and form metastases. While tumors isolated from different patients may exhibit genomic or epigenetic changes in distinct gene subsets, these alterations can be categorized as targeting a few key pathways that promote tumor cell survival and growth. Based on the known or hypothetical connections between these pathways and myosin functions, it may be possible to propose how changes in myosin expression or activity could affect the key cancer regulatory networks. Some of these connections have been experimentally identified in cancer cells, as will be discussed in this review, while others may offer a theoretical framework that can be used when considering potential mechanisms that may link myosin genomic or epigenetic alterations and cancer.
Enhanced proliferation and avoidance of growth suppression
The uncontrolled growth of transformed cells may be achieved via an upregulation of growth factor receptor expression or receptor presentation on the cell surface, mutations that lead to constitutive activation of growth factor signaling pathways (including activating mutations in the transmembrane receptors or in the intracellular components of the pathway), disruption of proliferation-controlling mechanisms such as growth suppressor pathways (governed by the p53 and Rb proteins) or changes in the contact inhibition of cell division. Since many myosins have been linked to endocytic, exocytic, and recycling pathways, it is easy to envision that the disruption of myosin activity may interfere with the mechanisms that regulate growth factor receptor signaling via receptor internalization and recycling. Additionally, deficiency in myosins that are involved in the establishment and maintenance of cell-cell adhesion could decrease the ability of epithelial cells to undergo contact inhibition. Somewhat unexpectedly, recent findings concerning myosin IIA role as a tumor suppressor have also identified a connection between myosin IIA and p53 stability [Schramek et al., 2014], as will be discussed in the section on myosin II, indicating that myosins may directly affect signaling pathways that induce growth suppression in response to stress.
Replicative immortality
Unlike non-transformed cells, which are typically able to undergo only a limited number of cell divisions, cancer cells acquire the ability to continuously replicate their genome and divide without undergoing senescence. This ability has been linked to the upregulation of telomerase activity as well as the disruption of p53-mediated control of genomic integrity, which allows cells to divide in spite of the damage to the chromosomes and the loss of telomeres [Bernardes de Jesus and Blasco, 2013; Hanel and Moll, 2012]. The lack of the genomic quality control in p53-deficient cancer cells plays an important role in the accumulation of carcinogenic mutations; thus, myosin IIA loss may contribute to tumor progression via a number of p53-mediated pathways.
Defects in programmed cell death pathways
Programmed cell death, or apoptosis, plays an important role in limiting the proliferation of tumor cells and in mediating the cytotoxic effects of anti-cancer therapies. The ability to evade programmed death pathways that are normally induced in response to DNA damage or other stressors leads to both an uncontrolled growth of tumors and their resistance to chemotherapy [Adams and Cory, 2007; Hanahan and Weinberg, 2011]. Myosins that are known to interact with the components of apoptosis-regulating pathways, such as the antiapoptotic proteins belonging to the Bcl-2 family (see the section on myosin V), can modulate cell death program.
Loss of epithelial cell polarity
The loss of cell polarity may contribute to tumor initiation via disruption of asymmetrical cell division, a process that supports epithelial tissue organization by maintaining distinct populations of epithelial stem cells and differentiating epithelial cells [Royer and Lu, 2011]. Loss of cell polarity may also occur during the late stages of epithelial tumor progression in the course of the epithelial-mesenchymal transition (EMT) [Thiery, 2002]. EMT is characterized by the downregulation of epithelial differentiation markers and the loss of cell polarity and tissue organization and is generally associated with poor prognosis in patients with invasive cancers. Many oncogenes and tumor suppressors regulate key cell polarity pathways, and mutations or changes in the expression level of core polarity proteins are frequently observed in cancer [Huang and Muthuswamy, 2010]. Mutations in myosins that are involved in the establishment or maintenance of cell polarity may, therefore, be linked to cancer progression.
Angiogenesis
The growth of new blood vessels that supply tumors with nutrients and oxygen represents one of the key steps in tumor progression. The induction of angiogenesis may be a required step for progression from a microscopic, asymptomatic tumor to a larger neoplasm [Naumov et al., 2006]. VEGF signaling, which acts via a tyrosine kinase receptor, plays a key role in promoting angiogenesis, and also contributes to other aspects of tumor development. Thus, as discussed above, myosin motors that regulate receptor-mediated endocytosis may contribute to suppression of angiogenesis under normal circumstances by downregulating VEGF receptor signaling. Moreover, angiogenesis depends on cell migration and cell shape changes, two processes that require myosin activity. In particular, migration of endothelial cells during angiogenesis relies on filopodia formation at the leading edge, which is associated with Myo10 activity [Almagro et al., 2010; Pi et al., 2007], while branching of blood vessels is controlled by myosin II [Farber et al., 2011; Fischer et al., 2009].
Inflammation
Recent studies have led to an appreciation of a link between tumors and inflammation [Grivennikov et al., 2010]. Some inflammatory diseases, such as colitis/inflammatory bowel disease, have been linked to an increased risk of cancer [Waldner and Neurath, 2009]. On the other hand, growing tumors elicit an inflammatory response that promotes establishment of a tumor-permissive microenvironment. Thus, myosins that affect inflammatory pathways, such as those that modulate macrophage activity [Maravillas-Montero and Santos-Argumedo, 2012] or have been linked to inflammatory disorders such as Crohn’s disease and colitis [van Bodegraven et al., 2006], may influence tumor progression.
Evasion of the immune response
Studies in immunodeficient mouse models and in immunosuppressed patients indicate that tumors may develop more readily in the absence of a robust immune response, and that the immune system may play a role in determining tumor phenotype by selecting for less immunogenic tumor cells, a process known as immunoediting [Kim et al., 2007; Teng et al., 2008]. Specific cell types implicated in tumor control include T cells and NK cells. Myosins that contribute to the immune system functions and modulate immune response to tumor antigens could be important for controlling tumor growth.
Tumor cell invasion and metastasis
Switch from the primary tumor growth to metastasis requires acquisition of novel properties by tumor cells. These include the ability to invade through the extracellular matrix and stroma, enter blood vessels, travel to distant sites, and successfully colonize the new sites. Tumor cells can utilize a variety of migration modes to achieve successful invasion and spreading. These include collective cell migration, which is characteristic of many solid tumors of epithelial origin that have retained cadherin-based cell-cell junctions, and single-cell migration, which is exhibited by leukemia and lymphoma cells as well as cells derived from solid tumors that have undergone epithelial-mesenchymal transition (EMT) and lost cadherin expression [Mierke, 2013; Sahai, 2005].
Furthermore, cells that migrate individually can exhibit one of the two types of migratory behavior, specifically, either mesenchymal or amoeboid migration [Sahai, 2005]. Mesenchymal migration, similar to that observed in fibroblasts in culture, is characterized by the spindle-like shape of the cell, the presence of a well-defined leading edge and trailing tail, formation of numerous filopodia and lamellipodia at the leading edge and focal adhesions and stress fibers in the cell body, and a relatively low speed of migration. This mode of migration requires matrix degradation and, therefore, relies on the activity of matrix metalloproteases that may be secreted by the tumor cells or tumor-associated macrophages. Matrix metalloproteases secretion is often concentrated at invadopodia, actin-rich, integrin anchored adhesive structures that are important for matrix degradation and tumor cell invasion [Eckert and Yang, 2011; Linder et al., 2011; Sibony-Benyamini and Gil-Henn, 2012]. On the other hand, amoeboid migration is reminiscent of the mode of movement of Dictyostelium amoeba and cells such as leukocytes, with the cells maintaining a rounded shape and undergoing repeated cycles of contraction and relaxation. Cells using the amoeboid migration mode are able to squeeze through the ECM without degrading it. Tumor cells exhibit surprising plasticity in their ability to switch between mesenchymal and amoeboid modes of migration, which makes the task of disrupting migration of cancer cells particularly challenging. Both types of individual migration rely on cell contractility; therefore, myosin activity is likely to be important for both mesenchymal and amoeboid migration, although differential regulation of myosin isoforms may be important for selection of a specific migration mode.
Collective cell migration, observed in many epithelial solid tumors, may utilize pathways similar to those involved in collective migration during normal development and morphogenesis; however, the precise mechanisms driving collective migration of cancer cells remain to be identified [Friedl et al., 2012]. Moreover, different tumor types may utilize distinct modes of collective migration. In some cases, the migrating cell sheet develops distinct leader cells, which form actin-rich protrusions at the leading edge and secrete proteases to digest the ECM; the “follower” cells then invade into the partially degraded matrix and widen the areas of matrix depletion [Wolf et al., 2007]. In other cases, migrating cells form a unified front without distinct leaders or protrusions; this is observed during branching morphogenesis in normal mammary glands as well as in breast tumors [Ewald et al., 2008]. Both types of collective migration require dynamic reorganization of cell-cell junctional complexes and associated cytoskeletal structures in order to allow cells to change their positions without losing cell-cell contacts. Some myosins, such as myosins II, VI and IX, have been implicated in collective cell migration in in vitro and in vivo experimental models; thus, it is likely that they may contribute to collective migration in some cancer types.
Myosin functions: motors, anchors, and tethers
In order to understand how changes in myosin expression and activity may affect cell behavior, it is important to determine the contribution of myosin motor activity and myosin-generated tension to the processes that lead to neoplastic transformation and metastasis. Motor activity is likely important for the functions of myosin II, which may exert its effects on cell contractility by actively moving actin filaments relative to each other. Similarly, processive myosins that are responsible for long-range transport (for example, myosin V) clearly rely on the motor activity for their functions. On the other hand, some myosins may act as anchors, rather than as active motors, by promoting organelle or protein accumulation at specific sites via anchoring of the cargo to actin filaments. Given the presence of multiple protein and lipid interaction motifs in many myosins, one could also envision some myosins acting simply as adaptor or scaffolding proteins, bridging multiple interacting partners together and linking the resulting multimolecular complexes to actin. For example, class I myosins that contain membrane binding motifs may be responsible for tethering the plasma membrane to actin filaments and maintaining the shape of membrane-bound protrusions such as microvilli or stereocilia. This function may not necessarily require myosin motor activity since rigor binding of the motor domain to actin filaments may be sufficient for tethering.
Myosins and cancer
In pinpointing the connections between myosin upregulation or inactivation and cancer, it is important to distinguish between the data from in vitro studies examining the effects of myosin overexpression, depletion, or inhibition on cell transformation and motility in culture and the findings from the screens for genes or transcripts affecting metastasis or patient survival in vivo. In this review, we will discuss the findings from both in vitro and in vivo studies. In many cases, a combination of data from the genetic, epigenetic and transcriptomic studies of tumor samples and in vitro tests of myosin effects on cell transformation and invasion provides strong support for the role of specific myosin isoforms in tumor suppression or tumor progression. In vivo data, such as mutations and epigenetic modifications in myosin heavy chain genes, associations observed in gene signature screens, and transcriptomic analysis of tumor samples are summarized in Table 1, while proposed functions for myosins in cells are depicted in Figure 2. Analysis of the frequency of tumor-associated somatic mutations in myosin genes (Table 1) shows that myosins that may function as tumor suppressors are mutated in 2–45% of tumor samples, depending on the myosin and the type of cancer. This mutation frequency is comparable to that of other tumor suppressor genes. For example, TP53 gene, an important tumor suppressor, is mutated in 5–50% of tumors, depending on the cancer type [Olivier et al., 2010].
Table I.
Known associations between myosin expression level or genetic or epigenetic modifications and cancer
| Myosin | Mutations or allelic loss associated with cancer (frequency of mutations shown in parentheses) |
Epigenetic modifications |
Transcriptomic data (gene signature, RT- PCR, immunohistochemistry, and microarray studies) |
Proposed function |
|---|---|---|---|---|
| Myo1a | Colorectal tumors [Mazzolini et al., 2012] (>30% of tumors with microsatellite instability), gastric tumors [Mazzolini et al., 2013] (>45% of tumors with microsatellite instability). |
Methylation [Mazzolini et al., 2012; Mazzolini et al., 2013]. |
Tumor suppression |
|
| Myo1e | Component of the gene signature for invasive breast cancer [Hallett et al., 2012]. |
Cell invasion | ||
| Myo1f | Fused to MLL in infant acute leukemia [Duhoux et al., 2011; Taki et al., 2005]. |
Modifying MLL activity |
||
| MYH9 (myosin IIA) |
Squamous cell carcinoma and other cancers [Schramek et al., 2014]. |
Tumor suppression | ||
| Myosin Va | Elevated in colorectal cancer (qRT-PCR) [Lan et al., 2010]; decreased in gastric cancer (IHC) [Dong et al., 2012]. |
|||
| Myosin VI | Elevated in ovarian (IHC) and prostate (IHC and cDNA microarrays) cancers [Dunn et al., 2006; Yoshida et al., 2004]. |
Promotes collective migration of tumor cells |
||
| Myosin IXb | Polymorphic allele associated with esophageal cancer [Menke et al., 2012]. |
|||
| Myosin X | Elevated in breast cancer [Arjonen et al., 2011; Cao et al., 2014]. |
Cell invasion | ||
| Myosin XVIIIb | Mutations in lung (13%), colorectal (2%), ovarian (∼6%) [Nakano et al., 2005; Nishioka et al., 2002; Yanaihara et al., 2004]. |
Hypermethylation, histone acetylation [Nakano et al., 2005; Nishioka et al., 2002; Yanaihara et al., 2004]. |
Tumor suppression |
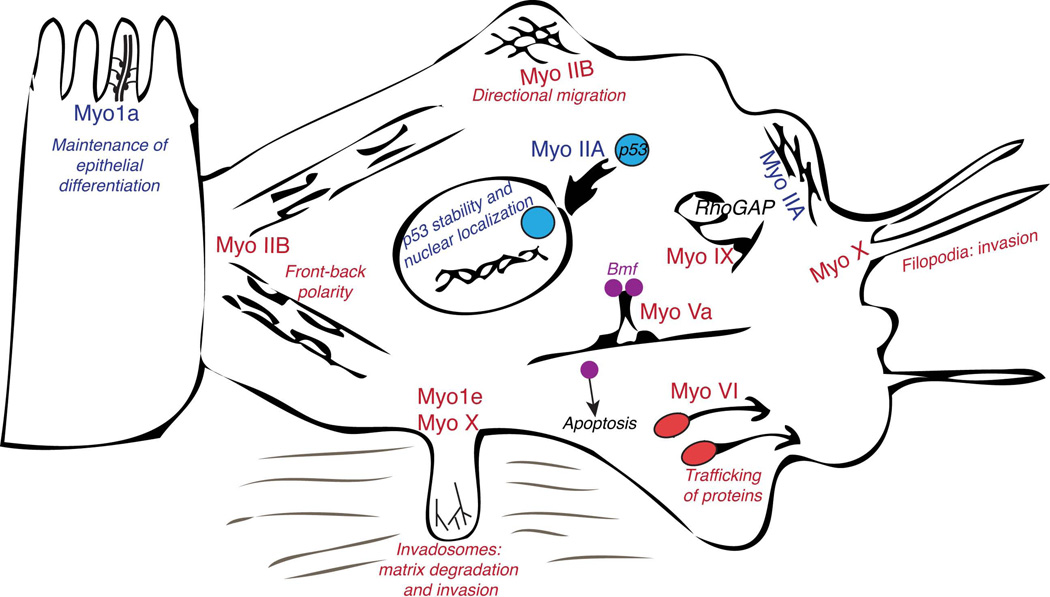
Figure 2.
Schematic representation of some of the roles that have been proposed for the various members of myosin superfamily during tumor progression and metastasis. Myosins that may contribute to tumor suppression are indicated by the blue font while myosins that may promote tumor cell migration and invasion are designated by the red font.
The discussion is this review has been limited to those myosin classes whose connections to cancer have been relatively well established. As discussed above, myosins are involved in many aspects of cell physiology that affect tumor progression and metastasis; thus, as research progresses, other myosin classes may be implicated in cancer biology.
Myosin I
Class I myosin heavy chains are relatively short compared to other myosins and do not contain dimerization motifs. All class I myosins contain membrane binding tail homology 1 (TH1) domains, which are capable of binding to negatively charged phospholipids. Within the TH1 domain lies a pleckstrin homology (PH) domain, which allows class I myosins to bind to specific phosphoinositides [Hokanson et al., 2006; Patino-Lopez et al., 2010], as well as additional basic or basic-hydrophobic motifs important for membrane interactions [Brzeska et al., 2008; Feeser et al., 2010; Mazerik and Tyska, 2012]. These characteristics provide class I myosins with the ability to bind to the plasma membrane or to the membrane-bound organelles. For this reason, class I myosins may serve as linkers tethering the plasma membrane to the underlying cortical actin and may act as a scaffold for membrane protrusions as well as contribute to membrane remodeling during endo- and exocytosis and cell migration [Greenberg and Ostap, 2013; McConnell and Tyska, 2010]. Some class I myosins (Myo1c, Myo1e) are broadly expressed in a variety of cell types while others (Myo1a) are more restricted in their expression pattern. Class I myosins associate with calmodulin-like light chains, which may allow them to be regulated by calcium [Greenberg and Ostap, 2013].
Myosin 1a
Myosin 1a, also known as brush border myosin I or Myo1a, is highly expressed in the intestinal and gastric epithelium, particularly in the intestinal brush border consisting of actin-rich microvilli [Garcia et al., 1989; Mazzolini et al., 2013; Mooseker and Coleman, 1989]. Studies utilizing Myo1a knockout mice and expression of the dominant-negative Myo1a construct show that Myo1a is important for maintaining the molecular composition and overall organization of the intestinal microvilli; in addition, Myo1a also regulates vesicle release at the tips of microvilli, which may contribute to gut immunity [McConnell and Tyska, 2007; Tyska et al., 2005; Tyska and Mooseker, 2004]. Myo1a function relies on its membrane binding ability via the TH1 domain, and the TH1 domain is sufficient for Myo1a localization to microvilli in cultured cells [Mazerik and Tyska, 2012; Tyska and Mooseker, 2002]. Enterocytes of Myo1a–null mice exhibit a partial disruption of apico-basal polarity, as characterized by the redistribution of apical proteins (sucrase-isomaltase and CFTR) to the basolateral cell surface and mislocalization of basolateral Myo1c to the brush border, although they still form apical microvilli [Kravtsov et al., 2012; Tyska et al., 2005].
Since Myo1a contributes to maintaining apico-basal polarity in intestinal epithelial cells, it may be important for resisting the EMT that occurs during neoplastic transformation. Thus, loss of function of Myo1a may be expected to promote cancer progression. Indeed, both frequent mutations and enhanced promoter methylation in the MYO1A gene have been found in patients with colorectal cancer, leading to the hypothesis that Myo1a acts as a tumor suppressor in intestinal epithelial cells [Mazzolini et al., 2012]. Knockdown of Myo1a in colon cancer cell lines resulted in decreased polarization and differentiation, enhancement of anchorage-independent growth, and increased tumorigenicity [Mazzolini et al., 2012]. Similarly, genetically- or chemically-induced tumors showed faster progression in Myo1a–knockout mice than in the animals expressing Myo1a [Mazzolini et al., 2012]. In patients with colorectal cancer, lower level of Myo1a expression was associated with faster tumor progression and poor prognosis [Mazzolini et al., 2012]. Similar findings (enhanced mutation frequency and promoter hypermethylation of MYO1A) were observed in patients with gastric tumors [Mazzolini et al., 2013]. Thus, several lines of evidence point to Myo1a serving as an important tumor suppressor in colorectal cancers both in vitro and in vivo and loss of Myo1a function may also serve as a prognostic indicator in colorectal and gastric cancer.
Myosin 1e
Myosin 1e (Myo1e) is a broadly expressed myosin that is necessary for normal renal filtration [Krendel et al., 2009] and formation of cell-cell adhesions between glomerular epithelial cells [Bi et al., 2013]. In addition to its role in kidney functions, Myo1e contributes to receptor-mediated endocytosis [Cheng et al., 2012; Krendel et al., 2007] and has been identified as a component of cell-substrate adhesions in proteomic studies [Cervero et al., 2012; Schiller et al., 2011]. Thus, Myo1e performs a variety of functions that impinge on the interactions between cell surface receptors, plasma membrane, and the underlying actin cytoskeleton. Recently, we identified Myo1e as a component of invadosomes in RSV-transformed fibroblasts [Ouderkirk and Krendel, 2014]. Inhibition of Myo1e activity through dominant-negative tail expression led to mislocalization of the newly forming invadosomes in RSV-transformed cells. In cancer cells, invadosomes promote cell migration and matrix degradation, therefore correct localization and assembly of these structures is likely important for tumor cell invasion. Thus, loss or inhibition of Myo1e activity may prevent metastasis of tumor cells that rely on invadosomes for their migration while upregulation of Myo1e expression may serve as a marker of highly invasive tumors.
Indeed, Myo1e expression has been shown to correlate with poor prognosis in patients with invasive breast cancer [Hallett et al., 2012]. This study examined gene expression in patients with basal-like breast cancer – a subtype of breast cancer characterized by the expression of the markers typical of basal, or myoepithelial, cells of mammary glands, rather than luminal epithelial cells. Basal cells exhibit mesenchymal features and are more contractile and motile than luminal epithelial cells. A number of gene expression profiling studies of various breast cancer samples has identified gene signatures characteristic of basal-like breast cancers (aggressive, highly metastatic, often triple-negative) and luminal-like breast cancers, which have a better prognosis [Hu et al., 2006; Rakha et al., 2008]. Investigation of gene expression signatures in a sample set of basal-like breast tumors showed a correlation between Myo1e expression level and poor prognosis in patients with basal-like breast cancer [Hallett et al., 2012]. We hypothesize that upregulation of Myo1e in cancer cells may promote migration and invasion through extracellular matrix, thereby enhancing metastasis, whereas the loss of Myo1e activity may be associated with decreased migration, as shown for glomerular epithelial cells expressing mutant/non-functional Myo1e [Mele et al., 2011]. In addition, Myo1e may potentially play a role in regulating growth factor receptor internalization via receptor-mediated endocytosis, although this has not been shown directly.
Myosin 1f
Myo1f is an SH3-domain containing myosin similar to Myo1e. It is highly expressed in some immune cells and has been identified as a component of the macrophage podosomes [Cervero et al., 2012]. Further, Myo1f is essential for neutrophil migration, and Myo1f–null neutrophils showed increased adhesion as a result of upregulated integrin exocytosis [Kim et al., 2006]. These observations suggest that Myo1f could potentially contribute to changes in cancer cell adhesion and migration properties or to modulation of the immune response and inflammation in cancer, although no direct evidence of this has been found so far.
Myo1f has also been identified as a fusion partner of the MLL (Mixed Lineage Leukemia) gene in some patients with acute leukemia [Duhoux et al., 2011; Taki et al., 2005]. In patients with acute infant myeloid leukemia, the MLL gene is often affected by chromosomal translocations that lead to fusions with other genes [Slany, 2009]. MLL gene encodes a histone methyltransferase. Some of the MLL fusions with nuclear proteins convert MLL to a broad specificity transcription factor, leading to abnormal activation of gene expression and subsequent transformation of cells. Other MLL fusion partners contain dimerization motifs, such as coiled-coil domains, that lead to MLL dimerization. The mechanism linking dimerization of MLL to neoplastic transformation is unknown, although it has been shown that artificially dimerizing MLL is sufficient to induce cell immortalization [Martin et al., 2003]. It is unclear how fusions with Myo1f could affect MLL activity, although it has been proposed that the SH3 domain of Myo1f could interact with the proline-rich motifs within MLL, inducing oligomerization of MLL [Duhoux et al., 2011].
Diverse roles for Class I myosins in cancer progression
As discussed above, different members of myosin class I may play distinct and even opposing roles in cancer progression. While Myo1a may help maintain epithelial cells in a differentiated state, thus functioning as a tumor suppressor, other class I myosins that promote cell motility, such as Myo1e, may be associated with tumor cell de-differentiation and metastasis (Figure 2). Indeed, expression of Myo1a (brush border myosin I) and Myo1e (formerly known as Myo1c) within intestinal epithelium correlates with the differentiation state of epithelial cells, with the immature cells of the crypts having high level of expression of Myo1e and the mature cells of the villi expressing mostly Myo1a [Skowron et al., 1998].
Although other class I myosins have not been implicated in cancer progression, many of them are capable of serving as scaffolds between the actin cytoskeleton and the plasma membrane, and, therefore, may contribute to cell shape changes involved in migration, polarization, and/or metastasis. Furthermore, some class I myosins, such as Myo1b and Myo1c, regulate post-Golgi protein trafficking or exocytosis of transmembrane proteins and, therefore, may contribute to regulation of growth factor receptor presentation on the cell surface [Almeida et al., 2011; Tiwari et al., 2013]. In addition, Myo1c has been implicated in regulating epithelial cell-cell adhesion, an important property of normal epithelial cells [Tokuo and Coluccio, 2013].
Myosin II
Class 2 myosins include muscle myosins, responsible for the contraction of the sarcomeres, and non-muscle myosins, implicated in force generation and actin crosslinking that regulates cell migration [Aguilar-Cuenca et al., 2014; Vicente-Manzanares et al., 2009; Wang et al., 2011]. Like muscle myosin 2, non-muscle myosin 2 heavy chains form dimers that can further assemble into filaments [Billington et al., 2013]. Mammals have three different isoforms of non-muscle myosin II heavy chains, each with a distinct localization and functions. The genes encoding non-muscle myosin II heavy chains in humans are MYH9 (myosin IIA), MYH10 (myosin IIB), and MYH14 (myosin IIC). Class II myosin heavy chains associate with 2 types of light chains, known as regulatory and essential light chain. Rho GTPases, which play important roles in regulation of cancer cell migration, regulate myosin II activity by modulating phosphorylation of the regulatory light chain. During cell migration, the activity of class II myosins may contribute to several processes important for cell translocation, such as regulating the traction applied by the cell to the substrate, promoting directional migration by limiting formation of lamellipodia to the leading edge, and tail retraction. Cell deformation, which is necessary for tumor cell migration through a dense 3-D matrix, such as the neuropil in the brain, has also been shown to depend on the activity of class II myosins, and inhibition of myosin II interfered with glioma cell migration through the matrix [Beadle et al., 2008; Ivkovic et al., 2012]. In addition, class II myosins participate in regulation of cell-cell adhesion, as evidenced by severe adhesion defects and early embryonic lethality in mice lacking myosin IIA [Conti et al., 2004] and by the hydrocephalus observed in mice lacking functional myosin IIB, which has been linked to a cell-cell adhesion defect [Ma et al., 2007]. Since the loss of cell-cell adhesion often represents one of the key steps in EMT, depletion of myosin II from epithelial cells may promote cell dispersion and metastasis. Myosin II isoforms differ in their mechanochemical properties, with myosin IIB having a high duty ratio and myosins IIA and IIC having low duty ratio. Thus, the effects of class II myosin isoforms on cancer cell migration, invasion, and metastasis are likely to be complex and isoform-, context- and cell type-dependent.
Non-muscle myosin IIA
Non-muscle myosin IIA localizes to actin stress fibers [Maupin et al., 1994] and has been implicated in regulation of cell contractility and stress fiber organization in cell-based in vitro assays. Depletion of myosin IIA in carcinoma cell lines using siRNA decreased the number of stress fibers and focal adhesions and increased the rate of cell migration in a wound-healing assay [Sandquist et al., 2006]. Myosin IIA ablation in ES cells resulted in a marked decrease in contractility and an increase in cell migration velocity [Even-Ram et al., 2007]. RNAi experiments show that myosin IIA appears to function as the key negative regulator of cell spreading whereas myosin IIB depletion has little effect on spreading [Cai et al., 2006]. Thus, myosin IIA may act as an inhibitor of cell migration, and, therefore, the loss of myosin IIA may promote cancer metastasis (Figure 2).
The notion of myosin IIA as a tumor progression inhibitor is supported by findings from a recent large-scale RNAi-based in vivo screen for tumor suppressors, in which myosin IIA was identified as a potent tumor suppressor of squamous cell carcinomas [Schramek et al., 2014]. However, intriguingly, myosin IIA function as a tumor suppressor may not be limited to its ability to regulate cell migration; in addition to influencing cell migration and invasion, myosin IIA appeared to affect tumor progression via a novel pathway, which regulates p53 stability and nuclear accumulation [Schramek et al., 2014]. The same study also identified mutations in the MYH9 gene in squamous cell carcinoma samples, including mutations in the conserved regions of the motor domain involved in ATP binding. This finding, together with the observation that myosin II inhibitor blebbistatin disrupted p53-stabilizing activity in tumor cell lines, suggests that myosin II motor activity may be important for its tumor suppressor function. Somatic mutations in the MYH9 gene have also been identified in breast cancer samples [Ellis et al., 2012].
Other studies show that the level of myosin IIA expression may correlate with the increased migration and invasion of tumor cells [Derycke et al., 2011]. Upon EMT, a process that facilitates cell migration, myosin IIA phosphorylation is increased, indicating a role for myosin IIA activity in promoting a motile phenotype [Beach et al., 2011]. Myosin IIA is also regulated by S100A4, a protein associated with metastasis [Li and Bresnick, 2006]. Further studies will be needed to dissect and reconcile the paradoxical findings on the role of myosin IIA (tumor suppressor activity in squamous cell carcinomas vs. invasion promoting activity in some cancer cell lines).
Non-muscle myosin IIB
Localization of non-muscle myosin IIB may depend on the cell type, although it exhibits at least partial colocalization with stress fibers in various cell types. In Xenopus A6 kidney epithelial cells, myosin IIB is enriched at the leading edge [Kelley et al., 1996], while in cultured endothelial cells it is concentrated at the trailing edge of the cell [Kolega, 1998]. Mouse embryonic fibroblasts with decreased expression of myosin IIB display defects in directionality and response to mechanical stimulation, as evidenced by formation of unstable protrusions and rapid changes in cell shape and direction of movement [Lo et al., 2004]. Myosin IIB may be necessary to establish the front to back cell polarity associated with efficient cell migration and to define the position of the cell rear/tail (Figure 2) [Lo et al., 2004; Vicente-Manzanares et al., 2007]. Myosin IIB was able to partially compensate for contractility defects but not migration defects in myosin IIA null fibroblasts [Even-Ram et al., 2007]. Thus, myosins IIA and IIB may have some shared roles in stress fiber assembly and contraction but perform distinct functions during cell migration.
Beach et al. [2011] found that basal-like breast cancer cell lines and myoepithelial (basal) cells in sections of mouse mammary glands expressed primarily myosin IIB while luminal epithelial cells and luminal-like breast cancer cell lines expressed mostly myosins IIA and IIC. It has been proposed that, as a high duty ratio motor, myosin IIB may be well adapted for maintaining prolonged contractile forces, which may be an important property for the contractile myoepithelial cells of the mammary glands [Beach et al., 2011]. Further, when EMT was induced in mammary epithelial cells (NMuMG cell line) using TGF-beta, the authors observed isoform switching: upregulation of myosin IIB expression and downregulation of myosin IIC [Beach et al., 2011]. Depletion of myosin IIB did not affect 2-D migration but reduced invasion through a Transwell filter. Thus, myosin IIB may contribute to cancer cell invasion in a 3-D setting by maintaining cell contractility, which is required for squeezing through the pores in the ECM. In conclusion, myosin IIB is required for normal cell migration and may contribute to tumor invasion.
Non-muscle myosin IIC
While myosin IIB is highly expressed in metastatic basal cancer breast cell lines, such as MDA-MB-231 cells, myosin IIC expression is observed in less aggressive “luminal” breast cell lines [Beach et al 2011]. In fact, a decrease in myosin IIC is observed during EMT, suggesting that it is not important for cancer cell migration and may even act as a negative regulator during metastasis.
Myosin V
There are three class V myosin isoforms in vertebrates (5a, 5b, 5c). Class V myosins are highly processive motors that are well suited for vesicular trafficking and long-range transport of proteins, mRNA, and organelles [Hammer and Sellers, 2012; Hammer and Wagner, 2013]. At least two of the binding partners/cargoes of Myo5a are proteins with known connections to cancer. One is PTEN, a lipid phosphatase that serves as a tumor suppressor by antagonizing PI3-kinase-mediated growth regulatory pathways [van Diepen et al., 2009], and the other is Bmf, a pro-apoptotic protein [Puthalakath et al., 2001]. Both PTEN and Bmf bind to Myo5a tail, with Bmf binding being mediated by the dynein light chain DLC2, which interacts with the tail of Myo5a. Myo5a interaction with PTEN has been shown to contribute to the regulation of neuronal size by modulating PTEN signaling pathways [van Diepen et al., 2009]; this study also indicated that Myo5a and 5b may play redundant roles in PTEN regulation. Whether regulation of PTEN by Myo5 may also contribute to cancer progression remains to be determined.
In case of Bmf-Myo5a complex, sequestration of Bmf to the actin cytoskeleton via binding to Myo5a may prevent apoptosis, allowing cancer cell survival. Expression of a Myo5a tail fragment that disrupts Bmf sequestration resulted in enhanced apoptosis of melanoma cells and decreased melanoma growth in mice, indicating that Myo5a may regulate tumor cell death [Izidoro-Toledo et al., 2013]. Further, Myo5a is highly expressed in metastatic cancer cell lines derived from various tissue types, and its knockdown disrupted tumor cell spreading and migration in vitro and decreased pulmonary metastasis of tumor cells in a chick embryo chorioallantoic membrane model in vivo [Lan et al., 2010]. The expression of Myo5a was also elevated in metastatic colorectal cancers [Lan et al., 2010]. On the other hand, decreased expression of Myo5b isoform was found in gastric cancer [Dong et al., 2012]; the direct connection between Myo5b and cancer has not been established.
Myosin VI
Myo6 is distinguished from other myosin motors by its ability to move towards the actin filament pointed end (minus end), rather than the barbed end (plus end). It has diverse functions within cells, from endocytosis to cell migration [Buss et al., 2004]. Some of Myo6 intracellular functions may rely on its unusual directionality, which allows it to transport endocytic vesicles away from the cell periphery, towards actin filament minus ends, and to tether the plasma membrane at the base of membrane protrusions, such as stereocilia and microvilli. Myo6 localizes to the leading edge, the Golgi apparatus, and endosomal and clathrin-coated vesicles. Myo6 was first shown to be necessary for collective cell migration in the Drosophila border cell migration model [Geisbrecht and Montell, 2002], in which border cells of the ovary undergo collective migration. As discussed in the Introduction, collective cell migration plays an important role in tumor progression and spreading in some cancers [Friedl and Gilmour, 2009], therefore, Myo6 role in collective migration could also contribute to tumor invasion. Indeed, the level of expression of Myo6 has been shown to correlate with the aggressiveness of human ovarian cancers, while antisense inhibition of Myo6 expression in metastatic ovarian cancer cells decreased both their migration in vitro and spreading and metastasis throughout the abdominal cavity in vivo in a mouse model [Yoshida et al., 2004]. Microarray comparison of healthy and cancerous prostate specimens identified Myo6 overexpression in tumor tissue [Dunn et al., 2006], although, unlike the ovarian cancer study where Myo6 expression directly correlated with tumor aggressiveness, the highest level of Myo6 expression was observed in medium-grade prostate tumors, rather than in the most aggressive tumors. Knockdown of Myo6 in prostate cancer cells resulted in defects in the in vitro migration and growth [Dunn et al., 2006].
The precise mechanism leading to Myo6 effects on cell migration is unknown, although it may involve regulation of anterograde trafficking of proteins involved in cell migration, such as EGF receptors, to the leading edge [Chibalina et al., 2010]. Knockdown of Myo6 in prostate cancer cell line LNCaP led to misregulated protein secretion, with no obvious defects in endocytosis [Puri et al., 2010]. Thus, it is possible that Myo6 regulates cancer cell metastasis via its effects on protein transport to the cell surface, which, in turn, may affect cell migration. In addition to its effects on protein trafficking, Myo6 has been shown to influence adherens junctions organization in epithelial cells [Maddugoda et al., 2007]; therefore, Myo6 effects on collective migration could also be explained by its role in cell-cell adhesion.
Myosin VII
Myo7a and Myo7b are dimeric motors that have been implicated in cell adhesion, hearing and vision loss, and melanosome distribution in retinal pigmented epithelium [Maniak, 2001; Williams and Lopes, 2011]. A link between Myo7a polymorphism and malignant melanoma risk has been observed [Fernandez et al., 2009]; it remains to be determined whether it is related to Myo7a role in pigmentation or in regulation of cell-cell adhesion.
Myosin IX
Class IX myosins include a GTPase activating (GAP) domain within their tail region [Muller et al., 1997; Post et al., 1998]. The GAP domain promotes GTP hydrolysis by the small GTPases belonging to the Rho family of signaling proteins, resulting in downregulation of the activity of RhoA, RhoB, and RhoC. Rho GTPases act as important regulators of the actin cytoskeleton, cell adhesion, and cell motility; thus, class IX myosins are likely to play an important role in coordinating cell migration and adhesion via regulation of Rho-mediated pathways. Indeed, Myo9a is required for collective migration of epithelial cells [Omelchenko and Hall, 2012]. As discussed above, collective cell migration plays an important role in progression and metastasis of cancers of epithelial origin [Friedl and Gilmour, 2009]; therefore, similarly to Myo6, Myo9a may prove to serve as a potent promoter of metastasis.
Myo9b shows the highest level of expression in leukocytes [Wirth et al., 1996]. Genetic studies have linked polymorphisms in the MYO9B gene to elevated risk of celiac disease and other types of inflammatory bowel disease [Monsuur et al., 2005; van Bodegraven et al., 2006]. It remains to be determined whether Myo9b variants increase susceptibility to inflammatory bowel disease via their functions in the immune system (elevated inflammatory or autoimmune response) or their role in regulation of intestinal permeability, although Myo9b has already been shown to regulate wound healing response and junctional permeability in intestinal epithelial cells in culture [Chandhoke and Mooseker, 2012]. Intriguingly, genetic variants of MYO9B have been linked to esophageal adenocarcinoma and a precancerous lesion, Barrett’s esophagus [Menke et al., 2012]. As discussed in the Introduction, both altered immune response to tumor antigens and upregulation of pro-inflammatory pathways may contribute to cancer progression. Therefore, Myo9b mutations may affect tumorigenesis either via its role in the immune system functions and regulation of inflammation or via its effects on cell adhesion and migration.
Myosin X
Myo10 is important in phagocytosis and filopodia formation [Sousa and Cheney, 2005]. Myo10 is primarily localized to the tips of filopodia and is proposed to function in filopodia formation and extension either by regulating integrin-dependent adhesion [Zhang et al., 2004] or by stabilizing filopodia through integrin-independent mechanisms [Bohil et al., 2006]. Additionally, Myo10 localizes to lamellipodia at the leading edge of migrating cells [Berg et al., 2000] and is required for macrophage phagocytosis [Cox et al., 2002]. An increase in filopodia number is associated with increased cancer progression, and upregulation of genes that regulate filopodia formation, including MYO10, is associated with the aggressive subtypes of breast cancer [Arjonen et al., 2011; Cao et al., 2014]. Moreover, Myo10 is necessary for breast cancer cell growth, migration, and invasion in vitro and in vivo [Arjonen et al., 2014; Cao et al., 2014; Shibue et al., 2012]. Myo10 lacking its integrin-binding domain is unable to support breast cancer cell migration [Arjonen et al., 2014]. At least two mechanisms linking Myo10-mediated extension of filopodia to breast cancer metastasis may be envisioned, and the two might act in concert to promote tumor proliferation and invasion. One mechanism is based on the direct contribution of filopodia to cell invasion and migration [Arjonen et al., 2014] while the other involves formation of specialized signaling complexes at the tips of filopodia-like protrusions [Shibue et al., 2012]. In addition to its key role in filopodia extension, Myo10 has also been shown to contribute to formation of invadosomes/invadopodia, which may facilitate cancer cell metastasis and invasion [Cao et al., 2014; Schoumacher et al., 2010]. Moreover, Myo10 has been shown to interact with calmodulin-like protein (CLP), which can serve as Myo10 light chain and whose expression is downregulated in tumors [Rogers and Strehler, 2001].
Myosin XVIII
Myo18b was identified in the course of a search for a lung cancer tumor suppressor gene. Many human lung cancers are characterized by allelic loss on chromosome 22. Mapping the region altered in lung cancer led to isolation of a novel cDNA [Nishioka et al., 2002] encoding a myosin-like protein. The protein encoded by this gene was named myosin XVIIIb [Berg et al., 2001]. MYO18B gene, located at chromosome 22q12.1, was frequently mutated or hypermethylated in lung cancers and expression of Myo18b in lung cancer cells suppressed anchorage-independent growth [Nishioka et al., 2002]. The suppression of anchorage-independent growth was enhanced by co-expression of Myo18b binding partner, adaptor protein Homer2 [Ajima et al., 2007]. MYO18B mutations and epigenetic modifications have also been detected in ovarian and colorectal cancer [Nakano et al., 2005; Yanaihara et al., 2004].
Myo18b has been found to interact with the Golgi-associated protein GOLPH3; together, Myo18b and GOLPH3 contribute to Golgi vesiculation and dispersal [Dippold et al., 2009] and cooperate in promoting the dispersal of the Golgi in response to DNA damage [Farber-Katz et al., 2014]. Overexpression of GOLPH3 promotes cell survival after DNA damage [Farber-Katz et al., 2014], and this observation is in agreement with the finding that GOLPH3 can act as an oncogene [Scott et al., 2009]. Since Myo18b and GOLPH3 play cooperating roles in DNA damage response, it could be expected that genes encoding both of these proteins would act as oncogenes, promoting tumor survival in spite of DNA damage. Thus, it is somewhat puzzling that analysis of human cancers indicates that while GOLPH3 appears to act as an oncogene, Myo18b may serve as a tumor suppressor. The cell biological basis for this seeming contradiction remains to be determined.
In addition to MYO18B, chromosome 22 also contains the MYH9 gene at q13.1; thus, at least two myosins that act as tumor suppressors are located in the 22q12–22q13 chromosomal region. Another member of the same class, MYO18A, has been found to interact with important regulators and effectors of Rho GTPase signaling (PAK2 kinase and βPIX/GIT1 complex) and may regulate cell migration by modulating GIT1 and PAK2 localization [Hsu et al., 2010]. Interestingly, a fusion between MYO18A gene and FGFR1 gene has been found in a patient with myeloproliferative/myelodisplastic disease (a chronic myelogenous leukemia-like disease that likely results from constitutive activation of FGF receptor kinase) [Walz et al., 2005].
Members of myosin class 18 possess structural similarity to myosin II and may share a common evolutionary origin with myosin II [Foth et al., 2006]. Like myosin II, myo18 contains extensive coiled-coil regions in the C-terminal tail domain, which may allow Myo18 to dimerize or assemble into filaments. At the same time, members of this myosin class have some distinctive functional features, such as the lack of ATP hydrolysis activity and the presence of both ATP-dependent and ATP-independent actin-binding sites in some isoforms [Guzik-Lendrum et al., 2011; Taft et al., 2013]. Based on these properties, it appears likely that Myo18 may serve as a tethering protein or an actin cross-linker, rather than an actual motor protein.
Conclusions and Perspectives
Non-muscle myosins have been implicated in cancer progression through their roles in cell migration and invasion as well as their tumor suppressor functions. Genetic and epigenetic modifications of genes encoding myosin heavy chains are found in many types of cancer. In some cases, alterations in myosin expression serve as predictors of patient survival; thus, some members of the myosin superfamily may have potential use as cancer biomarkers. In addition, myosins represent attractive targets for drug development since small molecule inhibitors or activators of myosin activity can be readily screened using ATPase activity assays. Indeed, a number of small molecule myosin inhibitors and activators have been identified [Bond et al., 2013]. Further identification of compounds suitable for potential in vivo use, as well as studies in cancer models to test how myosin activation or inhibition affects tumor progression and metastasis, will be needed in order to harness the full therapeutic potential of myosin-targeting drug candidates.
Acknowledgements
Work reported in this publication was supported by the NIDDK of the National Institutes of Health under award number R01DK083345. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We would like to thank Drs. Tatiana Omelchenko and David Pruyne for the critical discussion of the manuscript and the anonymous reviewers for their valuable suggestions.
Abbreviations
- EMT
epithelial-mesenchymal transition
- IHC
immunohistochemistry
- PH
pleckstrin homology
- TH
tail homology
REFERENCES
- Adams JM, Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26(9):1324–1327. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Cuenca R, Juanes-Garcia A, Vicente-Manzanares M. Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell Mol Life Sci. 2014;71(3):479–492. doi: 10.1007/s00018-013-1439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajima R, Kajiya K, Inoue T, Tani M, Shiraishi-Yamaguchi Y, Maeda M, Segawa T, Furuichi T, Sutoh K, Yokota J. HOMER2 binds MYO18B and enhances its activity to suppress anchorage independent growth. Biochem Biophys Res Commun. 2007;356(4):851–856. doi: 10.1016/j.bbrc.2007.03.060. [DOI] [PubMed] [Google Scholar]
- Almagro S, Durmort C, Chervin-Petinot A, Heyraud S, Dubois M, Lambert O, Maillefaud C, Hewat E, Schaal JP, Huber P, et al. The motor protein myosin-X transports VE-cadherin along filopodia to allow the formation of early endothelial cell-cell contacts. Mol Cell Biol. 2010;30(7):1703–1717. doi: 10.1128/MCB.01226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida CG, Yamada A, Tenza D, Louvard D, Raposo G, Coudrier E. Myosin 1b promotes the formation of post-Golgi carriers by regulating actin assembly and membrane remodelling at the trans-Golgi network. Nat Cell Biol. 2011;13(7):779–789. doi: 10.1038/ncb2262. [DOI] [PubMed] [Google Scholar]
- Arjonen A, Kaukonen R, Ivaska J. Filopodia and adhesion in cancer cell motility. Cell Adh Migr. 2011;5(5):421–430. doi: 10.4161/cam.5.5.17723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjonen A, Kaukonen R, Mattila E, Rouhi P, Hognas G, Sihto H, Miller BW, Morton JP, Bucher E, Taimen P, et al. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Invest. 2014 doi: 10.1172/JCI67280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach JR, Hussey GS, Miller TE, Chaudhury A, Patel P, Monslow J, Zheng Q, Keri RA, Reizes O, Bresnick AR, et al. Myosin II isoform switching mediates invasiveness after TGF-beta-induced epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2011;108(44):17991–17996. doi: 10.1073/pnas.1106499108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell. 2008;19(8):3357–3368. doi: 10.1091/mbc.E08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associates with regions of dynamic actin. J Cell Sci. 2000;113(Pt 19):3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- Berg JS, Powell BC, Cheney RE. A millennial myosin census. Mol Biol Cell. 2001;12(4):780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardes de Jesus B, Blasco MA. Telomerase at the intersection of cancer and aging. Trends Genet. 2013;29(9):513–520. doi: 10.1016/j.tig.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J, Chase SE, Pellenz CD, Kurihara H, Fanning AS, Krendel M. Myosin 1e is a component of the glomerular slit diaphragm complex that regulates actin reorganization during cell-cell contact formation in podocytes. Am J Physiol Renal Physiol. 2013;305(4):F532–F544. doi: 10.1152/ajprenal.00223.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington N, Wang A, Mao J, Adelstein RS, Sellers JR. Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem. 2013;288(46):33398–33410. doi: 10.1074/jbc.M113.499848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci U S A. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond LM, Tumbarello DA, Kendrick-Jones J, Buss F. Small-molecule inhibitors of myosin proteins. Future Med Chem. 2013;5(1):41–52. doi: 10.4155/fmc.12.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzeska H, Hwang KJ, Korn ED. Acanthamoeba myosin IC colocalizes with phosphatidylinositol 4,5-bisphosphate at the plasma membrane due to the high concentration of negative charge. J Biol Chem. 2008;283(46):32014–32023. doi: 10.1074/jbc.M804828200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- Cai Y, Biais N, Giannone G, Tanase M, Jiang G, Hofman JM, Wiggins CH, Silberzan P, Buguin A, Ladoux B, et al. Nonmuscle myosin IIA-dependent force inhibits cell spreading and drives F-actin flow. Biophys J. 2006;91(10):3907–3920. doi: 10.1529/biophysj.106.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11(4):237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Chen J, Zhang X, Zhai Y, Qing X, Xing W, Zhang L, Malik YS, Yu H, Zhu X. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br J Cancer. 2014 doi: 10.1038/bjc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervero P, Himmel M, Kruger M, Linder S. Proteomic analysis of podosome fractions from macrophages reveals similarities to spreading initiation centres. Eur J Cell Biol. 2012 doi: 10.1016/j.ejcb.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Chandhoke SK, Mooseker MS. A role for myosin IXb, a motor-RhoGAP chimera, in epithelial wound healing and tight junction regulation. Mol Biol Cell. 2012;23(13):2468–2480. doi: 10.1091/mbc.E11-09-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Grassart A, Drubin DG. Myosin 1E coordinates actin assembly and cargo trafficking during clathrin-mediated endocytosis. Mol Biol Cell. 2012;23(15):2891–2904. doi: 10.1091/mbc.E11-04-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibalina MV, Poliakov A, Kendrick-Jones J, Buss F. Myosin VI and optineurin are required for polarized EGFR delivery and directed migration. Traffic. 2010;11(10):1290–1303. doi: 10.1111/j.1600-0854.2010.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti MA, Even-Ram S, Liu C, Yamada KM, Adelstein RS. Defects in cell adhesion and the visceral endoderm following ablation of nonmuscle myosin heavy chain II-A in mice. J Biol Chem. 2004;279(40):41263–41266. doi: 10.1074/jbc.C400352200. [DOI] [PubMed] [Google Scholar]
- Cox D, Berg JS, Cammer M, Chinegwundoh JO, Dale BM, Cheney RE, Greenberg S. Myosin X is a downstream effector of PI(3)K during phagocytosis. Nat Cell Biol. 2002;4(7):469–477. doi: 10.1038/ncb805. [DOI] [PubMed] [Google Scholar]
- Derycke L, Stove C, Vercoutter-Edouart AS, De Wever O, Dolle L, Colpaert N, Depypere H, Michalski JC, Bracke M. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int J Dev Biol. 2011;55(7–9):835–840. doi: 10.1387/ijdb.113336ld. [DOI] [PubMed] [Google Scholar]
- Dippold HC, Ng MM, Farber-Katz SE, Lee SK, Kerr ML, Peterman MC, Sim R, Wiharto PA, Galbraith KA, Madhavarapu S, et al. GOLPH3 bridges phosphatidylinositol-4-phosphate and actomyosin to stretch and shape the Golgi to promote budding. Cell. 2009;139(2):337–351. doi: 10.1016/j.cell.2009.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong W, Chen X, Chen P, Yue D, Zhu L, Fan Q. Inactivation of MYO5B promotes invasion and motility in gastric cancer cells. Dig Dis Sci. 2012;57(5):1247–1252. doi: 10.1007/s10620-011-1989-z. [DOI] [PubMed] [Google Scholar]
- Duhoux FP, Ameye G, Libouton JM, Bahloula K, Iossifidis S, Chantrain CF, Demoulin JB, Poirel HA. The t(11;19)(q23;p13) fusing MLL with MYO1F is recurrent in infant acute myeloid leukemias. Leuk Res. 2011;35(9):e171–e172. doi: 10.1016/j.leukres.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Dunn TA, Chen S, Faith DA, Hicks JL, Platz EA, Chen Y, Ewing CM, Sauvageot J, Isaacs WB, De Marzo AM, et al. A novel role of myosin VI in human prostate cancer. Am J Pathol. 2006;169(5):1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert MA, Yang J. Targeting invadopodia to block breast cancer metastasis. Oncotarget. 2011;2(7):562–568. doi: 10.18632/oncotarget.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486(7403):353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even-Ram S, Doyle AD, Conti MA, Matsumoto K, Adelstein RS, Yamada KM. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat Cell Biol. 2007;9(3):299–309. doi: 10.1038/ncb1540. [DOI] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14(4):570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber MJ, Rizaldy R, Hildebrand JD. Shroom2 regulates contractility to control endothelial morphogenesis. Mol Biol Cell. 2011;22(6):795–805. doi: 10.1091/mbc.E10-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farber-Katz SE, Dippold HC, Buschman MD, Peterman MC, Xing M, Noakes CJ, Tat J, Ng MM, Rahajeng J, Cowan DM, et al. DNA damage triggers Golgi dispersal via DNA-PK and GOLPH3. Cell. 2014;156(3):413–427. doi: 10.1016/j.cell.2013.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeser EA, Ignacio CM, Krendel M, Ostap EM. Myo1e binds anionic phospholipids with high affinity. Biochemistry. 2010;49(43):9353–9360. doi: 10.1021/bi1012657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez LP, Milne RL, Pita G, Floristan U, Sendagorta E, Feito M, Aviles JA, Martin-Gonzalez M, Lazaro P, Benitez J, et al. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol. 2009;18(7):634–642. doi: 10.1111/j.1600-0625.2009.00846.x. [DOI] [PubMed] [Google Scholar]
- Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19(3):260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proc Natl Acad Sci U S A. 2006;103(10):3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- Friedl P, Locker J, Sahai E, Segall JE. Classifying collective cancer cell invasion. Nat Cell Biol. 2012;14(8):777–783. doi: 10.1038/ncb2548. [DOI] [PubMed] [Google Scholar]
- Garcia A, Coudrier E, Carboni J, Anderson J, Vandekerkhove J, Mooseker M, Louvard D, Arpin M. Partial deduced sequence of the 110-kD-calmodulin complex of the avian intestinal microvillus shows that this mechanoenzyme is a member of the myosin I family. J Cell Biol. 1989;109(6 Pt 1):2895–2903. doi: 10.1083/jcb.109.6.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbrecht ER, Montell DJ. Myosin VI is required for E-cadherin-mediated border cell migration. Nat Cell Biol. 2002;4(8):616–620. doi: 10.1038/ncb830. [DOI] [PubMed] [Google Scholar]
- Greenberg MJ, Ostap EM. Regulation and control of myosin-I by the motor and light chain-binding domains. Trends Cell Biol. 2013;23(2):81–89. doi: 10.1016/j.tcb.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzik-Lendrum S, Nagy A, Takagi Y, Houdusse A, Sellers JR. Drosophila melanogaster myosin-18 represents a highly divergent motor with actin tethering properties. J Biol Chem. 2011;286(24):21755–21766. doi: 10.1074/jbc.M111.218669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallett RM, Dvorkin-Gheva A, Bane A, Hassell JA. A gene signature for predicting outcome in patients with basal-like breast cancer. Sci Rep. 2012;2:227. doi: 10.1038/srep00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat Rev Mol Cell Biol. 2012;13(1):13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- Hammer JA, 3rd, Wagner W. Functions of class V myosins in neurons. J Biol Chem. 2013;288(40):28428–28434. doi: 10.1074/jbc.R113.514497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hanel W, Moll UM. Links between mutant p53 and genomic instability. J Cell Biochem. 2012;113(2):433–439. doi: 10.1002/jcb.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman MA, Spudich JA. The myosin superfamily at a glance. J Cell Sci. 2012;125(Pt 7):1627–1632. doi: 10.1242/jcs.094300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodge T, Cope MJ. A myosin family tree. J Cell Sci. 2000;113(Pt 19):3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- Hokanson DE, Laakso JM, Lin T, Sept D, Ostap EM. Myo1c binds phosphoinositides through a putative pleckstrin homology domain. Mol Biol Cell. 2006;17(11):4856–65. doi: 10.1091/mbc.E06-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu RM, Tsai MH, Hsieh YJ, Lyu PC, Yu JS. Identification of MYO18A as a novel interacting partner of the PAK2/betaPIX/GIT1 complex and its potential function in modulating epithelial cell migration. Mol Biol Cell. 2010;21(2):287–301. doi: 10.1091/mbc.E09-03-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Muthuswamy SK. Polarity protein alterations in carcinoma: a focus on emerging roles for polarity regulators. Curr Opin Genet Dev. 2010;20(1):41–50. doi: 10.1016/j.gde.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivkovic S, Beadle C, Noticewala S, Massey SC, Swanson KR, Toro LN, Bresnick AR, Canoll P, Rosenfeld SS. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol Biol Cell. 2012;23(4):533–542. doi: 10.1091/mbc.E11-01-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izidoro-Toledo TC, Borges AC, Araujo DD, Mazzi DP, Nascimento Junior FO, Sousa JF, Alves CP, Paiva AP, Trindade DM, Patussi EV, et al. A myosin-Va tail fragment sequesters dynein light chains leading to apoptosis in melanoma cells. Cell Death Dis. 2013;4:e547. doi: 10.1038/cddis.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley CA, Sellers JR, Gard DL, Bui D, Adelstein RS, Baines IC. Xenopus nonmuscle myosin heavy chain isoforms have different subcellular localizations and enzymatic activities. J Cell Biol. 1996;134(3):675–687. doi: 10.1083/jcb.134.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber ML, Cheney RE. Myosin-X: a MyTH-FERM myosin at the tips of filopodia. J Cell Sci. 2011;124(Pt 22):3733–3741. doi: 10.1242/jcs.023549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim R, Emi M, Tanabe K. Cancer immunoediting from immune surveillance to immune escape. Immunology. 2007;121(1):1–14. doi: 10.1111/j.1365-2567.2007.02587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SV, Mehal WZ, Dong X, Heinrich V, Pypaert M, Mellman I, Dembo M, Mooseker MS, Wu D, Flavell RA. Modulation of cell adhesion and motility in the immune system by Myo1f. Science. 2006;314(5796):136–139. doi: 10.1126/science.1131920. [DOI] [PubMed] [Google Scholar]
- Kolega J. Cytoplasmic dynamics of myosin IIA and IIB: spatial 'sorting' of isoforms in locomoting cells. J Cell Sci. 1998;111(Pt 15):2085–2095. doi: 10.1242/jcs.111.15.2085. [DOI] [PubMed] [Google Scholar]
- Kravtsov DV, Caputo C, Collaco A, Hoekstra N, Egan ME, Mooseker MS, Ameen NA. Myosin Ia is required for CFTR brush border membrane trafficking and ion transport in the mouse small intestine. Traffic. 2012;13(8):1072–1082. doi: 10.1111/j.1600-0854.2012.01368.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Kim SV, Willinger T, Wang T, Kashgarian M, Flavell RA, Mooseker MS. Disruption of Myosin 1e promotes podocyte injury. J Am Soc Nephrol. 2009;20(1):86–94. doi: 10.1681/ASN.2007111172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Mooseker MS. Myosins: tails (and heads) of functional diversity. Physiology (Bethesda) 2005;20:239–251. doi: 10.1152/physiol.00014.2005. [DOI] [PubMed] [Google Scholar]
- Krendel M, Osterweil EK, Mooseker MS. Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett. 2007;581(4):644–650. doi: 10.1016/j.febslet.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Han H, Zuo H, Chen Z, Du Y, Zhao W, Gu J, Zhang Z. Upregulation of myosin Va by Snail is involved in cancer cell migration and metastasis. Int J Cancer. 2010;126(1):53–64. doi: 10.1002/ijc.24641. [DOI] [PubMed] [Google Scholar]
- Li ZH, Bresnick AR. The S100A4 metastasis factor regulates cellular motility via a direct interaction with myosin-IIA. Cancer Res. 2006;66(10):5173–5180. doi: 10.1158/0008-5472.CAN-05-3087. [DOI] [PubMed] [Google Scholar]
- Linder S, Wiesner C, Himmel M. Degrading devices: invadosomes in proteolytic cell invasion. Annu Rev Cell Dev Biol. 2011;27:185–211. doi: 10.1146/annurev-cellbio-092910-154216. [DOI] [PubMed] [Google Scholar]
- Lo CM, Buxton DB, Chua GC, Dembo M, Adelstein RS, Wang YL. Nonmuscle myosin IIb is involved in the guidance of fibroblast migration. Mol Biol Cell. 2004;15(3):982–989. doi: 10.1091/mbc.E03-06-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Bao J, Adelstein RS. Loss of cell adhesion causes hydrocephalus in nonmuscle myosin II-B-ablated and mutated mice. Mol Biol Cell. 2007;18(6):2305–2312. doi: 10.1091/mbc.E07-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddugoda MP, Crampton MS, Shewan AM, Yap AS. Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell cell contacts in mammalian epithelial cells. J Cell Biol. 2007;178(3):529–540. doi: 10.1083/jcb.200612042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniak M. Cell adhesion: ushering in a new understanding of myosin VII. Curr Biol. 2001;11(8):R315–R317. doi: 10.1016/s0960-9822(01)00173-7. [DOI] [PubMed] [Google Scholar]
- Maravillas-Montero JL, Santos-Argumedo L. The myosin family: unconventional roles of actin-dependent molecular motors in immune cells. J Leukoc Biol. 2012;91(1):35–46. doi: 10.1189/jlb.0711335. [DOI] [PubMed] [Google Scholar]
- Martin ME, Milne TA, Bloyer S, Galoian K, Shen W, Gibbs D, Brock HW, Slany R, Hess JL. Dimerization of MLL fusion proteins immortalizes hematopoietic cells. Cancer Cell. 2003;4(3):197–207. doi: 10.1016/s1535-6108(03)00214-9. [DOI] [PubMed] [Google Scholar]
- Maupin P, Phillips CL, Adelstein RS, Pollard TD. Differential localization of myosin-II isozymes in human cultured cells and blood cells. J Cell Sci. 1994;107(Pt 11):3077–3090. doi: 10.1242/jcs.107.11.3077. [DOI] [PubMed] [Google Scholar]
- Mazerik JN, Tyska MJ. Myosin-1A targets to microvilli using multiple membrane binding motifs in the tail homology 1 (TH1) domain. J Biol Chem. 2012;287(16):13104–13115. doi: 10.1074/jbc.M111.336313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini R, Dopeso H, Mateo-Lozano S, Chang W, Rodrigues P, Bazzocco S, Alazzouzi H, Landolfi S, Hernandez-Losa J, Andretta E, et al. Brush border myosin Ia has tumor suppressor activity in the intestine. Proc Natl Acad Sci U S A. 2012;109(5):1530–1535. doi: 10.1073/pnas.1108411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini R, Rodrigues P, Bazzocco S, Dopeso H, Ferreira AM, Mateo-Lozano S, Andretta E, Woerner SM, Alazzouzi H, Landolfi S, et al. Brush border myosin Ia inactivation in gastric but not endometrial tumors. Int J Cancer. 2013;132(8):1790–1799. doi: 10.1002/ijc.27856. [DOI] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Myosin-1a powers the sliding of apical membrane along microvillar actin bundles. J Cell Biol. 2007;177(4):671–681. doi: 10.1083/jcb.200701144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Tyska MJ. Leveraging the membrane - cytoskeleton interface with myosin-1. Trends Cell Biol. 2010;20(7):418–426. doi: 10.1016/j.tcb.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele C, Iatropoulos P, Donadelli R, Calabria A, Maranta R, Cassis P, Buelli S, Tomasoni S, Piras R, Krendel M, et al. MYO1E mutations and childhood familial focal segmental glomerulosclerosis. N Engl J Med. 2011;365(4):295–306. doi: 10.1056/NEJMoa1101273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menke V, Van Zoest KP, Moons LM, Pot RG, Siersema PD, Kuipers EJ, Kusters JG. Myo9B is associated with an increased risk of Barrett’s esophagus and esophageal adenocarcinoma. Scand J Gastroenterol. 2012;47(12):1422–1428. doi: 10.3109/00365521.2012.722673. [DOI] [PubMed] [Google Scholar]
- Michael M, Yap AS. The regulation and functional impact of actin assembly at cadherin cell-cell adhesions. Semin Cell Dev Biol. 2013;24(4):298–307. doi: 10.1016/j.semcdb.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Mierke CT. Physical break-down of the classical view on cancer cell invasion and metastasis. Eur J Cell Biol. 2013;92(3):89–104. doi: 10.1016/j.ejcb.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Monsuur AJ, de Bakker PI, Alizadeh BZ, Zhernakova A, Bevova MR, Strengman E, Franke L, van’t Slot R, van Belzen MJ, Lavrijsen IC, et al. Myosin IXB variant increases the risk of celiac disease and points toward a primary intestinal barrier defect. Nat Genet. 2005;37(12):1341–1344. doi: 10.1038/ng1680. [DOI] [PubMed] [Google Scholar]
- Mooseker MS, Coleman TR. The 110-kD protein-calmodulin complex of the intestinal microvillus (brush border myosin I) is a mechanoenzyme. J Cell Biol. 1989;108(6):2395–2400. doi: 10.1083/jcb.108.6.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller RT, Honnert U, Reinhard J, Bahler M. The rat myosin myr 5 is a GTPase-activating protein for Rho in vivo: essential role of arginine 1695. Mol Biol Cell. 1997;8(10):2039–2053. doi: 10.1091/mbc.8.10.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Tani M, Nishioka M, Kohno T, Otsuka A, Ohwada S, Yokota J. Genetic and epigenetic alterations of the candidate tumor-suppressor gene MYO18B, on chromosome arm 22q, in colorectal cancer. Genes Chromosomes Cancer. 2005;43(2):162–171. doi: 10.1002/gcc.20180. [DOI] [PubMed] [Google Scholar]
- Naumov GN, Akslen LA, Folkman J. Role of angiogenesis in human tumor dormancy: animal models of the angiogenic switch. Cell Cycle. 2006;5(16):1779–1787. doi: 10.4161/cc.5.16.3018. [DOI] [PubMed] [Google Scholar]
- Nishioka M, Kohno T, Tani M, Yanaihara N, Tomizawa Y, Otsuka A, Sasaki S, Kobayashi K, Niki T, Maeshima A, et al. MYO18B, a candidate tumor suppressor gene at chromosome 22q12.1, deleted, mutated, and methylated in human lung cancer. Proc Natl Acad Sci U S A. 2002;99(19):12269–12274. doi: 10.1073/pnas.192445899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell CB, Tyska MJ, Mooseker MS. Myosin at work: motor adaptations for a variety of cellular functions. Biochim Biophys Acta. 2007;1773(5):615–630. doi: 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Odronitz F, Kollmar M. Drawing the tree of eukaryotic life based on the analysis of 2,269 manually annotated myosins from 328 species. Genome Biol. 2007;8(9):R196. doi: 10.1186/gb-2007-8-9-r196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2(1):a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko T, Hall A. Myosin-IXA regulates collective epithelial cell migration by targeting RhoGAP activity to cell-cell junctions. Curr Biol. 2012;22(4):278–288. doi: 10.1016/j.cub.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostap EM. Tropomyosins as discriminators of myosin function. Adv Exp Med Biol. 2008;644:273–282. doi: 10.1007/978-0-387-85766-4_20. [DOI] [PubMed] [Google Scholar]
- Ouderkirk JL, Krendel M. Myosin 1e is a component of the invadosome core that contributes to regulation of invadosome dynamics. Experimental Cell Research. 2014 doi: 10.1016/j.yexcr.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patino-Lopez G, Aravind L, Dong X, Kruhlak MJ, Ostap EM, Shaw S. Myosin 1G is an abundant class I myosin in lymphocytes whose localization at the plasma membrane depends on its ancient divergent pleckstrin homology (PH) domain (Myo1PH) J Biol Chem. 2010;285(12):8675–8686. doi: 10.1074/jbc.M109.086959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pi X, Ren R, Kelley R, Zhang C, Moser M, Bohil AB, Divito M, Cheney RE, Patterson C. Sequential roles for myosin-X in BMP6-dependent filopodial extension, migration, and activation of BMP receptors. J Cell Biol. 2007;179(7):1569–1582. doi: 10.1083/jcb.200704010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard LW, Lord M. Getting myosin-V on the right track: tropomyosin sorts transport in yeast. Bioarchitecture. 2014;4(1):35–38. doi: 10.4161/bioa.28204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post PL, Bokoch GM, Mooseker MS. Human myosin-IXb is a mechanochemically active motor and a GAP for rho. J Cell Sci. 1998;111(Pt 7):941–950. doi: 10.1242/jcs.111.7.941. [DOI] [PubMed] [Google Scholar]
- Pruyne D. Tropomyosin function in yeast. Adv Exp Med Biol. 2008;644:168–186. doi: 10.1007/978-0-387-85766-4_14. [DOI] [PubMed] [Google Scholar]
- Puri C, Chibalina MV, Arden SD, Kruppa AJ, Kendrick-Jones J, Buss F. Overexpression of myosin VI in prostate cancer cells enhances PSA and VEGF secretion, but has no effect on endocytosis. Oncogene. 2010;29(2):188–200. doi: 10.1038/onc.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puthalakath H, Villunger A, O'Reilly LA, Beaumont JG, Coultas L, Cheney RE, Huang DC, Strasser A. Bmf: a proapoptotic BH3-only protein regulated by interaction with the myosin V actin motor complex, activated by anoikis. Science. 2001;293(5536):1829–1832. doi: 10.1126/science.1062257. [DOI] [PubMed] [Google Scholar]
- Rakha EA, Reis-Filho JS, Ellis IO. Basal-like breast cancer: a critical review. J Clin Oncol. 2008;26(15):2568–2581. doi: 10.1200/JCO.2007.13.1748. [DOI] [PubMed] [Google Scholar]
- Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436(7054):1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- Rogers MS, Strehler EE. The tumor-sensitive calmodulin-like protein is a specific light chain of human unconventional myosin X. J Biol Chem. 2001;276(15):12182–12189. doi: 10.1074/jbc.M010056200. [DOI] [PubMed] [Google Scholar]
- Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18(9):1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahai E. Mechanisms of cancer cell invasion. Curr Opin Genet Dev. 2005;15(1):87–96. doi: 10.1016/j.gde.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Sandquist JC, Swenson KI, Demali KA, Burridge K, Means AR. Rho kinase differentially regulates phosphorylation of nonmuscle myosin II isoforms A and B during cell rounding and migration. J Biol Chem. 2006;281(47):35873–35883. doi: 10.1074/jbc.M605343200. [DOI] [PubMed] [Google Scholar]
- Schiller HB, Friedel CC, Boulegue C, Fassler R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 2011;12(3):259–266. doi: 10.1038/embor.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189(3):541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 2014;343(6168):309–313. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KL, Kabbarah O, Liang MC, Ivanova E, Anagnostou V, Wu J, Dhakal S, Wu M, Chen S, Feinberg T, et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature. 2009;459(7250):1085–1090. doi: 10.1038/nature08109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, Brooks MW, Inan MF, Reinhardt F, Weinberg RA. The outgrowth of micrometastases is enabled by the formation of filopodium-like protrusions. Cancer Discov. 2012;2(8):706–721. doi: 10.1158/2159-8290.CD-11-0239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibony-Benyamini H, Gil-Henn H. Invadopodia: the leading force. Eur J Cell Biol. 2012;91(11–12):896–901. doi: 10.1016/j.ejcb.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Skowron JF, Bement WM, Mooseker MS. Human brush border myosin-I and myosin-Ic expression in human intestine and Caco-2BBe cells. Cell Motil Cytoskeleton. 1998;41(4):308–324. doi: 10.1002/(SICI)1097-0169(1998)41:4<308::AID-CM4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Slany RK. The molecular biology of mixed lineage leukemia. Haematologica. 2009;94(7):984–993. doi: 10.3324/haematol.2008.002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa AD, Cheney RE. Myosin-X: a molecular motor at the cell’s fingertips. Trends Cell Biol. 2005;15(10):533–539. doi: 10.1016/j.tcb.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Stark BC, Sladewski TE, Pollard LW, Lord M. Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol Biol Cell. 2010;21(6):989–1000. doi: 10.1091/mbc.E09-10-0852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szent-Gyorgyi AG. The early history of the biochemistry of muscle contraction. J Gen Physiol. 2004;123(6):631–641. doi: 10.1085/jgp.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft MH, Behrmann E, Munske-Weidemann LC, Thiel C, Raunser S, Manstein DJ. Functional characterization of human myosin-18A and its interaction with F-actin and GOLPH3. J Biol Chem. 2013;288(42):30029–30041. doi: 10.1074/jbc.M113.497180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki T, Akiyama M, Saito S, Ono R, Taniwaki M, Kato Y, Yuza Y, Eto Y, Hayashi Y. The MYO1F, unconventional myosin type 1F, gene is fused to MLL in infant acute monocytic leukemia with a complex translocation involving chromosomes 7, 11, 19 and 22. Oncogene. 2005;24(33):5191–5197. doi: 10.1038/sj.onc.1208711. [DOI] [PubMed] [Google Scholar]
- Teng MW, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: an equilibrium with cancer. J Leukoc Biol. 2008;84(4):988–993. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- Tiwari A, Jung JJ, Inamdar SM, Nihalani D, Choudhury A. The myosin motor Myo1c is required for VEGFR2 delivery to the cell surface and for angiogenic signaling. Am J Physiol Heart Circ Physiol. 2013;304(5):H687–H696. doi: 10.1152/ajpheart.00744.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Schevzov G, Hotulainen P, Naumanen P, Martin C, Gunning PW, Lappalainen P. A molecular pathway for myosin II recruitment to stress fibers. Curr Biol. 2011;21(7):539–550. doi: 10.1016/j.cub.2011.03.007. [DOI] [PubMed] [Google Scholar]
- Tokuo H, Coluccio LM. Myosin-1c regulates the dynamic stability of E-cadherin-based cell-cell contacts in polarized Madin-Darby canine kidney cells. Mol Biol Cell. 2013;24(18):2820–2833. doi: 10.1091/mbc.E12-12-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mackey AT, Huang JD, Copeland NG, Jenkins NA, Mooseker MS. Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell. 2005;16(5):2443–2457. doi: 10.1091/mbc.E04-12-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mooseker MS. MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys J. 2002;82(4):1869–1883. doi: 10.1016/S0006-3495(02)75537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mooseker MS. A role for myosin-1A in the localization of a brush border disaccharidase. J Cell Biol. 2004;165(3):395–405. doi: 10.1083/jcb.200310031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Warshaw DM. The myosin power stroke. Cell Motil Cytoskeleton. 2002;51(1):1–15. doi: 10.1002/cm.10014. [DOI] [PubMed] [Google Scholar]
- van Bodegraven AA, Curley CR, Hunt KA, Monsuur AJ, Linskens RK, Onnie CM, Crusius JB, Annese V, Latiano A, Silverberg MS, et al. Genetic variation in myosin IXB is associated with ulcerative colitis. Gastroenterology. 2006;131(6):1768–1774. doi: 10.1053/j.gastro.2006.09.011. [DOI] [PubMed] [Google Scholar]
- van Diepen MT, Parsons M, Downes CP, Leslie NR, Hindges R, Eickholt BJ. MyosinV controls PTEN function and neuronal cell size. Nat Cell Biol. 2009;11(10):1191–1196. doi: 10.1038/ncb1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10(11):778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M, Zareno J, Whitmore L, Choi CK, Horwitz AF. Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J Cell Biol. 2007;176(5):573–580. doi: 10.1083/jcb.200612043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vindin H, Gunning P. Cytoskeletal tropomyosins: choreographers of actin filament functional diversity. J Muscle Res Cell Motil. 2013;34(3–4):261–274. doi: 10.1007/s10974-013-9355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldner MJ, Neurath MF. Colitis-associated cancer: the role of T cells in tumor development. Semin Immunopathol. 2009;31(2):249–256. doi: 10.1007/s00281-009-0161-8. [DOI] [PubMed] [Google Scholar]
- Walz C, Chase A, Schoch C, Weisser A, Schlegel F, Hochhaus A, Fuchs R, Schmitt-Graff A, Hehlmann R, Cross NC, et al. The t(8;17)(p11;q23) in the 8p11 myeloproliferative syndrome fuses MYO18A to FGFR1. Leukemia. 2005;19(6):1005–1009. doi: 10.1038/sj.leu.2403712. [DOI] [PubMed] [Google Scholar]
- Wang A, Ma X, Conti MA, Adelstein RS. Distinct and redundant roles of the non-muscle myosin II isoforms and functional domains. Biochem Soc Trans. 2011;39(5):1131–1135. doi: 10.1042/BST0391131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CL, Coluccio LM. New insights into the regulation of the actin cytoskeleton by tropomyosin. Int Rev Cell Mol Biol. 2010;281:91–128. doi: 10.1016/S1937-6448(10)81003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DS, Lopes VS. The many different cellular functions of MYO7A in the retina. Biochem Soc Trans. 2011;39(5):1207–1210. doi: 10.1042/BST0391207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth JA, Jensen KA, Post PL, Bement WM, Mooseker MS. Human myosin-IXb, an unconventional myosin with a chimerin-like rho/rac GTPase-activating protein domain in its tail. J Cell Sci. 1996;109(Pt 3):653–661. doi: 10.1242/jcs.109.3.653. [DOI] [PubMed] [Google Scholar]
- Wolf K, Wu YI, Liu Y, Geiger J, Tam E, Overall C, Stack MS, Friedl P. Multi-step pericellular proteolysis controls the transition from individual to collective cancer cell invasion. Nat Cell Biol. 2007;9(8):893–904. doi: 10.1038/ncb1616. [DOI] [PubMed] [Google Scholar]
- Woolner S, Bement WM. Unconventional myosins acting unconventionally. Trends Cell Biol. 2009;19(6):245–252. doi: 10.1016/j.tcb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaihara N, Nishioka M, Kohno T, Otsuka A, Okamoto A, Ochiai K, Tanaka T, Yokota J. Reduced expression of MYO18B, a candidate tumor-suppressor gene on chromosome arm 22q, in ovarian cancer. Int J Cancer. 2004;112(1):150–154. doi: 10.1002/ijc.20339. [DOI] [PubMed] [Google Scholar]
- Yang C, Svitkina T. Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr. 2011;5(5):402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Cheng W, Hung J, Montell D, Geisbrecht E, Rosen D, Liu J, Naora H. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proc Natl Acad Sci U S A. 2004;101(21):8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Berg JS, Li Z, Wang Y, Lang P, Sousa AD, Bhaskar A, Cheney RE, Stromblad S. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat Cell Biol. 2004;6(6):523–531. doi: 10.1038/ncb1136. [DOI] [PubMed] [Google Scholar]