Abstract
BACKGROUND
BRAF mutation status, and therefore eligibility for BRAF inhibitors, is currently determined by sequencing methods. We assessed the validity of VE1, a monoclonal antibody against the BRAF V600E mutant protein, in the detection of mutant BRAF V600E melanomas as classified by DNA pyrosequencing.
METHODS
The cases were 76 metastatic melanoma patients with only one known primary melanoma who had had BRAF codon 600 pyrosequencing of either their primary (n=19), metastatic (n=57) melanoma, or both (n=17). All melanomas (n=93) were immunostained with the BRAF VE1 antibody using a red detection system. The staining intensity of these specimens was scored from 0 – 3+ by a dermatopathologist. Scores of 0 and 1+ were considered as negative staining while scores of 2+ and 3+ were considered positive.
RESULTS
The VE1 antibody demonstrated a sensitivity of 85% and a specificity of 100% as compared to DNA pyrosequencing results. There was 100% concordance between VE1 immunostaining of primary and metastatic melanomas from the same patient. V600K, V600Q, and V600R BRAF melanomas did not positively stain with VE1.
CONCLUSIONS
This hospital-based study finds high sensitivity and specificity for the BRAF VE1 immunostain in comparison to pyrosequencing in detection of BRAF V600E in melanomas.
INTRODUCTION
Forty to sixty percent of all cutaneous melanomas harbor mutations in the BRAF oncogene, which regulates cellular growth signals.(1, 2) Alterations within BRAF often occur as somatic point mutations in the activating segment at amino acid 600, with the V600E alteration resulting in a missense substitution of valine by glutamic acid.(1, 3–5) This V600E mutation accounts for 69 – 94% of BRAF mutations in melanoma.(1, 6, 7) Two BRAF inhibitors are FDA approved for treatment of unresectable or metastatic melanoma patients; vemurafenib in patients with V600E mutant melanoma and dabrafenib in patients with a V600E or V600K mutant melanoma.(8–10) Current methods of detection of a BRAF mutation are DNA-based assays.(11, 12) These methods often take weeks for completion and require meticulous selection of a specimen with predominantly viable tumor.(12–14) Treatment with BRAF inhibitors often results in rapid clinical improvement, and a delay in therapy could be detrimental to patient care.(13)
Treating patients without a known mutation status with BRAF inhibitors carries the risk of further acceleration of melanoma tumor growth in NRAS mutant cases due to paradoxical activation of MAPK signaling.(15–18) With the use of current molecular methods, the potential for enhanced tumor growth must be weighed against harmful delays in treatment. Recently, a monoclonal antibody against mutant BRAF V600E protein (VE1) has been developed.(11, 19–22) Initial studies indicate high sensitivity and specificity of this antibody as compared to DNA sequencing.(11, 14,19–24) Use of immunohistochemistry for VE1 could potentially allow for a quick and efficient method of detection of BRAF mutation status. In this study, we attempt to validate the VE1 antibody using a different immunostaining platform and protocol as compared to previous investigators, test the antibody against different BRAF mutations, measure interobserver differences in scoring VE1 staining, examine the heterogeneity of VE1 staining within melanomas, and determine concordance of BRAF V600E status between primary and metastatic lesions.
MATERIALS AND METHODS
Case Selection
Following institutional review board approval, 97 primary and metastatic melanomas were retrieved from a case series of 79 patients treated at UNC Healthcare with known BRAF mutational status determined for clinical purposes in the UNC Molecular Genetics Laboratory using a CLIA-certified method of DNA pyrosequencing.(9, 25) H&E slides from these cases were reviewed for presence of sufficient tumor. One primary and three metastatic melanomas were excluded because of insufficient melanoma tissue in the block for recuts as determined by the study dermatopathologist. The remaining 93 primary and metastatic melanomas from 76 patients with a sufficient amount of tumor tissue for immunohistochemistry were analyzed.
Immunohistochemistry
Immunohistochemistry for mutant BRAF V600E protein was performed using the monoclonal mouse antibody VE1 (Spring Bioscience, Pleasanton, CA). Immunostaining was performed in the UNC Department of Dermatology Dermatopathology Laboratory. In this study, all tissue was fixed in neutral buffered formalin purchased commercially. Most samples had between 6 and 48 hours of total formalin fixation time prior to tissue processing. Our routine overnight tissue processing cycle includes the following: formalin for 60 minutes, 70% alcohol for 55 minutes, 95% alcohol for 35 minutes, 95% alcohol for 55 minutes, 100% alcohol for 30 minutes, 100% alcohol for 40 minutes, 100% alcohol for 55 minutes, xylene for 45 minutes, xylene for 55 minutes, paraffin for 30 minutes, paraffin for 30 minutes, paraffin for 30 minutes, and paraffin for 45 minutes. The original block used for genetic analysis was accessible and immunostained for all but 3 of the specimens. A tissue block adjacent to the original block was chosen for these three specimens. Freshly cut 4-µm thick sections of formalin-fixed and paraffin-embedded melanoma tissue blocks were stained using the fully automated Leica Bond III system. Pretreatment was performed using an onboard heat-induced epitope retrieval in EDTA buffer (ER2) for 30 minutes. Incubation with the VE1 antibody at a 1:100 dilution was done for 30 minutes at room temperature. Chromogenic detection was performed using the Leica Refined Red polymer detection system (Leica Microsystems). Incubation with hematoxylin for 10 minutes was used for counterstaining. Melanomas with documented BRAF mutational status were used as internal controls.
Pathology scoring
Immunostained slides were subsequently evaluated by a dermatopathologist (D.C.Z.) blinded to all genetic and clinical data. Specimens were analyzed for their degree of cytoplasmic immunostaining (0–3+). When cytoplasmic staining was scored as 0 or 1+ (with 1+ representing a weak cytoplasmic blush of staining), cases were scored as negative; while scores of 2+ and 3+ (indicating strong cytoplasmic staining) were considered positive results. Additionally, the dermatopathologist commented on whether there was VE1 nuclear staining independent of cytoplasmic staining. The percent of tumor that stained was also scored. To measure interobserver differences, a second dermatopathologist (P.A.G) similarly scored the metastatic melanomas with pyrosequencing results (n=57) independently.
Statistical analysis
The sensitivity and specificity of the VE1 immunostain as compared to DNA pyrosequencing were determined. A Pearson chi-squared test was used to determine the relationship of VE1 status with patient sex. An unpaired Student’s t-test was used to determine the association of VE1 status with age at diagnosis of the primary melanoma. P-values are two-sided. Statistical analyses were implemented in SAS (SAS Institute, Cary, NC) version 9.3.
RESULTS
Patients
The study included a series of 76 melanoma patients with known BRAF mutational status in their metastatic or primary melanoma and sufficient tumor for immunostaining with the VE1 antibody. Of these 76 patients, 27 had a BRAF V600E mutation and another 9 had an alternate BRAF V600 mutation, (V600K, V600R, or V600Q) on initial analysis. Of these specimens with pyrosequencing results, 19 were primary melanomas while 57 were metastatic melanomas. The age at diagnosis of the primary melanoma, sex, and AJCC TNM stage of these patients at the time of pyrosequencing of their melanoma is provided in Table 1. Additionally, 17 patients in this series with sufficient tumor had an additional matched primary (n=13) or metastatic melanoma (n=4) that was stained for VE1.
Table 1.
Patient Case Series
| Characteristic | Value | N (%) | VE1+ (%) | VE1− (%) |
|---|---|---|---|---|
| Total patients | 76 (100) | 22(29) | 54(71) | |
| Age at Diagnosis of Primary Melanoma | ||||
| Mean | 59 | |||
| Median | 60 | |||
| Range | 19–92 | |||
| Sex | ||||
| Male | 44 (58) | 9 (20) | 35 (80) | |
| Female | 32 (42) | 13 (41) | 19 (59) | |
| Overall AJCC TNM Stage at Pyrosequencing | ||||
| I | 9 (14) | 4 (44) | 5 (55) | |
| II | 20 (31) | 3 (15) | 17 (85) | |
| III | 33 (51) | 12 (36) | 21 (64) | |
| IV | 3 (5) | 0 (0) | 3 (100) | |
| DNA Pyrosequencing Results | ||||
| Wild type | 40 (53) | |||
| V600E | 27 (36) | |||
| V600K | 7 (9) | |||
| V600R | 1 (1) | |||
| V600Q | 1 (1) | |||
| Melanoma Type Tested for Pyrosequencing | ||||
| Metastatic | 57 (75) | 16 (28) | 41 (72) | |
| Primary | 19 (25) | 6 (32) | 13 (68) | |
| Primary Location | ||||
| Skin | 16 (84) | 6 (38) | 10 (63) | |
| Mucosal | 3 (16) | 0 (0) | 3 (100) | |
| Metastatic Location | ||||
| Skin | 19 (33) | 6 (32) | 13 (68) | |
| Lymph node | 28 (49) | 9 (32) | 19 (68) | |
| Muscosal | 4 (7) | 1 (25) | 3 (75) | |
| Soft tissue | 2 (4) | 0 (0) | 2 (100) | |
| Brian | 2 (4) | 0 (0) | 2 (100) | |
| Muscle | 1 (2) | 0 (0) | 1 (100) | |
| Lung | 1 (2) | 0 (0) | 1 (100) | |
| VE1 Cytoplasmic Staining Intensity | ||||
| 0 | 48 (63) | |||
| 1+ | 6 (8) | |||
| 2+ | 9 (12) | |||
| 3+ | 13 (17) |
Staining Intensity scored from 0 to 3+ (0=negative staining, 1+=weak background staining, 2+=moderately positive staining, 3+ strongly positive staining) with 0 and 1+ considered negative scores and 2+ and 3+ considered positive scores.
Interobserver Differences of BRAF VE1 Staining and Concordance of VE1 Immunohistochemistry Between Matched Primary and Metastatic Lesions
The kappa statistic for agreement for VE1 positivity between the two dermatopathologists was 1.00. All 17 matched pairs of metastatic and primary melanomas with sufficient tissue for immunohistochemistry demonstrated 100% concordance for VE1 staining. Three of these matched cases had positive immunohistochemistry results while 14 stained negatively.
VE1 Immunohistochemistry Compared with Genetic Analysis and Re-review of Pyrosequencing on Melanomas with Discrepant Results
According to DNA pyrosequencing, 40 of the 76 cases were determined as BRAF wild type while 36 cases had a BRAF mutation, comprised of BRAF V600E (n=26), V600K (n=8), V600R (n=1), or V600Q (n=1) mutations (Table 2). Twenty-two specimens immunostained positively (scores of 2+ and 3+) for VE1, while 54 specimens immunostained negatively (scores of 0 and 1+) (Figures 1, 2).
Table 2.
Discordant Cases of Immunohistochemical Analysis and Genetic BRAF Analysis Results
| DNA Pyrosequencing Result |
BRAF Cytoplasmic VE1 Staining Intensity |
Percent Tumor Stained in Cytoplasm |
BRAF Nuclear VE1 Staining Intensity |
Percent Tumor Stained in Nucleus |
Pyrogram Results for BRAF V600E mutation |
|---|---|---|---|---|---|
| V600E* | 0 | 100 | 0 | 0 | V600K |
| V600E | 0 | 100 | 0 | 0 | Low Allele Frequency |
| V600E | 0 | 100 | 0 | 0 | High Allele Frequency |
| V600E | 1+ | 100 | 1+ | 50 | High Allele Frequency |
| V600E | 1+ | 100 | 3+ | 75 | Low Allele Frequency |
This case indicates the discrepant case on re-review of molecular analysis.
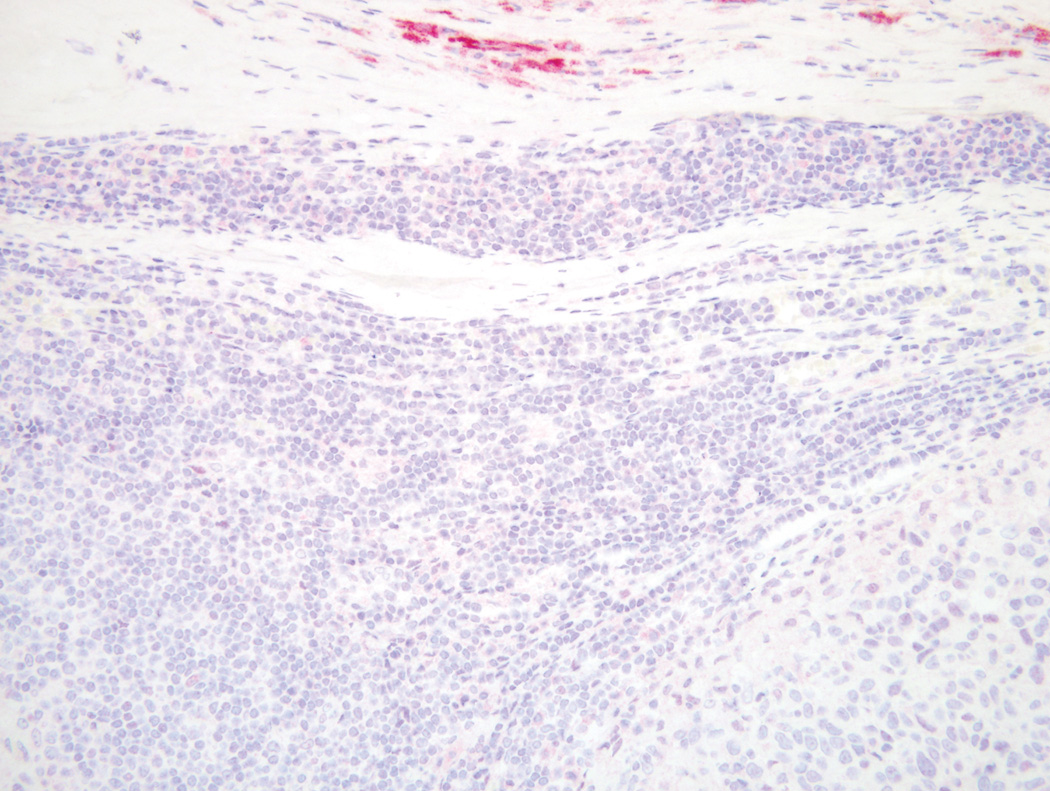
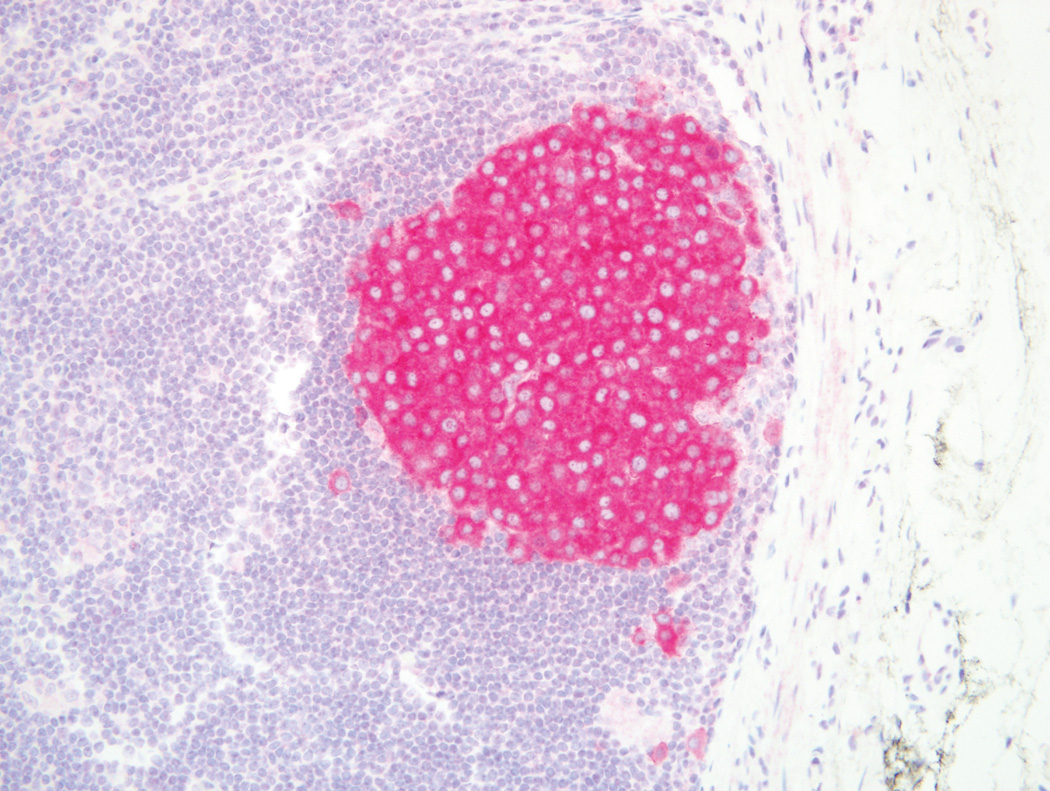
Figure 1.
BRAF wild type melanoma in a lymph node that is negative for VE1 immunostain alongside a 3+ positive capsular melanocytic nevus (Original magnification ×100).
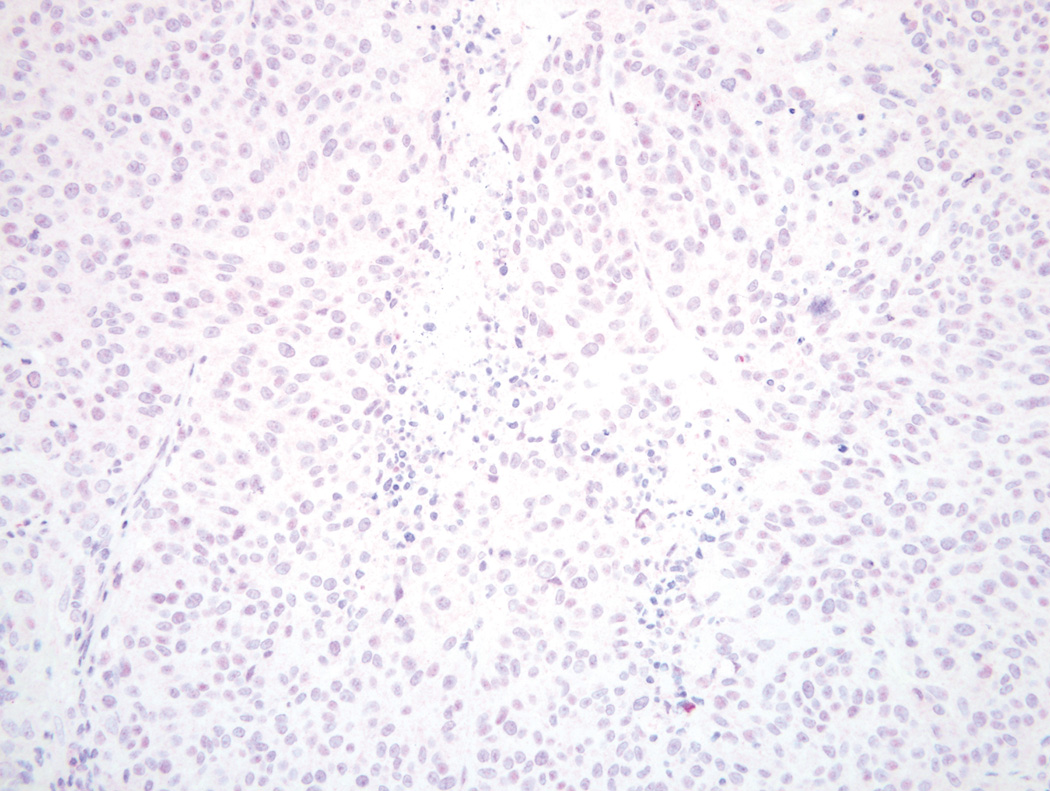
Figure 2.
BRAF wild type melanoma with 1+ weak cytoplasmic and nuclear blush on VE1 immunostain (Original magnification ×200).
In this study there were 5 (6.6%) discordant cases, all of which were 0 (n=3) or 1+ (n=2) by VE1 immunostaining but DNA pyrosequencing showed a V600E mutation (Table 2). Three of the discordant cases were metastatic lesions while the remaining 2 were primary lesions. Two of the discordant metastatic lesions had a matched primary lesion that was also tested. In both of those cases, the VE1 immunostain results for the primary and metastatic lesions were concordant with each other but were discordant with the positive DNA pyrosequencing result.
The pyrograms and interpretations from the five discrepant cases that were BRAF V600E mutation positive by pyrosequencing, but negative by IHC, were reviewed. Two of the cases showed a BRAF V600E mutation at a high allele frequency, and the high level of mutant DNA was consistent with the high estimated tumor percentage on the reviewed H&E section. Two of the cases showed a BRAF V600E mutation at an allele frequency lower than expected, possibly indicating tumor heterogeneity for the mutation. These two cases may represent true false negatives by IHC or false positives by pyrosequencing. The fifth case was found on review to contain a BRAF V600K mutation that was originally misinterpreted as a V600E mutation.
Sensitivity and Specificity
Compared to pyrosequencing (which was used as the gold standard for this study) after rereview of the discrepant cases, one case was reclassified as V600K rather than V600E. After reclassification of this case, immunohistochemical analysis with VE1 demonstrated a sensitivity of 85% (22/26) and a specificity of 100% (50/50) for the BRAF V600E mutation (Table 3). Specimens with the V600K, V600R, and V600Q mutations were not immunoreactive with the VE1 antibody.
Table 3.
Comparison of VE1 Immunostain Results and Pyrosequencing Results After Pyrosequencing Re-review
| BRAF Mutation Type |
Pyrosequence Positive |
Pyrosequence Negative |
VE1 Positive |
VE1 Negative |
Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| V600E | 26* | 40 | 22 | 44 | 85% | 100% |
| V600K | 8* | 0 | 0 | 8 | - | - |
| V600R | 1 | 0 | 0 | 1 | - | - |
| V600Q | 1 | 0 | 0 | 1 | - | - |
After molecular reanalysis, one discrepant case was determined to be V600K by pyrosequencing as opposed to V600E. Therefore, this case was counted as a V600K metastatic melanoma.
Heterogeneity of VE1 Staining Within Melanomas
Only 2 specimens were determined to be heterogeneous in immunohistochemical staining. One of these cases stained 99% of the tumor. Upon initial scoring, the other case was scored as homogeneously negative; however, upon re-examination, this tumor was determined to be 10% positive. This specimen was also one of the 5 discordant cases reported, and we did not change its classification because it was originally scored as negative. No other heterogeneity in cytoplasmic staining of the melanoma tumor cells was found.
Primary Melanomas
Including the matched pairs, 33 of the 93 specimens with VE1 immunostaining results were primary melanomas. Ten of these primaries stained positively while 23 stained negatively. These melanomas varied in stage, site, and classification. In this study, there was positive staining for superficial spreading (6/12), nodular (2/11), acral lentiginous (1/3), and spitzoid (1/1) melanoma subtypes. Primary melanoma subtypes tested with negative immunostaining results included mucosal (n=4), desmoplastic (n=1), and lentigo maligna (n=1) melanoma. Additionally, primary melanomas of different AJCC clinical stages were tested with the VE1 antibody. AJCC Stage T1 (0/2), Stage T2 (1/4), Stage T3 (5/9), and Stage T4 (4/18) melanomas demonstrated positive immunostain results. Primary melanomas with and without ulceration were used in this study, showing positive immunostaining in ulcerated (3/17) and non-ulcerated (7/16) melanomas (see Fig. 3).
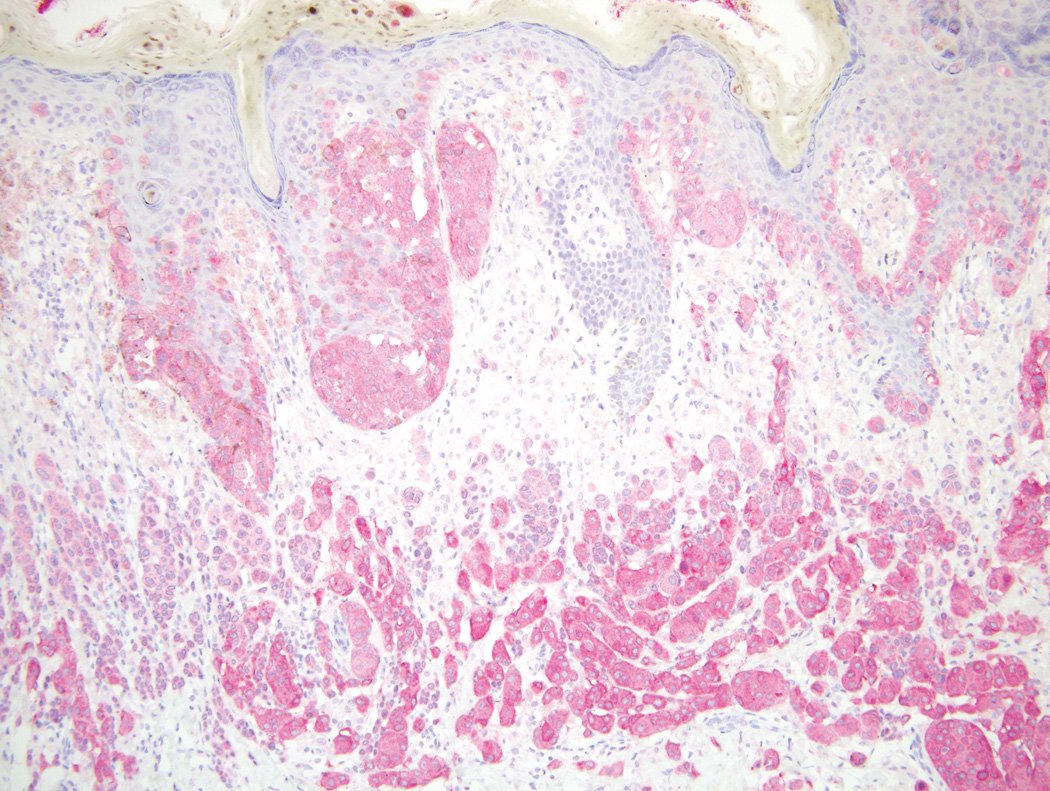
Figure 3.
A, BRAF V600E mutant primary melanoma with positive staining for VE1 (×200). B, Corresponding BRAF V600E metastatic melanoma with positive staining for VE1 in the same patient (Original magnification ×100).
Metastatic Melanomas
Our results demonstrated associations between immunostaining with VE1 in metastatic melanomas and certain clinical characteristics. Younger age at diagnosis of the primary melanoma was found to be associated with increased positive immunostaining (p=0.002). There was a borderline but non-significant association with sex between positive and negative VE1 results (p=0.06). Melanoma metastases from a variety of locations were tested with the VE1 immunostain, with positive results in cutaneous (6/19), lymph node (9/28), and mucosal (1/4) locations. Other metastases with negative immunostaining results had soft tissue (n=2), brain (n=2), muscle (n=1), or lung (n=1) location.
DISCUSSION
Our study demonstrated a sensitivity of 85% and a specificity of 100% for the BRAF VE1 immunohistochemistry using a red chromagen as a method of detection of BRAF V600E mutant protein as compared to DNA pyrosequencing for detection of BRAF V600E mutant DNA in routinely processed formalin-fixed paraffin-embedded melanoma tumor tissues. No melanoma with a BRAF V600K, V600R, or V600Q mutation stained with the VE1 antibody. Also the antibody had little variability in strength or intensity across our 93 specimens. After gaining experience with the antibody, we noticed that in nearly every case the tumor is either strongly positive or it is negative/very weak blush. The majority of specimens had homogeneous VE1 melanoma staining. Our study also demonstrated 100% concordance between matched primary and metastatic melanomas. We also demonstrated high interobserver agreement for scoring of the VE1 antibody staining, similar to Marin et al.(26) We demonstrated the utility of VE1 on a variety of metastatic and primary melanomas, including different primary melanoma histologic subtypes and metastases from a variety of cutaneous, lymph node, and visceral locations. Furthermore, the VE1 immunostain was associated with younger patient age at diagnosis of the primary melanoma, supporting the association of BRAF mutated melanoma and younger age.(27)
There are several differences in our methods as compared to most previous investigators. These include the use of the Mixed Red Refine reagent from Leica Microsystems as opposed to the Ventana OptiView and Ultraview Universal DAB Detection kits. This difference allows for improved differentiation of staining in melanoma specimens with a red chromagen as opposed to a brown chromagen. Additionally, our study is unique in using the Leica Bond III system as opposed to the Ventana Benchmark system.(11, 19,22, 28, 29) Other differences in protocol as compared to previous investigators include dilution of the antibody used, method of epitope retrieval, and time and temperature of incubation with the immunostain. We used a 1:100 dilution of the antibody, while other studies used higher concentrations including dilutions of 1:50, 1:5, or undiluted solutions.(11, 19,24, 29) We also performed epitope retrieval with EDTA for 30 minutes as opposed to Ventana Cell Conditioning solution for 64 minutes.(24) Additionally, we incubated the antibody for 30 minutes at room temperature as opposed to 37 degrees Celsius for 32 minutes.(11, 19, 29) Therefore, our study serves to provide additional validation of this antibody with a different immunostaining platform and protocol.
We used a graded system of staining as a method of assessment, similar to initial studies (11, 28), to determine the variability in the strength and intensity of VE1 staining. Other investigators did not use a graded system.(19, 20, 23) Our results showed only occasional heterogeneity of BRAF VE1 staining, similar to some other studies.(19, 24) Like us, some previous investigators scored isolated nuclear staining as negative.(19, 20, 24) We characterized the nuclear staining as part of the process of validating the antibody but did not find any clinical significance of nuclear staining in retrospective analysis. In accordance with the antibody specifications sheet, the VE1 immunostain is a cytoplasmic stain, consistent with BRAF being a cytoplasmic protein.
Our analysis is consistent with prior studies indicating a high sensitivity and specificity of the VE1 antibody for identifying BRAF V600E mutant melanomas, although some studies have achieved higher sensitivity (Table 4).(11, 19–22) Several reasons could account for the differences in sensitivity. Methods of VE1 staining may have played a major role. Additionally, previous investigators have used different gold standards to determine the presence of the BRAF mutation. Colomba et al. compared different methods of detection of BRAF mutations in melanoma specimens and found pyrosequencing to be the most efficient. We used pyrosequencing as our reference. It is also possible that preanalytical variables played a part in the discrepancy between our results and previous investigators. However, our results might also approximate what will be found in widespread clinical practice as many different labs start offering this test. Commonly, when immunostains are first published, it is often difficult for subsequent investigators to achieve the same high results as the initial publications.
Table 4.
Comparison of Our Results with Initial VE1 Studies
| Study | Sensitivity | Specificity | Number of Primary Melanomas Tested |
Number of Metastatic Melanoma Tested |
Evaluation of VE1 Staining* |
Method of Molecular Analysis |
|---|---|---|---|---|---|---|
| Our Study | 22/26 (85%) | 50/50 (100%) | 19 | 57 | Quantitative | Pyrosequencing |
| Long, 2013 | 37/38 (97%) | 58/59 (98%) | 10 | 87 | Qualitative | PCR-HRM, PCR-mass spectrometry |
| Colomba, 2013 | 40/40 (100%) | 39/39 (100%) | 0 | 79 | Qualitative | Sanger Sequencing, Real-time PCR, Pyrosequencing |
| Menzies et al. (2014) | 30/30 (100%) | 34/34 (100%) | 7 | 57 | Quantitative | PCR-HRM |
| Busam, 2012 | 22/22 (100%) | 22/22 (100%) | 7** | 44 | Quantitative | PCR-mass spectrometry |
| Capper, 2011 | 16/16 (100%) | 27/27 (100%) | 0 | 43 | Quantitative | Direct Sequencing |
Quantitative assessment of VE1 staining indicates a graded system for the evaluation of the VE1 staining while qualitative assessment indicates either the presence or absence of staining.
These 7 cases were not included in the statistical analysis published in this article.
Five of our cases were discordant on initial review, where DNA pyrosequencing was positive for the BRAF V600E mutation but immunostaining of the tissue with the VE1 antibody was negative. However on reanalysis of the pyrosequencing results, one case that was originally interpreted as V600E was found on review to represent a V600K mutation. The pyrosequencing assay used in the UNC Clinical Molecular Genetics Laboratory is designed to quantitatively interrogate mutations at position 1799. While the assay is only semiquantitative when applied to tumors with varying amounts of mixed stromal cells, the percentage of T or A alleles at that position can be estimated. This assay is meant to identify the V600E c.1799T>A mutation and other clinically important variants at or near nucleotide position 1799, such as variants at position 1798 or 1800. While surrounding base cells are represented by peaks on the pyrogram, the level of those nucleotide peaks are not quantitatively interrogated by the software. The alternate peak patterns may be difficult to interpret, especially if the mutant allele frequency is low. Previous investigators have also indicated discrepant results in molecular analysis.(14, 19) While BRAF inhibitors have demonstrated improved rates of overall and progression-free survival in patients with the BRAF V600E mutation in a phase 3 randomized clinical trial, some sensitivity has been shown against other mutations.(30) Of the five discordant cases, only one patient has been treated with targeted BRAF treatment. This metastatic melanoma patient is currently being treated with BRAF/MEK combination therapy (dabrafenib and trametinib) and, while improved since baseline, demonstrates slow progression.
Mismatches between immunohistochemistry and DNA pyrosequencing may also be due to sampling errors, tissue necrosis, or a decreased sensitivity of immunohistochemistry for the BRAF mutation. These differential results may also be due to intratumoral heterogeneity regarding the presence of a BRAF mutation,(31) although the majority of the melanomas in our study had homogenous VE1 staining. Generally homogenous VE1 staining with occasional or no heterogeneity in BRAF V600E expression has been demonstrated in previous studies.(11, 20,22, 32, 33) This heterogeneity has not been shown to correlate with survival and may be due to pre-analytical factors.(11, 32, 34) Heterogeneity within a tumor for the presence of the BRAF mutation status must be further studied.
Our results corroborate preservation of the BRAF V600E mutation between paired primary and metastatic melanomas from the same patient,(35) and we extend these results to concordance of BRAFV600E at the protein level. These findings are consistent with the early occurrence of BRAF mutations within melanoma pathogenesis.(35) Additionally, we show strong VE1 staining of a capsular melanocytic nevus in Figure 1. This finding supports the presence of BRAF V600E mutant protein in nevi, indicating that BRAF mutations alone are not sufficient for melanoma pathogenesis.(3) Therefore, this immunostain should not be considered diagnostic for melanoma. Together, these results suggest that in cases where metastatic lesions are inaccessible or unavailable, the primary lesion could potentially be used for testing. In this situation, patients with the BRAF mutation may have improved access to BRAF inhibitors.
Immunostaining with the VE1 antibody for the BRAF V600E mutation did not produce any false positive results. This finding combined with the high sensitivity of VE1 for this BRAF mutation supports the use of an algorithm incorporating both the VE1 antibody and DNA mutational analysis. In this model, immunohistochemistry initially could be used in patients with insufficient melanoma tissue for genetic analysis. This subset of patients would otherwise not be analyzed for the BRAF mutation using existing methods of detection. In patients with sufficient melanoma tissue, immunohistochemistry first could be used to quickly and inexpensively detect BRAF V600E mutations. Cases with negative VE1 results should be tested by a DNA mutational analysis assay to rule out a possible false negative result or a different BRAF mutation. The sequential use of these methods should allow for a highly sensitive and specific detection of the BRAF V600E mutation.(9, 14) Alternatively, if immunostaining were done in addition to mutational analysis on melanomas, the combined results might increase overall diagnostic accuracy for V600 mutational subtype if discrepant cases were reevaluated for their mutational status.
In the era of personalized medicine, BRAF mutation status has become a key piece of information in the clinical management of melanoma patients. In this study, we found the VE1 monoclonal antibody as method of detection of the BRAF V600E mutation in our institution to have high sensitivity and specificity with generally homogenous staining. Based on these results, immunostaining with the VE1 antibody seems to be an effective and efficient screening tool in the assessment of BRAF V600E mutant status in melanoma patients. In addition, VE1 staining seems to be complementary with mutational screening, as reevaluation of discrepant cases may improve diagnostic accuracy.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Greg Sturtz of the UNC Department of Dermatology for his contributions to this study.
Financial support:
R01 CA112243 and grants UL1TR000083, KL2TR000084, and TL1TR000085 from the National Center for Advancing Translational Sciences, NIH.
REFERENCES
- 1.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 2.Nissan MH, Solit DB. The "SWOT" of BRAF Inhibition in Melanoma: RAF Inhibitors, MEK Inhibitors or Both? Current Oncology Reports. 2011;13:479–487. doi: 10.1007/s11912-011-0198-4. [DOI] [PubMed] [Google Scholar]
- 3.Pollock PM, Harper UL, Hansen KS, Yudt LM, Stark M, Robbins CM, et al. High frequency of BRAF mutations in nevi. Nature Genetics. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 4.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Research. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busca R, Abbe P, Mantoux F, Aberdam E, Peyssonnaux C, Eychene A, et al. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. Embo Journal. 2000;19:2900–2910. doi: 10.1093/emboj/19.12.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rubinstein JC, Sznol M, Pavlick AC, Ariyan S, Cheng E, Bacchiocchi A, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. Journal of Translational Medicine. 2010;8 doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edlundh-Rose E, Egyhazi S, Omholt K, Mansson-Brahme E, Platz A, Hansson J, et al. NRAS and BRAF mutations in melanoma turnours in re ation to clinical characteristics: a study based on mutation screening by pyrosequencing. Melanoma Research. 2006;16:471–478. doi: 10.1097/01.cmr.0000232300.22032.86. [DOI] [PubMed] [Google Scholar]
- 8.Pazdur R. FDA Approval for Vemurafenib. Cancer Drug Information. National Cancer Institute at the National Institutes of Health. 2013 [Google Scholar]
- 9.Wilson MA, Nathanson KL. Molecular Testing in Melanoma. Cancer Journal. 2012;18:117–123. doi: 10.1097/PPO.0b013e31824f11bf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pazdur R. FDA Approval for Dabrafenib. Cancer Drug Information: National Cancer Institute at the National Institutes of Health. 2014 [Google Scholar]
- 11.Capper D, Preusser M, Habel A, Sahm F, Ackermann U, Schindler G, et al. Assessment of BRAF V600E mutation status by immunohistochemistry with a mutation-specific monoclonal antibody. Acta Neuropathologica. 2011;122:11–19. doi: 10.1007/s00401-011-0841-z. [DOI] [PubMed] [Google Scholar]
- 12.Grossmann AH, Grossmann KF, Wallander ML. Molecular testing in malignant melanoma. Diagnostic Cytopathology. 2012;40:503–510. doi: 10.1002/dc.22810. [DOI] [PubMed] [Google Scholar]
- 13.Nathan P, Sharma A, Lorigan P. Urgent treatment of patients with metastatic melanoma using braf inhibitors in the absence of braf mutation status. Annals of Oncology. 2013;24:2. doi: 10.1093/annonc/mdt167. [DOI] [PubMed] [Google Scholar]
- 14.Colomba E, Helias-Rodzewicz Z, Von Deimling A, Marin C, Terrones N, Pechaud D, et al. Detection of BRAF p.V600E Mutations in Melanomas Comparison of Four Methods Argues for Sequential Use of Immunohistochemistry and Pyrosequencing. Journal of Molecular Diagnostics. 2013;15:94–100. doi: 10.1016/j.jmoldx.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, et al. RAS Mutations in Cutaneous Squamous-Cell Carcinomas in Patients Treated with BRAF Inhibitors. New England Journal of Medicine. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatzivassiliou G, Song K, Yen I, Brandhuber BJ, Anderson DJ, Alvarado R, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature. 2010;464:431-U132. doi: 10.1038/nature08833. [DOI] [PubMed] [Google Scholar]
- 17.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464:427-U126. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Carvalho DD, Sharma S, You JS, Su SF, Taberlay PC, Kelly TK, et al. DNA Methylation Screening Identifies Driver Epigenetic Events of Cancer Cell Survival. Cancer Cell. 2012;21:655–667. doi: 10.1016/j.ccr.2012.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long GV, Wilmott JS, Capper D, Preusser M, Zhang YXE, Thompson JF, et al. Immunohistochemistry Is Highly Sensitive and Specific for the Detection of V600E BRAF Mutation in Melanoma. American Journal of Surgical Pathology. 2013;37:61–65. doi: 10.1097/PAS.0b013e31826485c0. [DOI] [PubMed] [Google Scholar]
- 20.Capper D, Berghoff AS, Magerle M, Ilhan A, Woehrer A, Hackl M, et al. Immunohistochemical testing of BRAF V600E status in 1,120 tumor tissue samples of patients with brain metastases. Acta Neuropathologica. 2012;123 doi: 10.1007/s00401-011-0887-y. [DOI] [PubMed] [Google Scholar]
- 21.Koperek O, Kornauth C, Capper D, Berghoff AS, Asari R, Niederle B, et al. Immunohistochemical Detection of the BRAF V600E-mutated Protein in Papillary Thyroid Carcinoma. American Journal of Surgical Pathology. 2012;36 doi: 10.1097/PAS.0b013e318246b527. [DOI] [PubMed] [Google Scholar]
- 22.Busam KJ, Sung J, Wiesner T, von Deimling A, Jungbluth A. Combined BRAF(V600E)-positive Melanocytic Lesions With Large Epithelioid Cells Lacking BAP1 Expression and Conventional Nevomelanocytes. American Journal of Surgical Pathology. 2013;37:193–199. doi: 10.1097/PAS.0b013e318263648c. [DOI] [PubMed] [Google Scholar]
- 23.Skorokhod A, Capper D, von Deimling A, Enk A, Helmbold P. Detection of BRAF V600E mutations in skin metastases of malignant melanoma by monoclonal antibody VE1. Journal of the American Academy of Dermatology. 2012;67 doi: 10.1016/j.jaad.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 24.Menzies AM, Lum T, Wilmott JS, et al. Intrapatient homogeneity of BRAFV600E expression in melanoma. Am J Surg Pathol. 2014;38:377. doi: 10.1097/PAS.0000000000000136. [DOI] [PubMed] [Google Scholar]
- 25.Gulley ML. UNC Healthcare System. Chapel Hill, NC: 2013. BRAF gene mutation test in cancer diagnosis, prognosis and prediction. [Google Scholar]
- 26.Marin C, Beauchet A, Capper D, Zimmermann U, Julié C, Ilie M, et al. Detection of BRAF p.V600E Mutations in Melanoma by Immunohistochemistry Has a Good Interobserver Reproducibility. Arch Pathol Lab Med. 2014;138:71–75. doi: 10.5858/arpa.2013-0031-OA. [DOI] [PubMed] [Google Scholar]
- 27.Schlaak M, Bajah A, Podewski T, Kreuzberg N, von Bartenwerffer W, Wardelmann E, et al. Assessment of clinical parameters associated with mutational status in metastatic malignant melanoma: a single-centre investigation of 141 patients. British Journal of Dermatology. 2013;168:708–716. doi: 10.1111/bjd.12140. [DOI] [PubMed] [Google Scholar]
- 28.Busam KJ, Hedvat C, Pulitzer M, von Deimling A, Jungbluth AA. Immunohistochemical analysis of BRAF(V600E) expression of primary and metastatic melanoma and comparison with mutation status and melanocyte differentiation antigens of metastatic lesions. Am J Surg Pathol. 2012;37:413. doi: 10.1097/PAS.0b013e318271249e. [DOI] [PubMed] [Google Scholar]
- 29.Andrulis M, Penzel R, Weichert W, von Deimling A, Capper D. Application of a BRAF V600E Mutation-specific Antibody for the Diagnosis of Hairy Cell Leukemia. American Journal of Surgical Pathology. 2012;36:1796–1800. doi: 10.1097/PAS.0b013e3182549b50. [DOI] [PubMed] [Google Scholar]
- 30.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved Survival with Vemurafenib in Melanoma with BRAF V600E Mutation. New England Journal of Medicine. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yancovitz M, Litterman A, Yoon J, Ng E, Shapiro RL, Berman RS, et al. Intra- and Inter-Tumor Heterogeneity of BRAF(V600E)Mutations in Primary and Metastatic Melanoma. Plos One. 2012;7 doi: 10.1371/journal.pone.0029336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilmott JS, Menzies AM, Haydu LE, Capper D, Preusser M, Zhang YE, et al. BRAF(V600E) protein expression and outcome from BRAF inhibitor treatment in BRAF(V600E) metastatic melanoma. British Journal of Cancer. 2013;108:924–931. doi: 10.1038/bjc.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeh I, von Deimling A, Bastian BC. Clonal BRAF mutations in melanocytic nevi and initiating role of BRAF in melanocytic neoplasia. J Natl Cancer Inst. 2013;105:917. doi: 10.1093/jnci/djt119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schindler G, Capper D, Meyer J, Janzarik W, Omran H, Herold-Mende C, et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra- cerebellar pilocytic astrocytoma. Acta Neuropathologica. 2011;121:397–405. doi: 10.1007/s00401-011-0802-6. [DOI] [PubMed] [Google Scholar]
- 35.Omholt K, Platz A, Kanter L, Ringborg U, Hansson J. NRAS and BRAF mutations arise early during melanoma pathogenesis and are preserved throughout tumor progression. Clinical Cancer Research. 2003;9:6483–6488. [PubMed] [Google Scholar]