Abstract
Background
Runt-related transcription factor 3 (RUNX3) is a member of the runt-domain family of transcription factors. Emerging evidence indicates that RUNX3 is a tumor suppressor gene in several types of human cancers including esophageal cancer. However, the association between RUNX3 promoter methylation and esophageal cancer remains unclear. Here we conducted a systematic review and meta-analysis to quantitatively evaluate the effects of RUNX3 promoter methylation on the incidence of esophageal cancer.
Methods
A detailed literature search was made on Medline, Pubmed and Web of Science for related research publications written in English and/or Chinese. Methodological quality of the studies was also evaluated. The data were extracted and assessed by two reviewers independently. Analysis of pooled data were performed, the odds ratios (OR) were calculated and summarized respectively.
Results
Final analysis of 558 patients from 9 eligible studies was performed. The result showed that RUNX3 methylation was significantly higher in esophageal cancer than in normal squamous mucosa from the proximal resection margin or esophageal benign lesions (OR = 2.85, CI = 2.01–4.05, P<0.00001). The prevalence of lymph node involvement, tumor size (T1–T2 vs T3–T4) and histological grade was significantly greater in RUNX3-negative cases (RUNX3 unmethylated groups) than in RUNX3-positive cases (OR = 0.25, CI = 0.14–0.43, P<0.00001). RUNX3 methylation was significantly higher in esophageal adenocarcinoma (EAC) than Barrett’s esophagus (OR = 0.35, CI = 0.20–0.59, P<0.0001). In addition, the pooled HR for overall survival (OS) showed that decreased RUNX3 expression was associated with worse survival in esophageal cancer (HR = 4.31, 95% CI = 2.57–7.37, P<0.00001).
Conclusions
The results of this meta-analysis suggest that RUNX3 methylation is associated with an increased risk, progression as well as worse survival in esophageal cancer. RUNX3 methylation, which induces the inactivation of RUNX3 gene, plays an important role in esophageal carcinogenesis.
Introduction
Esophageal cancer is the eighth most common cancer worldwide, esophageal squamous cell carcinoma (ESCC) and adenocarcinoma (EAC) are two major histopathological types of esophageal cancer [1], [2]. It has been reported that esophageal tumor currently affects more than 450, 000 people worldwide and the incidence is still increasing [1]. Surgery is the standard therapy for esophageal tumor [3]. However, the overall prognosis is far less satisfactory and pretty modest, with 5-year survival rates ranging between 15 and 50% [4], [5]. Therefore, investigating the mechanism of initiation and progression and finding out the therapeutic targets at biomolecular levels are highly desired for the treatment of esophageal cancer.
The Runt-related transcription factor 3 (RUNX3) gene is a tumor suppressor gene involved in the TGF-β signaling pathway, which was cloned and identified as a human runt-domain containing gene in 1994 [6]. Its precise function has been intensively studied in gastric cancer, with upregulation inducing cell cycle arrest, apoptosis, and down regulating cyclin D1 expression [7], [8], [9], [10], however, its role in esophageal cancer has not been thoroughly investigated and reviewed. Inactivation of RUNX3 by promoter methylation has been found to play an important role during normal tissue development and in tumorigenesis in esophagus [11]. In this study, we reviewed and performed a meta analysis on the published clinical studies regarding the effect of RUNX3 on patients with esophageal cancer.
Materials and Methods
Search strategy
Medline, Pubmed and Web of Science were searched in December 2013 using the search terms: “esophageal” and “cancer or tumor or neoplasm or carcinoma” and “RUNX3”. Studies identified through the approaches as described above were screened by titles first, then abstracts of the publications. After exclusion of non-relevant publications and identifications of duplicates from the different databases, the remaining papers were evaluated in the full text version for in- and exclusion criteria and for relevant articles in the reference lists. All clinical studies except case reports were chosen. The language of publication was restricted to English and Chinese. All searched data were retrieved. Authors’ bibliographies and references of selected studies were also searched for other relevant studies. The most complete study was chosen to avoid duplication if same patient populations were reported in several publications.
Selection criteria
We collected all eligible articles about relationship between RUNX3 methylation and/or expression and clinicopathological features and clinical outcomes in esophageal cancer in this meta-analysis. Studies meeting the following inclusion criteria were included: (1) RUNX3 methylation and/or expression evaluated in the circulation and/or primary esophageal cancer tissues, (2) studies revealing the relationship between RUNX3 methylation and/or expression and esophageal cancer clinicopathological parameters and prognosis, (3) RUNX3 methylation and/or expression examined by polymerase chain reaction (PCR), (4) articles published as a full paper in English, (5) studies providing sufficient information to estimate hazard ratio (HR) about overall survival (OS) and 95% confidence interval (CI). The exclusion criteria included the following: (1) letters, reviews, case reports, conference abstracts, editorials, expert opinion, non-English, non-Chinese language papers; (2) articles with no information on OS or insufficient information for calculation of HR; and (3) all publications regarding in vitro/ex vivo studies, cell lines and human xenografts.
Data extraction
Two investigators independently extracted data from eligible studies. Disagreements were resolved by discussion and consensus. Two investigators reviewed all of articles that fit inclusion and exclusion criteria. The following information was recorded for each study: the first author name, year of publication, sample source, number of cases, clinicopathological parameters, cancer with tumor node metastasis (TNM) stage, RUNX3 methylation and/or expression, and patient survival. Data for study characteristics and clinical response were summarized and turned the data into table format. Heterogeneity of investigation was evaluated to determine whether the data of the various studies were appropriate for a meta-analysis.
Statistical analysis
Analysis was conducted using the Stata 12.0 (Stata Corporation, TX, USA) and Review Manager 5.2 (Cochrane Collaboration, Oxford, UK). Comparisons of dichotomous measures were done by pooled estimates of odds ratios (ORs) as well as their 95% CIs. P value of <0.05 was considered to be statistically significant. Heterogeneity was examined by a chi-square test with significance set at P<0.10; the total variation among studies was estimated by I square. If there was heterogeneity among studies, we used a random effect model to pool the ORs; otherwise, a fixed effect model was chosen.
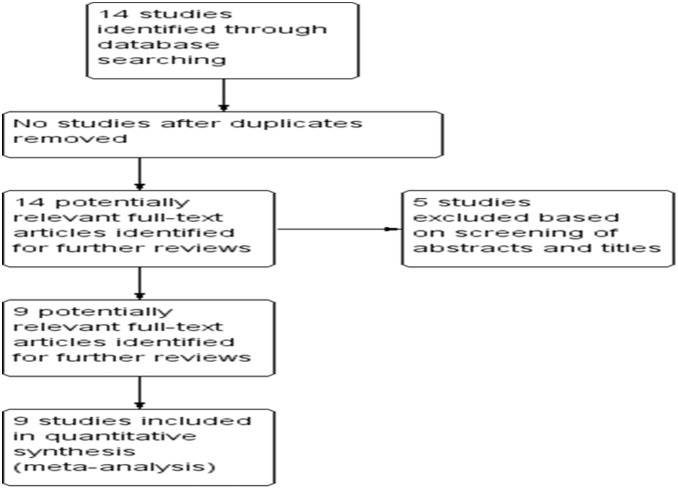
The database search generated 14 articles from Pubmed and the Web of Science. After initial screening of all titles, abstracts and eligibility, 9 full-text studies were retracted for more detailed assessment. The search of the article references did not produce additional publications. Eventually, 9 publications met the inclusion criteria for qualitative study and for meta-analysis. The article search and study selection is depicted in Figure 1.
Figure 1. Flow chart of study selection.
Results
Identification of relevant studies
Fourteen publications were identified by using the search method as described above. Five of those were excluded due to laboratory studies, non-original articles (review), or studies irrelevant to the current analysis. Eventually, there were nine studies included in the final meta-analysis [11], [12], [13], [14], [15], [16], [17], [18], [19] (Figure. 1).
Study characteristics
Nine studies published from 2005 to 2013 were eligible for meta-analysis. A total of 558 patients including esophageal squamous cell carcinomas (ESCCs) and esophageal adenocarcinomas (EACs) from China, Japan, Australia and USA were enrolled. A total of 140 cases of Barrett’s esophagus (BE) were also included in this analysis. Their basic characteristics are summarized in Table 1.
Table 1. Basic characteristics of the included studies.
| Study/Country | Patients | Methods | Primary Aim | Methylationsite | RUNX3 expression |
| Hiramatsu et al2005 [17] | 69 | RT-PCR,Western blot | Eexamine the expression of RUNX3protein in human esophageal mucosaand squamous cell carcinoma incomparison with clinicopathological profiles | + | |
| Schulmann et al2005 [13] | 77 | Methylationspecic PCR (MSP) | Determine at what stage,inactivation of RUNX3 occursand predicts prognosis | Promoter,CpG islands | + |
| Long et al2007 [19] | 42 | RT-PCR,MSP | Determine whether promotermethylation of the RUNX3gene correlates with ESCCtumor progression | Promoter,CpG islands | + |
| Sakakura et al2007 [34] | 51 | MSP | Determine whether RUNX3expression is associated withradioresistance and prognosis | Promoter,CpG islands | + |
| Tonomoto et al2007 [18] | 61 | qRT-PCR,MSP | Detemine whetherthe precise expression, prognosticimpact and methylation status ofRUNXs in esophageal squamouscell carcinoma. | Promoter,CpG islands | + |
| Smith et al2008 [15] | 37 | MSP | Determine at what stage,in the progression fromBE to EAC, methylation ofkey genes occurs. | Promoter,CpG islands | + |
| Sugiura et al2008 [12] | 70 | qRT-PCR,MSP | Determine whether RUNX3expression is correlated withESCC progression and prognosis | Promoter,CpG islands | + |
| Liu et al2011 [11] | 81 | MSP | Determine the effects of plasmaDNA methylation of RUNX3on recurrence of ESCC | Promoter,CpG islands | + |
| Zheng et al2011 [16] | 70 | MSP | Evaluate the diagnostic roleof RUNX3 gene methylation inserum DNA of esophageal squamouscell carcinoma (ESCC), gastriccancer (GC) and colorectalcancer (CRC) patients. | Promoter,CpG islands | + |
RUNX3 methylation and expression and clinicopathological features
1. Inactivation of RUNX3 through methylation in esophageal cancers
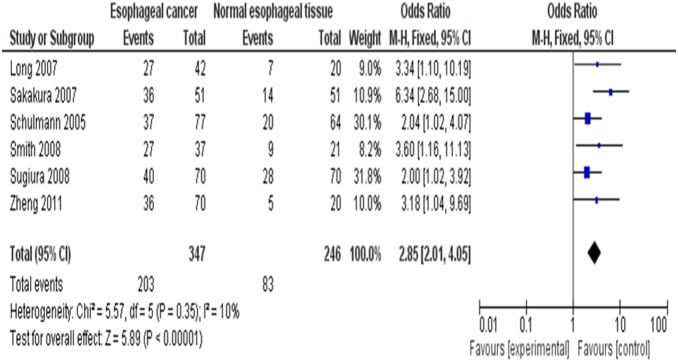
It was reported that the loss of RUNX3 mRNA expression was statistically correlated with the promoter hypermethylation in esophageal tumors (P<0.001) [18], [19]. We observed that RUNX3 methylation was significantly higher in ESCC/EAC than in normal squamous mucosa from the proximal resection margin or esophageal benign lesions. The pooled OR from 6 studies including 347 esophageal cancers and 246 normal squamous mucosa was shown in Figure 2 (OR = 2.85, CI = 2.01–4.05, P<0.00001), which indicated that RUNX3 inactivation through methylation plays an important role in the pathogenesis of esophageal cancers.
Figure 2. The pooled OR from 6 studies including 347 esophageal cancers and 246 normal squamous mucosa OR = 2.85, CI = 2.01–4.05, P<0.00001.
2. Role of RUNX3 methylation in esophageal cancer development
We analyzed 263 patients pooled in 4 studies to assess whether the aberrant RUNX3 methylation/expression in serum/cancer tissues DNA was associated with advanced stage, including tumor size (T1–T2 vs T3–T4), lymph node involvement, lymph and blood vessels metastasis, and recurrence in esophageal carcinomas. RUNX3 methylation/expression estimated in biopsy/blood samples and clinicopathological factors as described above were examined. The prevalence of lymph node involvement, tumor size (T1–T2 vs T3–T4) and histological grade was significantly greater in RUNX3-negative cases (RUNX3 unmethylated groups) than in RUNX3-positive cases (Figure 3), with OR = 0.25, CI = 0.14–0.43, P<0.00001. These results suggest that epigenetic silencing of RUNX3 gene expression by promoter hypermethylation may play an important role in esophageal cancer progression and development. With similarity in pathologic stage, Hiramatsu et al [17] showed that RUNX3 expression was significantly higher in 19 well-differentiated ESCCs than in 56 moderately or 69 poorly differentiated ESCCs (p<0.01).
Figure 3. Forest plot of odds ratio (OR) in 263 patients pooled in 4 studies in serum/cancer tissues DNA from advanced stage, including tumor size (T1–T2 vs T3–T4), lymph node involvement, lymph and blood vessels metastasis, and recurrence in esophageal carcinomas.
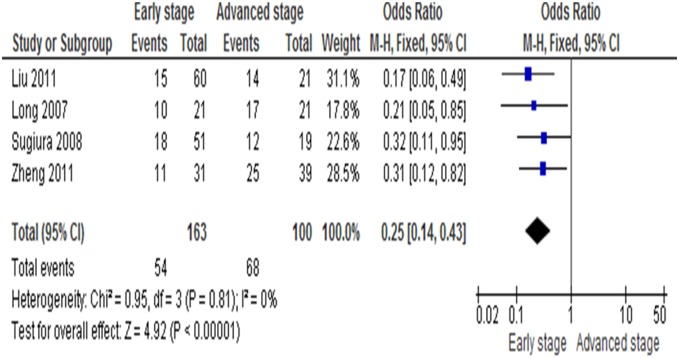
The prevalence of lymph node involvement, tumor size (T1–T2 vs T3–T4) and histological grade was significantly greater in RUNX3-negative cases (RUNX3 unmethylated group) than in RUNX3-positive cases, OR = 0.25, CI = 0.14–0.43, P<0.00001.
Barrett's esophagus (BE) is the metaplastic replacement of squamous with columnar epithelium in the esophagus, as a result of reflux. It is a major risk factor for the development of EAC [20], [21]. We observed that RUNX3 methylation was significantly higher in EAC than in BE as shown in Figure 4, OR = 0.35, CI = 0.20–0.59, P<0.0001. RUNX3 hypermethylation is an independent risk factor for progression of BE to high-grade dysplasia of esophagus and EAC [13], [22].
Figure 4. RUNX3 methylation significantly higher in esophageal adenocarcinoma (EAC) than Barrett’s esophagus (BE), OR = 0.35, CI = 0.20–0.59, P<0.0001.
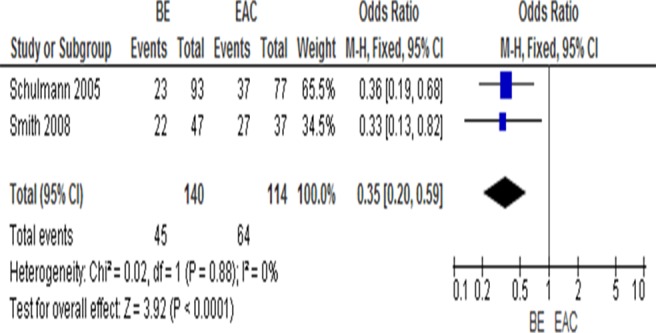
No heterogeneity was observed in the analysis of RUNX3 methylation/low expression in normal samples and esophageal patient samples (P = 0.81), in BE and EAC patients (P = 0.88). There is no heterogeneity of RUNX3 methylation with advanced stage (P = 0.81), so the fixed effect model was used.
3. RUNX3 as a prognostic factor for esophageal cancer
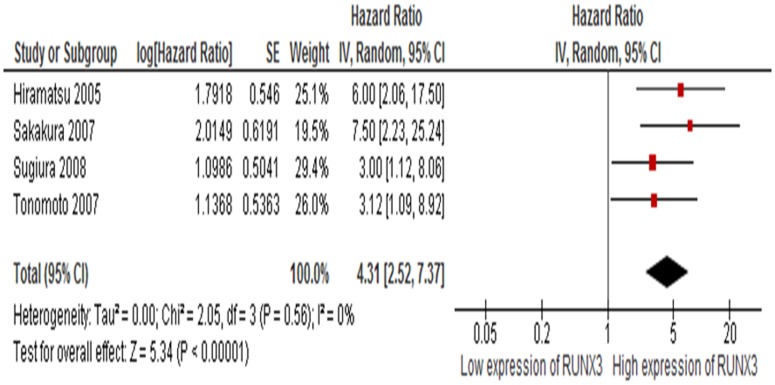
Four studies included investigated the relationship between OS and RUNX3 methylation/expression. The pooled HR for OS showed that decreased RUNX3 expression was associated with worse survival in esophageal cancer as shown in Figure 5 (HR = 4.31, 95% CI = 2.57–7.37, P<0.00001).
Figure 5. All four included studies estimated the relationship between OS and RUNX3 methylation/expression.
The pooled HR for OS showed that decreased RUNX3 expression was associated with worse survival in esophageal cancer, HR = 4.31, 95% CI = 2.57–7.37, P<0.00001.
4. Sensitivity analyses and publication bias
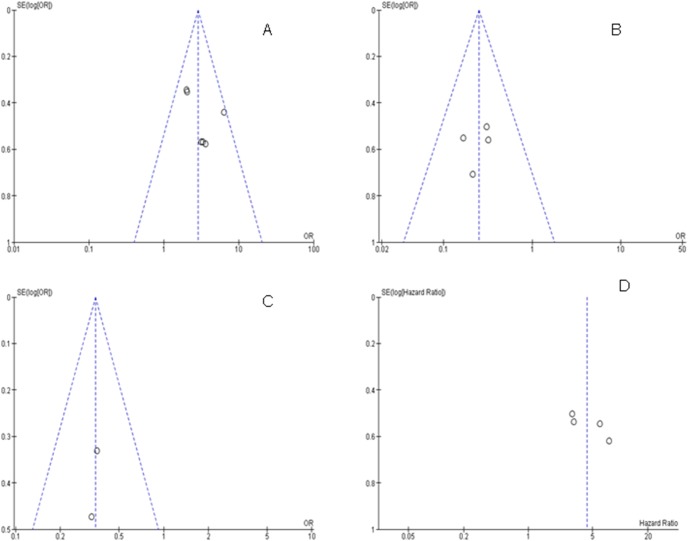
A sensitivity analysis, in which one study was removed at a time, was conducted to assess the result stability. The pooled ORs and HRs were not significantly changed, indicating the stability of our analyses. The funnel plots were largely symmetric (Figure 6) suggesting there were no publication biases in the meta-analysis of RUNX3 methylation/expression and clinicopathological features as well as overall survival respectively.
Figure 6. The funnel plots were largely symmetric suggesting there were no publication biases in the meta-analysis of RUNX3 methylation/expression and clinicopathological features as well as overall survival respectively.
The funnel plot from 6 studies comparing esophageal cancers and normal squamous mucosa (A). The funnel plot from 4 studies in determining RUNX3 hypermethylation in advanced stage (T3–T4) and early stage (T1–T2) (B). The funnel plot from 2 studies in determining RUNX3 hypermethylation in Barrett’s esophagus (BE) and esophageal adenocarcinoma (EAC) (C). The funnel plot from 4 studies in determining the relationship between RUNX3 hypermethylation and overall survival (OS) in esophageal cancer (D).
Discussion
RUNX3 belongs to the runt domain family of transcriptional factors that plays an important role during normal tissue development and in tumorigenesis [7], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32], [33]. RUNX3 inactivation is a crucial factor to determine cancer pathogenesis and clinical outcome in a variety of cancer types [23]. The different modes of RUNX3 inactivation in various tumor types include hemizygous deletion, mutations, hypermethylation, histone modifications and cytoplasmic mislocalization [23]. To date, there have been some studies describing the precise expression, prognostic impact and methylation status of RUNX3 in esophageal cell carcinoma [13], [16], [17], [19], [33], [34]. We conducted a meta-analysis to evaluate the correlation between RUNX3 promoter methylation/low expression and esophageal cancer. Analysis of the pooled data showed that 1) Esophageal cancers had a higher methylation rate than normal tissues, or well-differentiated cancer tissues; 2) The prevalence of lymph node involvement, tumor size (T1–T2 vs T3–T4) and histological grade was significantly greater in RUNX3-negative cases (RUNX3 unmethylated groups) than in RUNX3-positive cases; RUNX3 methylation was significantly higher in EAC than in BE; and 3) The pooled HR for OS showed that decreased RUNX3 expression was associated with worse survival in esophageal cancer.
ESCC and EAC are the two major histological types of esophageal carcinoma. ESCC occurs mostly in the endemic areas such as Asia and Africa, whereas EAC is the most common in North America and Europe [35]. Although these two esophageal cancers are different in pathogenesis, epidemiology, tumor biology, prognosis and therapeutic strategies among the pooled patients in this meta analysis, RUNX3 methylation/low expression was associated with the pathogenesis of both types of esophageal cancer, indicating that RUNX3 might be responsible for different patterns of tumorigenesis.
Patients with BE have an increased risk of developing EAC. The most established marker for the risk of developing EAC in BE is dysplasia [36]. Because of not well defined natural history of low grade dysplasia (LGD), not well characterized histological classification of dysplasia, moreover especially extremely high interobserver variability for LGD, molecular biomarkers are needed to improve the risk classification of BE patients [37], [38], [39]. It was accepted that BE is a precancerous tissue, and that aberrant promoter methylation occurs early in metaplasia before histological evidence of progression to cancer. Some underlying mechanisms for aberrant DNA methylation in Barrett’s metaplasia have been noted. Frequent RUNX3 inactivation through promoter hypermethylation was reported in ESCC, EAC, Barrett’s metaplasia and dysplasia [13], [15], [17], [18]. The result of this meta analysis indicated that the methylation of RUNX3 gene was significantly higher in both BE and EAC than in the squamous samples, that metaplasia BE was nearly as abnormal epigenetically as EAC. In other words, the aberrant methylation of RUNX3 gene is an early event, which most probably occurs independently of EAC [13], [15].
RUNX3 exerts pleiotropic effects during tumor suppression. It inhibits the oncogenic Wnt signaling pathway via formation of a complex with the TCF4-β-catenin complex and hampering it from binding to target genes such as c-myc and cyclin D1 [23], [40]. RUNX3 interacts with SMAD3/SMAD4 to regulate TGF-β-dependent inhibition of proliferation and apoptosis by activation of p21 and Bim. Sakakura et al [34] found that frequent silencing of RUNX3 by promoter hypermethylation in esophageal squamous cell carcinomas is associated with radioresistance and poor prognosis. In other words, RUNX3 gene expression promotes radiosensitivity, whereas its inactivation facilitates radioresistance [34]. They further confirmed that RUNX3 activates Bim expression and increases sensitivity to radiation and induces TGF-β-mediated apoptosis in ESCC cells, therefore functioning as a crucial determinant of radiosensitivity [34]. Thus, induction of RUNX3 expression by overcoming gene silencing may enhance radiosensitivity against tumor which may have crucial clinical impact for esophageal cancer patients. It is also exciting that measurement of RUNX3 expression status in pretreatment specimens may predict radiosensitivity [34].
Progression from BE to esophageal cancer appears to mirror the accumulation of genetic abnormalities, suggesting a stepwise progression of genetic changes in esophageal cancer. RUNX3, in combination of a panel of other genes that are inactivated by methylation, can be developed as biomarkers for various tissues. This is becoming a good strategy for risk stratification to predict neoplastic progression including esophageal cancer [14].
Consistent results were shown in sensitivity analyses, and no evidence of publication bias was found. However, this study has several potential limitations. First, the possibility of information and selection biases and unidentified confounders could not be completely excluded because all of the included studies were observational. Second, the searching strategy was restricted to articles published in English and Chinese. Articles with potentially high-quality data that were published in other languages were not included because of anticipated difficulties in obtaining accurate medical translation. Hence, cautions should be taken when our findings are interpreted among the general populations.
In conclusion, our meta-analysis showed RUNX3 may play an important role in esophageal cancer initiation and progression. Plasma levels of RUNX3 promoter hypermethylation may be a promising biomarker for the early diagnosis of esophagus squamous cell carcinoma [16]. In addition, RUNX3 methylation is associated with an increased risk and worse survival in esophageal cancer patients. Further large-scale studies, especially multi-center and well-matched cohort research will provide more insight into the role of RUNX3 in the prognosis and clinical implementation of esophageal cancer patients. The fact that silencing of RUNX3 gene at the transcriptional level and functional inactivation at the protein level in esophagus cancer are strongly correlated with poor prognosis and may occur in early phase of tumor initiation, making it a promising target for therapeutic approaches. Maintaining RUNX3 expression under microenvironment stress conditions, either directly or indirectly or reversing RUNX3 silencing could be a new direction for drug discovery for esophageal cancers. Further, RUNX3 was reported to control Notch signaling which is tightly linked to cancer stem cell (CSCs) [7]. Conventional chemotherapy can induce resistance to chemotherapeutic agents, and tumor regrowth mediated by CSCs, therefore targeting RUNX3 gene and its related signaling pathways could be another mechanism for therapeutic approaches for cancer treatment aiming at CSC elimination.
Supporting Information
PRISMA checklist.
(DOC)
PRISMA flowchart.
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors have no funding or support to report.
References
- 1.Li X, Gao C, Yang Y, Zhou F, Li M, et al.. (2013) Systematic review with meta-analysis: the association between human papillomavirus infection and oesophageal cancer. Aliment Pharmacol Ther. [DOI] [PubMed]
- 2.Wu J, Wu X, Liang W, Chen C, Zheng L, et al.. (2013) Clinicopathological and prognostic significance of chemokine receptor CXCR4 overexpression in patients with esophageal cancer: a meta-analysis. Tumour Biol. [DOI] [PubMed]
- 3. Gebski V, Burmeister B, Smithers BM, Foo K, Zalcberg J, et al. (2007) Survival benefits from neoadjuvant chemoradiotherapy or chemotherapy in oesophageal carcinoma: a meta-analysis. Lancet Oncol 8: 226–234. [DOI] [PubMed] [Google Scholar]
- 4. Rutegard M, Charonis K, Lu Y, Lagergren P, Lagergren J, et al. (2012) Population-based esophageal cancer survival after resection without neoadjuvant therapy: an update. Surgery 152: 903–910. [DOI] [PubMed] [Google Scholar]
- 5. van Hagen P, Hulshof MC, van Lanschot JJ, Steyerberg EW, van Berge Henegouwen MI, et al. (2012) Preoperative chemoradiotherapy for esophageal or junctional cancer. N Engl J Med 366: 2074–2084. [DOI] [PubMed] [Google Scholar]
- 6. Levanon D, Negreanu V, Bernstein Y, Bar-Am I, Avivi L, et al. (1994) AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics 23: 425–432. [DOI] [PubMed] [Google Scholar]
- 7. Shiraha H, Nishina S, Yamamoto K (2011) Loss of runt-related transcription factor 3 causes development and progression of hepatocellular carcinoma. J Cell Biochem 112: 745–749. [DOI] [PubMed] [Google Scholar]
- 8. Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, et al. (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124. [DOI] [PubMed] [Google Scholar]
- 9. Chi XZ, Yang JO, Lee KY, Ito K, Sakakura C, et al. (2005) RUNX3 suppresses gastric epithelial cell growth by inducing p21(WAF1/Cip1) expression in cooperation with transforming growth factor {beta}-activated SMAD. Mol Cell Biol 25: 8097–8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wei D, Gong W, Oh SC, Li Q, Kim WD, et al. (2005) Loss of RUNX3 expression significantly affects the clinical outcome of gastric cancer patients and its restoration causes drastic suppression of tumor growth and metastasis. Cancer Res 65: 4809–4816. [DOI] [PubMed] [Google Scholar]
- 11. Liu JB, Qiang FL, Dong J, Cai J, Zhou SH, et al. (2011) Plasma DNA methylation of Wnt antagonists predicts recurrence of esophageal squamous cell carcinoma. World J Gastroenterol 17: 4917–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sugiura H, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, et al. (2008) Decreased expression of RUNX3 is correlated with tumor progression and poor prognosis in patients with esophageal squamous cell carcinoma. Oncol Rep 19: 713–719. [PubMed] [Google Scholar]
- 13. Schulmann K, Sterian A, Berki A, Yin J, Sato F, et al. (2005) Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett’s-associated neoplastic progression and predicts progression risk. Oncogene 24: 4138–4148. [DOI] [PubMed] [Google Scholar]
- 14. Jin Z, Cheng Y, Gu W, Zheng Y, Sato F, et al. (2009) A multicenter, double-blinded validation study of methylation biomarkers for progression prediction in Barrett’s esophagus. Cancer Res 69: 4112–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith E, De Young NJ, Pavey SJ, Hayward NK, Nancarrow DJ, et al. (2008) Similarity of aberrant DNA methylation in Barrett’s esophagus and esophageal adenocarcinoma. Mol Cancer 7: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zheng Y, Zhang Y, Huang X, Chen L (2011) Analysis of the RUNX3 gene methylation in serum DNA from esophagus squamous cell carcinoma, gastric and colorectal adenocarcinoma patients. Hepatogastroenterology 58: 2007–2011. [DOI] [PubMed] [Google Scholar]
- 17. Hiramatsu T, Osaki M, Ito Y, Tanji Y, Tokuyasu N, et al. (2005) Expression of RUNX3 protein in human esophageal mucosa and squamous cell carcinoma. Pathobiology 72: 316–324. [DOI] [PubMed] [Google Scholar]
- 18. Tonomoto Y, Tachibana M, Dhar DK, Onoda T, Hata K, et al. (2007) Differential expression of RUNX genes in human esophageal squamous cell carcinoma: downregulation of RUNX3 worsens patient prognosis. Oncology 73: 346–356. [DOI] [PubMed] [Google Scholar]
- 19. Long C, Yin B, Lu Q, Zhou X, Hu J, et al. (2007) Promoter hypermethylation of the RUNX3 gene in esophageal squamous cell carcinoma. Cancer Invest 25: 685–690. [DOI] [PubMed] [Google Scholar]
- 20. Davila ML, Hofstetter WL (2013) Endoscopic management of Barrett’s esophagus with high-grade dysplasia and early-stage esophageal adenocarcinoma. Thorac Surg Clin 23: 479–489. [DOI] [PubMed] [Google Scholar]
- 21. Ek WE, Levine DM, D’Amato M, Pedersen NL, Magnusson PK, et al. (2013) Germline Genetic Contributions to Risk for Esophageal Adenocarcinoma, Barrett’s Esophagus, and Gastroesophageal Reflux. J Natl Cancer Inst 105: 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramaniam MM, Chan JY, Yeoh KG, Quek T, Ito K, et al. (2009) Molecular pathology of RUNX3 in human carcinogenesis. Biochim Biophys Acta 1796: 315–331. [DOI] [PubMed] [Google Scholar]
- 23. Chuang LS, Ito Y (2010) RUNX3 is multifunctional in carcinogenesis of multiple solid tumors. Oncogene 29: 2605–2615. [DOI] [PubMed] [Google Scholar]
- 24. Jang BG, Kim WH (2011) Molecular pathology of gastric carcinoma. Pathobiology 78: 302–310. [DOI] [PubMed] [Google Scholar]
- 25. Kohya N, Koga Y, Kitajima Y, Miyazaki K (2006) Aberrant promoter hypermethylation in biliary tract carcinoma. J Hepatobiliary Pancreat Surg 13: 296–305. [DOI] [PubMed] [Google Scholar]
- 26. Lee JH, Pyon JK, Kim DW, Lee SH, Nam HS, et al. (2011) Expression of RUNX3 in skin cancers. Clin Exp Dermatol 36: 769–774. [DOI] [PubMed] [Google Scholar]
- 27. Lee JM, Shin JO, Cho KW, Hosoya A, Cho SW, et al. (2011) Runx3 is a crucial regulator of alveolar differentiation and lung tumorigenesis in mice. Differentiation 81: 261–268. [DOI] [PubMed] [Google Scholar]
- 28. Lee YM (2011) Control of RUNX3 by histone methyltransferases. J Cell Biochem 112: 394–400. [DOI] [PubMed] [Google Scholar]
- 29. Levanon D, Brenner O, Otto F, Groner Y (2003) Runx3 knockouts and stomach cancer. EMBO Rep 4: 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shima K, Nosho K, Baba Y, Cantor M, Meyerhardt JA, et al. (2011) Prognostic significance of CDKN2A (p16) promoter methylation and loss of expression in 902 colorectal cancers: Cohort study and literature review. Int J Cancer 128: 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tamura G (2004) Promoter methylation status of tumor suppressor and tumor-related genes in neoplastic and non-neoplastic gastric epithelia. Histol Histopathol 19: 221–228. [DOI] [PubMed] [Google Scholar]
- 32. Tamura G (2006) Alterations of tumor suppressor and tumor-related genes in the development and progression of gastric cancer. World J Gastroenterol 12: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhao C, Bu X (2012) Promoter methylation of tumor-related genes in gastric carcinogenesis. Histol Histopathol 27: 1271–1282. [DOI] [PubMed] [Google Scholar]
- 34. Sakakura C, Miyagawa K, Fukuda KI, Nakashima S, Yoshikawa T, et al. (2007) Frequent silencing of RUNX3 in esophageal squamous cell carcinomas is associated with radioresistance and poor prognosis. Oncogene 26: 5927–5938. [DOI] [PubMed] [Google Scholar]
- 35. Siewert JR, Ott K (2007) Are squamous and adenocarcinomas of the esophagus the same disease? Semin Radiat Oncol 17: 38–44. [DOI] [PubMed] [Google Scholar]
- 36. Anaparthy R, Gaddam S, Kanakadandi V, Alsop BR, Gupta N, et al. (2013) Association between length of Barrett’s esophagus and risk of high-grade dysplasia or adenocarcinoma in patients without dysplasia. Clin Gastroenterol Hepatol 11: 1430–1436. [DOI] [PubMed] [Google Scholar]
- 37. Montgomery E, Bronner MP, Goldblum JR, Greenson JK, Haber MM, et al. (2001) Reproducibility of the diagnosis of dysplasia in Barrett esophagus: a reaffirmation. Hum Pathol 32: 368–378. [DOI] [PubMed] [Google Scholar]
- 38. Montgomery E, Bronner MP, Greenson JK, Haber MM, Hart J, et al. (2002) Are ulcers a marker for invasive carcinoma in Barrett’s esophagus? Data from a diagnostic variability study with clinical follow-up. Am J Gastroenterol 97: 27–31. [DOI] [PubMed] [Google Scholar]
- 39. Montgomery E, Goldblum JR, Greenson JK, Haber MM, Lamps LW, et al. (2001) Dysplasia as a predictive marker for invasive carcinoma in Barrett esophagus: a follow-up study based on 138 cases from a diagnostic variability study. Hum Pathol 32: 379–388. [DOI] [PubMed] [Google Scholar]
- 40. Voon DC, Wang H, Koo JK, Nguyen TA, Hor YT, et al. (2012) Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells 30: 2088–2099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PRISMA checklist.
(DOC)
PRISMA flowchart.
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.