Abstract
Sleep is an essential process and yet mechanisms underlying it are not well understood. Loss of the Drosophila quiver/sleepless (qvr/sss) gene increases neuronal excitability and diminishes daily sleep, providing an excellent model for exploring the underpinnings of sleep regulation. Here, we used a proteomic approach to identify proteins altered in sss brains. We report that loss of sleepless post-transcriptionally elevates the CG7433 protein, a mitochondrial γ-aminobutyric acid transaminase (GABAT), and reduces GABA in fly brains. Loss of GABAT increases daily sleep and improves sleep consolidation, indicating that GABAT promotes wakefulness. Importantly, disruption of the GABAT gene completely suppresses the sleep phenotype of sss mutants, demonstrating that GABAT is required for loss of sleep in sss mutants. While SSS acts in distinct populations of neurons, GABAT acts in glia to reduce sleep in sss flies. Our results identify a novel mechanism of interaction between neurons and glia that is important for the regulation of sleep.
Keywords: sleep, GABA transaminase, glia, mitochondria, quiver/sleepless, Drosophila
Introduction
Sleep remains among the most poorly understood of all biological phenomena, and disruptions of sleep are associated with many neuropsychiatric and neurological disorders (1). However, little is known about the molecular mechanisms that underlie sleep. Although wake-promoting and sleep-promoting networks in the brain appear to be involved in switching behavioral states (2), the mechanisms by which different inter-cellular signals interact with each other and/or respond to physiological states of the brain to drive wake or sleep is largely unknown.
With any such process about which little is known, the best approach is a random, unbiased one that makes no assumptions about the molecules that may be involved. To this end, researchers have undertaken forward genetic screens in model organisms to identify sleep-regulating genes. One such screen in Drosophila led to the identification of sleepless (sss), a small GPI-anchored protein whose loss results in dramatic reductions in daily sleep, frequently >80% (3). The sleepless mutation is an allele of the quiver1 locus identified previously by Wu and colleagues (4) in screening for mutants hypersensitive to reactive oxygen species. SSS protein was found to be a positive regulator of the voltage-gated potassium channel, Shaker, so in the absence of SSS, neural excitability is increased (5, 6). Loss of Shaker also increases excitability and reduces daily sleep (7). However, the Shaker mutants exhibit a much smaller decrease in sleep than sleepless mutants, suggesting that loss of Shaker function does not account for all the mechanisms underlying the sleep phenotype of sleepless.
We took an unbiased approach towards identifying the mechanisms underlying the sleep phenotype of sss mutants by conducting proteomic analysis of brain protein abundance. Our data reveal a novel connection between sss and the mitochondrial metabolism of γ-aminobutyric acid (GABA) by GABA transaminase (GABAT) in glial cells. sss fly brains display Increased GABAT and reduced GABA levels. We show that GABAT is a wake-promoting factor in Drosophila, and loss of GABAT completely suppresses the phenotype of the sss mutant and restores sleep. Tissue specific rescue experiments reveal that GABAT is required in glia to promote wakefulness in response to neuronal loss of sleepless. Interestingly, changes in GABAT are implicated in epilepsy and in neuropsychiatric disorders, which may account for some of the sleep abnormalities reported in these disorders (8).
Materials and Methods
Fly strains and DNA constructs
All fly stocks were maintained on standard molasses-cornmeal-yeast food. An isogenic w1118 (iso31) strain was used as wild type in this study(3). sssp1, Δsss (Δ40), and UAS-sss were described by Koh et al(3, 5). The Gad1-GAL4, VGAT-GAL4, and Repo-GAL4 were gifts from Gero Miesenböck, Julie Simpson, and Vanessa Auld. The F01602 insertion was ordered from the Exelixis Collection at Harvard Medical School and the PL00338 insertion from the Bloomington Stock Center. Both F01602 and PL00338 stocks were outcrossed into the iso31 background for 7 generations. Wild type control strains (Con PL00338 and Con F01602) were established from siblings of PL00338 and F01602 heterozygotes prior to the cross for homozygosity.
To generate the gGABATtvh genomic transgene, a 4.9 kb HpaI/HindIII DNA fragment from the Bac clone BACR13N10 (BACPAC Resources Center) containing CG7433 was inserted into a pattB vector (9) with flanking FRT sites (Fig. 1B). To disable the Tom20 gene in the construct, the Lys6 residue of Tom20 was mutated to an amber stop codon (AAA->TAA) by two primers, TomutF, 5’ GTA ATA TGA TTG AAA TGA ACT AAA CTG CAA TCG GCA TTG 3’, and tomutR, 5’ CAA TGC CGA TTG CAG TTT AGT TCA TTT CAA TCA TAT TAC 3’. A 29 amino acid thrombin-V5-His6 tandem tag (LVPRGSGKPI PNPLLGLDST RTGHHHHHH) was then added to the C-terminus of CG7433. The final pattB-FRT-GABATtvh construct was used to generate transgenic flies with the phiC31 technique (9), which targets the transgene to the zh-attP-96E locus on the third chromosome (Rainbow Transgenic Flies, Inc). Transformants were confirmed with respect to insertion sites at the zh-attP-96E locus by genomic DNA PCR (with two primers: attBlanding5’, 5’ GAT CCA CTA GTG TCG ACG ATG 3’ and ZH96E, 5’ CGA AAT GTC GGC ATA TTG TG 3’) and outcrossed to iso31 background for 7 generations.
Figure 1.
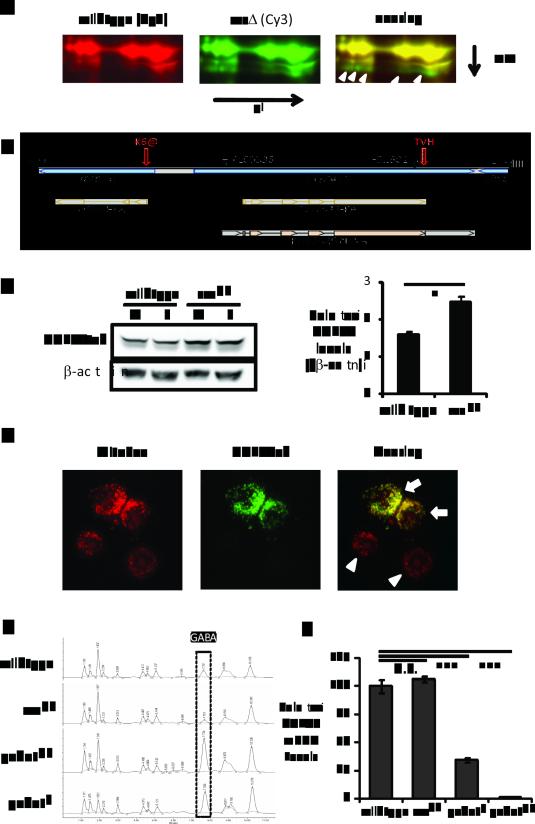
GABAT is increased and GABA is decreased in sss flies. (A) 2D-DIGE of sss and wild type brain proteins. Left, proteins in wild type brains stained with Cy3. Middle, proteins in sss mutants stained with Cy5. Right, overlay image of Cy5 and Cy3 staining. Shown in the image is only a zoom-in crop of the 2D-DIGE images. The yellow color denotes similar protein levels and the green color indicates proteins increased in sss mutants. White open arrows point to the protein spots that are increased in sss and identified by mass spectrometry as CG7433 positive. (B) Schematic of the genomic DNA structure around gene CG7433 (GABAT). Shown as a bar with a black border is the 4.9 kb genomic DNA from the Bac clone BACR13N10, flanked by HpaI and HindIII sites. The shaded diagram at the bottom depicts the proteins of the CG7433 and the neighboring Tom20 gene, as well as the CG7433 cDNA clone RH42429. Closed arrowheads show the positions of the two transposon insertions, PL00338 and F01602. Indicated by the red open arrows are two modifications to the gCG7433tvh genomic construct: a thrombin-V5-His6 tandem tag (TVH) added to the C-terminus of the CG7433 open reading frame (ORF) and the mutation of the Lys6 residue of TOM20 to an amber stop codon (K6@) to disable the Tom20 gene. (C) Western blot analysis (Left) and quantitation (Right) of GABATtvh protein derived from the gGABATtvh genomic transgene. V5 staining detected higher GABATtvh protein levels in the sssp1 mutants than in wild type for both male (M) and female (F) flies. Actin staining was used as a loading control. Shown in the right chart is the quantification of the left panel, the averages of GABAT/beta-actin ratios from both male and female samples in either a wild type or sss genetic background. *: P<0.05 (Student's t-test). (D) Localization of epitope tagged Drosophila GABAT to mitochondria in S2 cells. MitoSOX marks mitochondria (left panel, red) and V5 staining (middle panel, green) denotes the expression of GABATvh. Note that the lower two cells (white arrow heads) are negative control cells exhibiting only MitoSOX staining but not V5, indicating the cells were not transfected with pIZ-GABATvh. (E) HPLC amino acid analysis to compare GABA levels from fly brains. X-axis is elution time. Y-axis is absorbance at 254 nm, indicating the level of PITC-conjugated amino acids. The boxed peaks are GABA levels from the different genotypes shown on the left. (F) Reverse transcription quantitative PCR analyzing brain GABAT mRNA levels. GABAT mRNAs are first normalized to β-actin mRNA levels then to those in wild type flies which were set as 100%. GABAT mRNA levels from sssp1 were not different from those in wild type (P=0.385), while the GABAT mRNAs from gabatf and gabatpl were significantly reduced to 34% and 1%, respectively (P<0.001, One-way ANOVA with Tukey’s test).
To generate GABAT DNA constructs for GABAT constitutive overexpression in S2 cells, we slightly modified the pIZ-V5/His A vector (Life Technologies, Grand Island, NY) to generate a second NotI site after the V5-His6 tag to generate the DNA vector pIZ-VHn. The primers XbaI-BglII_CG7433f (5' aaa tct aga tct GAA ATT GAT AAA ATC CGA AC 3') and XbaI-CG7433r (5' aaa TCT AGA ATA CCT TGA AGA ACC TTG 3') were used to amplify the Drosophila CG7433 cDNA clone RH42429 (Drosophila Genomics Resource Center) and the XbaI fragment of the PCR product was inserted into pIZ-VHn to generate pIZ-GABATvh where the Drosophila GABAT is tagged with a V5-His6 tag at the C-terminus. To generate the UAS-GABATvh transgene, the 1.8 kb NotI DNA fragment from pIZ-GABATvh containing the entire V5-His6 tagged CG7433 cDNA was subcloned into the NotI site of pUAST-attB (9) to generate a UAS-GABATvh construct, which was used to generate UAS-GABATvh transgenic flies with the transgene targeted to the 3rd chromosome at the zh-attP-96E site.
Circadian behavior and sleep assays
To evaluate the locomotor activity rhythm, circadian period and sleep, flies were loaded in glass tubes containing standard molasses-cornmeal-yeast food, except for the experiments to rescue sss with GAL4 driven UAS-sss expression, where 5% sucrose with 2% agar was the food. For the gene-switch (GS) experiments or for the experiments where effects of drugs were tested, RU486 or drugs were added to the food as indicated. Flies were first entrained at 25°C in 12:12LD for two days and locomotor activity was recorded using the Drosophila Activity Monitoring System (Trikinetics, Waltham, MA) for 4 days in 12:12LD or > 7 days in constant darkness (DD) at 25°C. Activity counts were collected in 1 min bins and analyzed using ClockLab (Actimetrics, Wilmette, IL). Rhythmicity was determined initially by χ2 periodogram analysis for flies that had more than 5 days of activity data; flies were deemed arrhythmic if the activity data revealed no significant period and FFT (fast Fourier transform) values were lower than 0.01. For sleep analysis, only flies that survived the entire experiment were included in data analysis. pySolo (10) or a Matlab based custom software (3) was used for sleep analysis with the sleep definition being continued immobility of at least 5 minutes. Two methods were used to test for statistical difference in total (daily) sleep, daytime sleep, nighttime sleep, sleep bout number, sleep latency, and activity index in among genotypes. We used Student’s t-test for pair-wise comparison between two genotypes and One-way ANOVA with Tukey’s test for comparison among three or more genotypes. These tests were done using JMP 9 or Graphpad Prism 5 software. As sleep bout length is not normally distributed, it cannot be depicted as an average. Thus, we determine how sleep is distributed among bouts of different length. We use the Mann-Whitney U (MWU) test in Microsoft Excel to compare two genotypes in terms of how their sleep bout numbers are distributed among four different duration categories (5-15, 15-50, 50-150, and >150 minutes) and then use the Bonferroni method to adjust for multiple genotype comparison. Fraction of sleep time in each of the four categories of sleep bout duration is then plotted for qualitative comparison.
2D-DIGE proteomic analysis
4-6 days old wild type and sssΔ flies were entrained to 12:12LD at 25°C and collected in dark immediately before ZT0 (Zeitgeber Time 0, when lights are turned on). About 700 brains (~350 each from males and females) from each genotype were dissected in PBS buffer (pH7.4) on ice and stored at −80°C before protein extraction. Protein extraction, CyDye Fluor labeling, 2D-DIGE, data imaging, and statistical analysis of protein levels with DeCyder were performed at the Proteomic Core Facility of the University of Pennsylvania, as described by Marouga et al (11). Briefly, after protein extraction, a portion of protein was partially labeled (1-3% of total protein being labeled) with three CyDyes for each 2D-gel for imaging and statistical analysis: 50 μg wild type protein was labeled with Cy3, 50 μg sssΔ protein was labeled with Cy5, and 25 μg wild type plus 25 μg sssΔ protein was labeled with Cy2. To increase the protein amount for protein spot identification, 300 μg wild type plus 300 μg of sssΔ protein was loaded onto the third 2D-gel in addition to the CyDye labeled samples. The first dimension was run on pH3-10NL isoelectric focusing stripes (Amersham) and the second dimension on 30 cm x 30 cm SDS-PAGE gels (Jule Inc). After electrophoresis, Cy3, Cy5 and Cy2 images were scanned and the volumes of protein spots were analyzed using DeCyder software. The Cy2 signal was used as internal control across all three independent gels. The sssΔ:wild type volume ratios for each protein spot were determined relative to the Cy2 signal as internal standard and were used to calculate changes in average abundance and statistical probability by Student’s t-test across all three gels. Protein spots with an average of more than 1.5 fold difference and statistical significance (P<0.01) were marked and some of them were selected to be picked up by robot, trypsin-digested, and subjected to LC tandem mass-spectrometry for protein identification. The tandem mass spectrometry data were then analyzed by the Sequest software against the Drosophila proteome database to identify protein candidates corresponding to the spots. CG7433 was identified as a likely candidate gene from five spots.
Amino acid analysis
Flies were entrained at 12:12LD, 25°C and collected for brain dissection before the lights-on transition (ZT0). Twenty brains from each genotype were homogenized in 100 μl water on ice and subsequently deproteinized by passing through a filter with a 10,000-Dalton molecular weight cut-off (Millipore, Bedford, MA) via centrifugation. Flow-through for each genotype (40 μl) was then subjected to phenylisothiocyanate (PITC) derivatization. After the samples were cleaned up and dried, they were dissolved in 100 μl Pico-Tag diluent according to the Pico-Tag manual from the manufacturer (Waters, Milford, MA). GABA (10 nmole) and a few other L-amino acid standards were also treated in parallel with the brain samples to identify elution time of these amino acids. A Pico-Tag free amino acid analysis column (3.9 x 300 mm) was attached to a Waters HPLC system and run at 1 ml/min at ambient temperature. After equilibrating with Buffer A [70 mM sodium acetate (pH 6.45 with 10% acetic acid)–acetonitrile (975: 25)] (Waters), for each run, 50 μl of brain samples or standards were injected onto the column and eluted with standard gradients consisting of combinations of Buffer A and Buffer B [water–acetonitrile–methanol (40: 45: 15)], as recommended for physiologic amino acids by the manufacturer. At room temperature, GABA standards were eluted by 100% buffer A at 7.31 ± 0.06 minutes (mean ± stdev).
Immunohistochemistry
To staining for GABAT expression with MitoSOX in Schneider 2 cells, Drosophila Schneider 2 (S2) cells were cultured in Schneider’s Drosophila Medium (Invitrogen) at 25°C with standard techniques. Before transfection, cells were dislodged into fresh medium and diluted to 106 cells/ml. pIZ-GABATvh (50 ng/106 cells) were prepared to transfect the S2 cells with Effectene Transfection Reagent (Qiagen) according to the manufacturer’s protocol. After transfection, the cells were dispended 0.5×106 cells/well onto a Lab-Tek chambered coverglass (Fisher Scientific, Pittsburgh, PA). Forty eight hours later, the cells were then incubated with 1 μM MitoSOX in PBS buffer (pH 7.4) for 5 minutes to mark for mitochondria. After washing with PBS buffers, the cells were fixed with 4% para-formaldehyde (PFA) in PBS for 30 min at room temperature. After three 5 min washes with 0.3% Triton X-100 in PBS, samples were incubated with 5% normal donkey serum (NDS) in PBS-Triton X-100 and with primary antibodies rabbit anti-V5 (Bethyl Laboratories) in 5% NDS (1:1000 dilution) for 1 hour each at room temperature. After three 20 min washes, cells were incubated with donkey FITC anti-rabbit secondary antibody (1:300 dilution) (Jackson ImmunoResearch Laboratories) in 5% NDS for 2h at room temperature, followed by extensive washes. Cells were mounted with Vectashield mounting medium (Vector) and protein staining (FITC) was compared with MitoSOX staining using a Leica TCS SP5 confocal microscope. Untransfected cells without a FITC signal served as negative controls that only stain with MitoSOX.
To co-stain GABATvh and GAD1-YFP in the brains, GAD1CPTI000977/+;gGABATtvh/+ transheterozygous flies were dissected in PBS buffers. After fixing with 4% para-formaldehyde (PFA) in PBS for 30 min at room temperature, the brains were washed with PBS extensively and sequentially incubated with 5% NDS in PBS-Triton X-100 and with Alexa Fluor 647 conjugated V5 mouse monoclonal antibody (Invitrogen) (1:1000 dilution) in 5% NDS followed by extensive washes. The brains were mounted with Vectashield mounting medium (Vector) and V5 staining for GABATtvh (Alexa Fluor 647) was compared with the YFP signal from GAD1-YFP using a Leica TCS SP5 confocal microscope.
Quantitative real-time PCR and Western blot analysis
Flies were collected for brain dissection in 12:12LD, 25°C less than 30 minutes before the lights-on transition (ZT0). Brains were dissected on ice and total RNA was isolated using Trizol isolation system (Invitrogen) from the brain, and cDNAs were synthesized by using a high-capacity cDNA Archive kit (Life Technologies, Grand Island, NY). Quantitative real-time PCR (qPCR) was performed using an ABI prism 7100 with a SYBR Green kit (Life Technologies).
For Western blot analysis with fly heads, flies were collected on dry ice before lights-on (ZT0) in 12:12LD, 25°C and fly heads were separated for protein extraction using standard cell lysis protocol (12). After SDS-PAGE, proteins were transferred onto nitrocellulose membrane and processed for antibody incubation. Primary antibodies mouse anti-V5, and mouse anti-β-Actin were used at 1:3000 dilution. Following enhanced chemiluminescence, images were taken (12) via a Kodak image station.
Results
GABA transaminase is increased in sleepless mutants
To identify mechanisms underlying the sleep phenotype of sleepless (sss) flies, we screened for proteins mis-regulated in sss brains. We collected wild type and sss flies at the end of the night, following entrainment to 3 days of 12hr-light:12hr-dark (12:12LD) cycles, dissected ~700 brains for each genotype and subjected the extracts to two dimensional Differential In Gel Electrophoresis (2D-DIGE) (13). A >1.5 fold change and statistically significant (P<0.01) difference in abundance between the two genotypes was used as a cut-off to select 20 protein spots for identification by LC tandem mass spectrometry analysis (LC-MS/MS). A gene annotated CG7433 was identified as a leading candidate in five of these 20 spots, showing a 2-3 fold increase in the sss mutant (Fig. 1A). To confirm that the sss mutation leads to an increase in CG7433, we generated a transgene of the CG7433 genomic locus with a Thrombin-V5-His6 tandem tag (TVH) attached to the C-terminus of the coding sequence (Fig. 1B). This genomic transgene, termed gCG7433tvh, was introduced into wild type and sss mutant flies and levels of the epitope tagged transgenic CG7433 protein (CG7433tvh) were assayed by western blot analysis using an anti-V5 antibody. Levels of the CG7433tvh protein were increased in the brains of both male and female sss flies, based upon western blots (Fig. 1C, average of 54% increase, P<0.05) and protein purification from fly heads (data not shown), confirming the proteomic results that CG7433 is increased in sss mutants. However, CG7433tvh levels showed no detectable increase in flies deficient for Shaker (data not shown), which could be due to the weaker sleep phenotype of Shaker or a Shaker-independent effect of sleepless on CG7433.
Tissue specific mRNA profiling indicated that CG7433 expression is highly enriched in the adult brain and thoracic-abdominal ganglia (14). It is also broadly expressed in neuropil-associated glia and subperineurial glia during late embryonic stages (15). Analysis of the coding sequence revealed that CG7433 encodes a protein that has a 23-amino-acid N-terminal mitochondrial signal peptide (16) and its primary amino acid sequence is highly similar to both human and pig γ-aminobutyric acid transaminases (GABAT) (56% identity) (Fig. S1). Pig GABAT requires pyridoxal 5’-phosphate (PLP) as a co-factor for its enzymatic activity and it homodimerizes via two cysteines to form a [Fe2-S2] cluster (17). BLAST analysis indicates that all 8 amino acids required for PLP binding and the two cysteine residues required for homodimerization are conserved between CG7433, pig, and human GABATs (Fig. S1).
We next expressed CG7433 with a C-terminal V5-His6 tag in Drosophila S2 cells and confirmed that it colocalizes with a mitochondrial dye, MitoSox (Fig. 1D). GABATs typically break down GABA in the mitochondria by removing an amino group to form succinic semialdehyde (SSA). To further confirm the predicted function of CG7433 as a GABA transaminase, we performed amino acid analysis to evaluate GABA levels in brain extracts of wild type flies, sssp1 mutants and two putative mutant alleles of CG7433 (PL00338 and F01602). The PL00338 allele consists of a piggybac GAL4 enhancer trap insertion in the 5’ untranslated region of CG7433 and F01602 contains an Exelixis piggybac WH insertion in the last exon of CG7433, disrupting 20 amino acids prior to the C-terminus (Fig. 1B) (18, 19). The sssp1 mutation dramatically reduces brain GABA levels to about ~30% of those in wild type flies, while PL00338 and F01602 boost GABA levels ~2 and 3 fold respectively (Fig. 1E). These data support the identification of CG7433 as a GABA transaminase (GABAT) and so we refer to it as such hereafter, and refer to the genomic transgene gCG7433tvh and the two alleles, PL00338 and F01602, as gGABATtvh, gabatpl and gabatf, respectively.
Notably, GABAT is detected as multiple spots horizontally on 2D gels (Fig. 1A), likely due to post-translational modifications that lead to significant changes in isoelectric point but little change in molecular weight. Analysis of GABAT mRNA levels from brains by reverse transcription quantitative PCR revealed no significant difference between wild type and sss flies (Fig. 1F). This supports the idea that the altered GABAT levels in sss mutants also occur at a translational or post-translational level. On the other hand, GABAT mRNA levels are negligible in the gabatpl mutant (1% of wild type level) and reduced to 35% in gabatf. Thus, gabatpl is overall a more severe allele than gabatf. However, because the transposon in the gabatf allele is located in the last exon of the protein coding region, the reduced GABAT mRNA is predicted to produce a truncated protein. Through these experiments we established that GABAT levels are increased post-transcriptionally and GABA levels are decreased in the brains of sss flies. Conversely, disruption of the GABAT gene increases GABA in fly brains.
GABA transaminase is a negative regulator of sleep
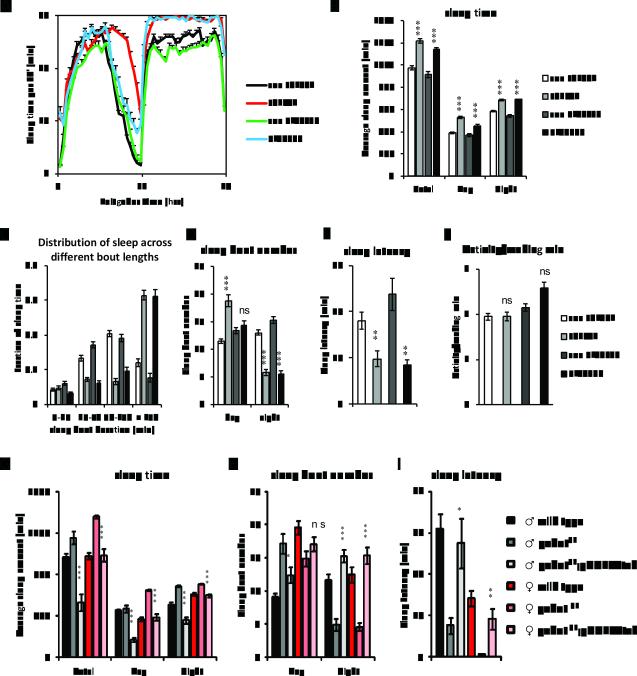
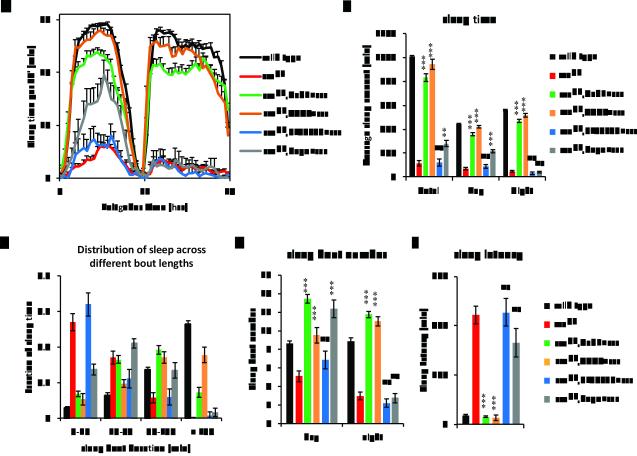
GABA is a major inhibitory neurotransmitter that promotes sleep in both mammals and Drosophila (2, 20-22). However, it is not known how a mitochondrial enzyme such as GABAT transaminase might affect sleep. Since gabatpl and gabatf mutants have increased GABA in the brain, we asked if they promote sleep. Sleep was assayed in the gabat mutant flies and their respective genetic background controls (con PL00338 and con F01602), derived from outcrossing at 25°C, in the presence of standard 12:12LD cycles (3). GABAT mutant flies showed significant increases in both daytime and nighttime sleep, amounting to an increase in total daily sleep time of >3 hrs for each of the two mutants, gabatf and gabatpl (Fig. 2A and 2B). Strikingly, mutant flies also showed a dramatic increase in the fraction of sleep spent in long (>150min/sleep episode) sleep episodes, with such episodes accounting for 50-60% of total daily sleep for the mutants compared with approximately 20% for wild type controls (Fig. 2C). In addition, the mutants exhibited a marked decrease in sleep episode number at night (Fig. 2D). Longer episode length and reduced episode number together indicate better consolidation of sleep at night in the gabat mutants. During the daytime, an increase in episode number also contributes to the increased sleep of gabatf flies. Lastly, we also measured sleep latency, or the time the flies take to fall asleep after lights-off at ZT12, an indicator of mechanism underlying sleep initiation (23). The GABAT mutant flies showed significantly reduced sleep latency compared with their respective wild type controls (Fig. 2E). However, when awake, these mutants showed at least as much locomotor activity as controls (Fig. 2F), suggesting that they are not hypoactive. To summarize, loss of GABA transaminase promotes daily sleep amount and sleep consolidation in Drosophila.
Figure 2.
Loss of GABAT promotes sleep. (A-F) Male GABAT mutants and control flies from outcrossing were monitored for sleep behavior in 12hr light:12hr dark cycles (12:12LD) at 25°C. Daily sleep profiles (A) and parameters of sleep behavior (B-F) were compared between each of the two GABAT mutants (gabatf and gabapl) and their respective wild type genetic background controls (Con F01602 and Con PL00338). (A) X-axis denotes the time of day, with Zeitgeber Time (ZT) 0-12 denoting lights-on and ZT12-24 denoting lights-off. The Y-axis shows the amount of sleep per 30 minute bins. (G-I) A gGABATtvh transgene was used to rescue the sleep phenotype of PL00338 under 12:12LD at 25°C. Sleep time (G), sleep bout number (H), and sleep latency (I) were compared among male (♂) and female (♀) flies from wild type control (iso31), gabat mutant (gabatpl), and genomic rescue (gabatpl;gGABATtvh) flies. For (B-F), Student’s t-test was performed between each GABAT mutant and its respective control for all sleep parameters, except for the distribution of sleep bouts based on their duration, where the Mann-Whitney U (MWU) test was used. The mutants exhibited increased average total, daytime, and night time sleep (all P<0.001) (B), increased proportion of sleep in longer sleep episodes (Mann-Whitney U test, both P<0.0001) (C), increased number of sleep bouts (daytime, F01602 vs Con F01602 P<0.001, PL00338 vs Con PL00337, not significant; nighttime, both P<0.001) (D) and decreased sleep latency (both P<0.01) (E). However, the waking activity (F) was not significantly different between mutants and controls. For (G-I),One-way ANOVA with Tukey’s test was used to examine the differences in sleep time, bout number, and latency for male and female flies respectively. Asterisks show statistical significance between gabatpl and gabatpl,gGABATtvh (***, P<0.001; **, P<0.01; *, P<0.05; ns, not significant).
Next we examined if the gGABATtvh genomic transgene could rescue the increased sleep in the gabatpl mutant. Indeed, introduction of the genomic GABAT transgene reduced sleep of gabatpl to levels similar to or lower than those of wild type controls (Fig. 2G). In addition, the mutants carrying the genomic transgene showed increased sleep bout number and reduced sleep bout duration, demonstrating a reduction in sleep consolidation (Fig. 2H and data not shown). Finally, sleep latency of the gabatpl mutant was also restored to wild type levels (Fig. 2I). These data demonstrate that loss of the GABAT protein accounts for the altered sleep time and sleep architecture in gabatpl flies. Thus, GABAT transaminase is a wake-promoting molecule in Drosophila melanogaster.
To exclude the possibility that a dysregulated circadian clock causes the sleep phenotypes in gabat mutants (24), we tested the locomotor behavior of gabat mutants in constant darkness. Although gabatf flies showed some arrhythmicity, most gabatpl flies were rhythmic and neither genotype showed statistical differences in circadian period from wild type controls (Fig. 3H and Table S1). The lack of a circadian phenotype in gabatpl , which is a very severe allele of gabat and has a strong sleep phenotype, indicates that clock disruption does not cause the sleep phenotype. More likely, the excessive sleep in some gabatf flies masks an underlying circadian rhythm. Double mutant analysis (see below) supported the idea that circadian rhythms are largely unimpaired in gabat flies. These findings also indicate that the GABAergic inputs required for maintenance of circadian rhythms in Drosophila (21, 25) are not directly impacted by loss of GABAT.
Figure 3.
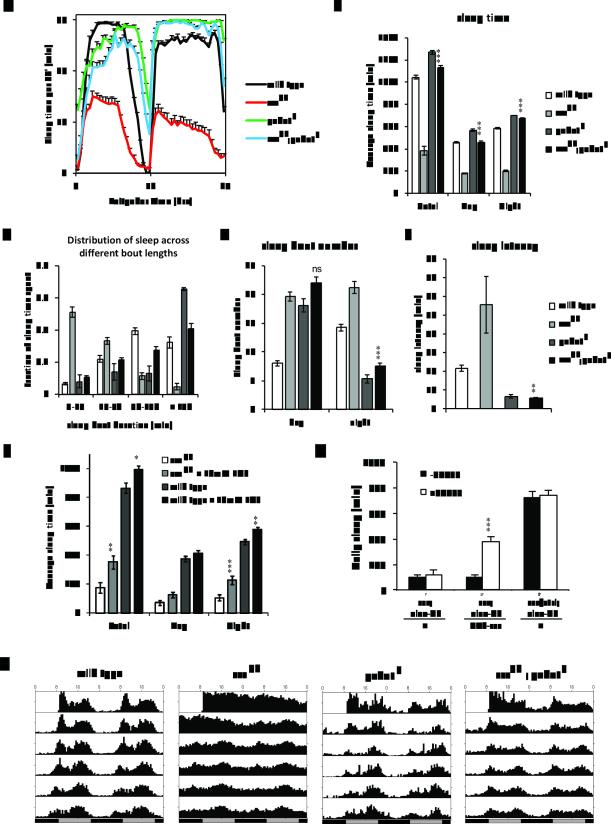
Disruption of gabat completely suppresses the short sleep phenotype of sssp1 flies. To determine the effect of the gabat mutation on the sss phenotype, sssp1;gabatf male flies were compared with sssp1, gabatf and wild type control flies for daily sleep profiles (A), sleep time (B), distribution of sleep with respect to the length of sleep episodes (C), number of sleep bouts (D), and sleep latency (E). (F) Inhibition of GABAT in adults partially rescues the sss mutant. We treated wild type and sss flies with an inhibitor specific for GABAT, EOS, and compared day, night, and total sleeptime. sssp1 and wild-type males (between 1-5 days of age) were fed either 10mM EOS or control food for 5 days. We averaged sleeptime of flies from days 3-5 to ensure that the drug was ingested. (G) The sssP1 phenotype results from adult and developmental effects. To restrict sss expression to adults, we used an elav-GS driver to conditionally rescue sssp1 adults. Daily sleep was monitored in females in the presence (white bar) or absence (black bar) of 500μM RU486. Heterozygous (sss/ctrl) flies were included as positive controls. (H) gabatf improves circadian rest:activity rhythms of sssp1. Male flies were first entrained to 12:12 Light:Dark cycles at 25°C for 4 days before the flies were transferred into constant darkness for assay of free-running rhythms. The figure shows average 30 minute activity bins from flies of the respective groups recorded for 7 days in continuous darkness (DD), starting from day 1 (DD1). One-way ANOVA with Tukey’s test was used to examine the differences in sleep time, bout number, and latency. Asterisks depict the results of statistical tests between sssp1 and sssp1;gabatf (B-E) or control and EOS-treated sssp1 and wild-type flies (F) ***, P<0.001; **, P<0.01; ns, not significant. MWU test was used to test for differences in sleep bout distribution. gabatf rescued the short sleep bout duration phenotype of sss (MWU test with Bonferroni adjustment, sssp1 vs sssp1;gabatf P<0.0001; gabatf vs sssp1;gabatf, P=0.92). Unpaired t-tests with Bonferroni correction were used to test for differences in daily sleep in (G). *** P<0.001.
Disruption of GABAT restores sleep specifically in sss mutants
To test if the severely reduced sleep in sss is due to elevated GABAT levels, we crossed gabat mutations into a sssP1 background to reduce GABAT. Indeed, the gabatf mutation suppressed the sleep phenotype of sssp1 mutants, completely restoring nighttime sleep to levels observed in the gabatf mutants alone (Fig. 3A and 3B). In addition, the gabatf;sssp1 double mutants showed an increase in the time spent in long sleep episodes (>150 min/sleep episode) and reduced nighttime sleep episode number, equivalent to that in gabatf flies (Fig. 3C and 3D, respectively). The increased sleep latency in sss flies was also completely suppressed in the sssp1;gabatf double mutant to the level of gabatf mutants (Fig. 3E). Even though stronger suppression of the short sleep phenotype was observed when the stronger gabatpl allele was coupled with sssp1 (Fig. S2), only the data for sssp1;gabatf double mutants are shown here for the sake of easier comparison with data from datfmn;gabatf double mutants (see details below, Fig. 4).
Figure 4.
Disruption of gabat does not restore wild type sleep in datfmn flies. The double mutant datfmn;gabatf male flies were compared with datfmn and gabatf flies for daily sleep profiles (A), sleep time (B), distribution of sleep with respect to the length of sleep episodes (C), number of sleep bouts (D), and sleep latency (E). Statistics was applied as in Figure 3. Asterisks indicate results of one-way ANOVA followed by a posthoc Tukey’s test. ***, P<0.001; **, P<0.01. While gabatf showed longer sleep bout duration than both datfmn and datfmn;gabatf flies (MWU test with Bonferroni adjustment, P<0.0001 for both), datfmn;gabatf did not show differences from datfmn in sleep bout duration (MWU test with Bonferroni adjustment, P=0.273).
To test whether the sss P1 phenotype is caused by high GABAT expression in adults, and not only during development, we conditionally inhibited the enzyme in adult male flies using the pharmacological agent ethanolamine O-sulfate (EOS), which is a specific, competitive, and catalytic inhibitor of GABAT (26). 10mM EOS treatment partially rescued nighttime sleep in sssp1 (Fig. 3F), suggesting that the sss p1 phenotype is at least in part due to increased GABAT activity in adults. Importantly, these data indicate that the relevant GABA/GABAT circuits are in place in sss adults for the drug to have an effect. Results from a genetic rescue experiment of sssp1, using an inducible panneuronal driver (elav-GS) to express sss specifically in adults, corroborated these findings. Partial rescue of the sss phenotype was obtained, supporting an adult function and perhaps also an effect during development (Fig. 3G).
Although sss flies have a functional clock (Fig. 3H), they display weak activity rhythms, most likely because of the reduced sleep (3). We found that gabatf also strengthens circadian activity rhythms of sssp1 and improves rhythmic activity in constant darkness with no apparent change in circadian period (Fig. 3H and Table S1). However, neither gabat mutant rescued the ether-induced leg shaking phenotype of sssp1 (data not shown), clearly indicating that different mechanisms underlie the leg shaking and loss of sleep phenotypes of sss.
Since gabat increases sleep and sss decreases sleep, independent and additive effects of the two mutants should produce only a moderate restoration of sleep in the gabatf;sssp1 double mutant compared with sss. The complete and epistatic suppression of night-time sleep and overall sleep quality of sss by mutations in gabat indicates that an additive effect is unlikely. In other words, the two genes appear to be in the same sleep-regulating pathway with the effect of the gabat mutant being epistatic to that of sss. Nevertheless, we investigated the possibility of additive effects by testing if gabat mutants restore sleep in a different severe short-sleeping mutant datfmn, which has a single nucleotide change that disrupts the dopamine transporter gene (27).
Since datfmn;gabatpl flies are homozygous-lethal, we compared the behavior of datfmn;gabatf flies with that of each of the two single mutants. Male datfmn;gabatf flies exhibited ~6 hrs of daily sleep, which represents a significant increase over ~1.5 hrs in datfmn flies, but only accounting for ~30-40% of the daily sleep seen in wild type control flies (Fig. 4A and 4B). Meanwhile, the increase in sleep in the datfmn;gabatf flies was largely during the daytime, with the increase in nighttime restricted mostly to the very early night; over the course of the night, sleep in the double mutant decreased to levels seen in datfmn single mutants (Fig. 4A). Furthermore, sleep in datfmn;gabatf flies was as or even more fragmented than in datfmn flies, in contrast to gabat mutants and sssp1;gabatf double mutants where long sleep bout durations are indicative of well-consolidated sleep (compare Fig. 3C with Fig. 4C). In fact, the increased sleep in datfmn;gabatf over datfmn was largely due to an increase in sleep bout number (Fig. 4D). Interestingly, although it was largely ineffective in rescuing other aspects of sleep architecture, gabatf rescued sleep latency in the datfmn flies (Fig. 4E). As latency to sleep is measured after lights-off in the early evening, increased sleep at this time may have accounted for the decreased latency in the double mutant. These data indicated that incorporation of gabatf in the short sleeping dat mutant only restores sleep latency and not any other sleep parameters.
The differential rescue of the various sleep parameters of datfmn mutants by gabatf indicates that genetic interactions between DAT and GABAT are complicated. In the double mutant datfmn;gabatf flies, the sleep promoting effect of gabatf mutation dominates to reduce sleep onset and sleep amount during the early evening (Fig. 4A). However, the wake promoting datfmn dominates in shortening the sleep bout length throughout the night and reducing sleep amount during the late evening. On the other hand, all sleep parameters of sss mutants are completely suppressed by the gabat mutants (Fig. 3). If high levels of GABAT (and low GABA) were always a consequence of short sleep, disruption of gabat would be expected to suppress all short-sleeping mutants. Given that GABAT promotes wakefulness (Fig. 2) and loss of GABAT strongly suppresses the short-sleep phenotype of sss (Fig. 3) but not that of datfmn (Fig. 4), we conclude that elevated GABAT is relevant for the sleep loss of sss mutants.
SSS likely acts in GABAergic neurons to promote sleep
To identify the cellular link between SSS and GABAT, we next examined whether restoring SSS expression in GABA producing neurons rescues sleep in sss mutants. In Drosophila, GABA is synthesized by glutamic acid decarboxylase 1 (Gad1) and packaged presynaptically into vesicles by the vesicular GABA transporter (VGAT). It is widely accepted that once GABA is released, it can be re-taken up by pre-synaptic neurons for recycling or by apposed glial cells for degradation (28).
We showed previously that sss cDNA, expressed panneuronally or in cholinergic cells with the UAS-GAL4 system, rescues the sleep phenotype of sss mutants (5). In light of the suppression of sss by gabatf, we drove expression of UAS-sss in GABAergic neurons of sssp1 animals using either a Gad1-GAL4 (29) or a VGAT-GAL4 transgene (30). Both GABAergic GAL4 drivers were able to significantly restore daily sleep to 80% and 90% that of wild type, respectively (Fig. 5A and 5B), with effects on both daytime and nighttime sleep. The restored sleep was due to both lengthened sleep bout duration and increased bout number (Fig. 5C and 5D). Lastly, SSS expression in GABAergic cells completely rescued sleep latency to wild type levels (Fig. 5E). Thus, directing UAS-sss expression to GABAergic cells using either Gad1-GAL4 or VGAT-GAL4 can rescue the sss mutant. The rescue by VGAT-GAL4 is consistently better than that by Gad1-GAL4 as it restores higher levels of nighttime sleep and longer sleep bout duration. Since expression of SSS in either GABAergic or cholinergic neurons almost completely rescues the sleep phenotypes of sss, we suggest that at least a subset of SSS expressing neurons are both GABAergic and cholinergic.
Figure 5.
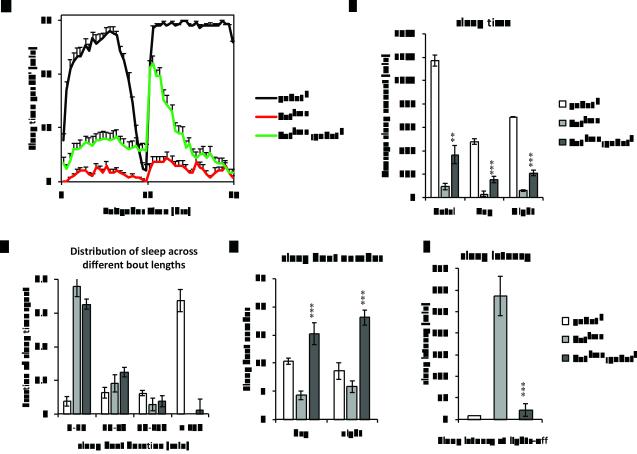
GABAergic expression of SSS rescues sssp1. To rescue sleep in sss flies, daily sleep behavior was tested at 12:12LD, 25°C in sssp1 animals with UAS-sss expression driven by various GAL4 drivers in either GABAergic neurons (Gad1-GAL4, green or VGAT-GAL4, orange), GABAT expressing neurons (PL00338, blue), or glial cells (Repo-GAL4, grey). Wild type (black) and sssp1 (red) animals were used as positive and negative controls, respectively. The average of 3 day behavior from three independent experiments was plotted with standard error of means. Shown in the figures are average daily sleep profiles (A), sleep time (B), distribution of sleep with respect to the length of sleep episodes (C), number of sleep bouts (D), and sleep latency (E). Statistics was applied as in Figure 3. Asterisks depict statistical comparisons with sssp1. ***, P<0.001; **, P<0.01; *, P<0.05; ns, not significant. Compared with sssp1 controls, overexpression of SSS with either Gad1-GAL4 or VGAT-GAL4 rescues sleep bout duration (MWU test with Bonferroni adjustment, P<0.0001 for both drivers), while neither PL00338 nor Repo-GAL4 do (MWU test with Bonferroni adjustment, P=0.771 and 0.118, respectively).
We also tried to rescue sss by driving expression of SSS in GABAT cells, using the GAL4 enhancer trap allele of GABAT (PL00338), and in glia, using Repo-GAL4. Other than a slight (2-hour) increase in daytime sleep driven by Repo-GAL4, neither driver rescued the sss short sleep phenotype. Both drivers failed to rescue the following sleep parameters of the sss mutant—nighttime sleep, sleep bout duration, nighttime sleep bout number, and sleep latency (Fig. 5A-E). Therefore, while SSS may function in GABAergic cells, it does not act to modulate sleep in GABAT cells, nor in glia. Thus, loss of sleepless may act non-cell-autonomously to up-regulate GABAT levels.
GABAT acts in glia to modulate sleep in sss mutants
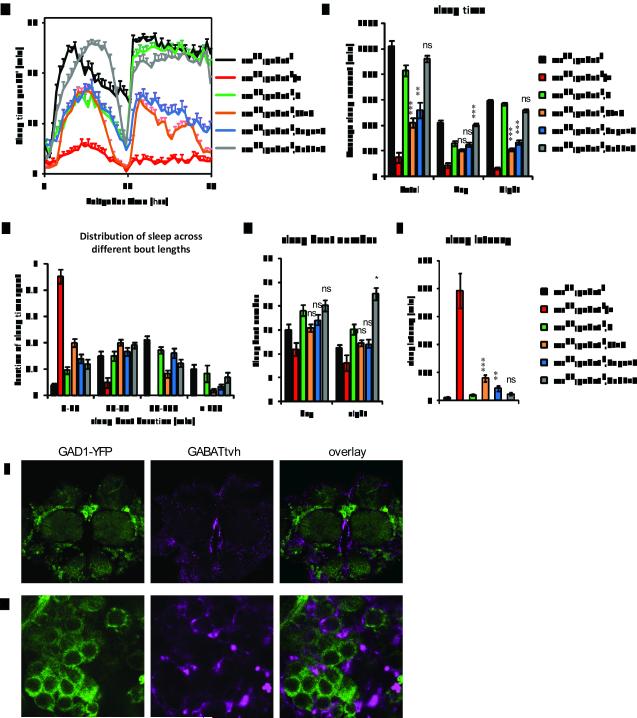
To further confirm the non-cell-autonomous regulation of GABAT by the loss of sleepless, we next asked where GABAT expression is required for the short sleep phenotype of sss mutants. To this end, we generated a UAS transgene of V5-His6 tagged GABAT (UAS-GABATvh) and drove its expression in different cells of the sssp1;gabatf double mutant using various GAL4 drivers. As expected, presence of the UAS-GABATvh transgene alone had minimal effects on sleep in sssp1;gabatf flies (Fig. 6). However, restoring the expression of GABAT in GABAT cells with the GAL4 enhancer trap allele, PL00338, diminished nighttime sleep to ~35% that of sss;gabat (Fig. 6A and 6B, compare orange with blackxs), indicating partial restoration of the sss phenotype (Fig 6A, compare orange with red). In addition to a reduction in total sleep, the sleep was more fragmented and sleep latency increased, even though the number of sleep bouts remained unchanged (Fig. 6C to 6E). These data demonstrate that expression of GABAT in its native pattern of expression can restore a sss-like phenotype in sss;gabat double mutants. Strikingly, restoring the expression of GABAT in glial cells with Repo-GAL4 was equally effective in reducing sleep of the sss;gabat double mutants (Fig. 6A and 6B, compare black, orange and blue). The restoration in glia was also accompanied by fragmented sleep bouts and increased sleep latency (Fig. 6C to 6E). On the other hand, expression of GABAT by the Gad1-GAL4 driver did not restore the short-sleeping phenotype of sssp1 (Fig. 6, grey), although Gad1-Gal4 driven expression of SSS is sufficient to rescue the sssp1 mutant (Fig. 5). These data indicate that GABAT expression in glia is required to promote wakefulness in the sss mutant. Thus, SSS is likely required in GABAergic neurons for normal sleep and loss of SSS non-cell-autonomously modulates GABA transaminase in glia, indicating that a neuron-glia interaction mediates the loss of sleep in sss.
Figure 6.
Glial expression of GABAT partially restores the sssp1 sleep phenotype in sssp1;gabatf flies. Daily sleep behavior was tested at 12:12LD, 25°C in sssp1;gabatF animals carrying UAS-GABATvh driven by GAL4 drivers in native GABAT expressing neurons (PL00338, orange), glial cells (Repo-GAL4, blue) , or GABAergic neurons (Gad1-GAL4, grey). sssp1;gabatf (black) and sssp1;gabatf, UAS-GABATvh (green) were used as negative controls, while sssp1;gabatf/+ (red) animals were used as positive controls. The average of 4 day behavior was plotted with standard error of means. Shown in the figures are average daily sleep profiles (A), sleep time (B), distribution of sleep with respect to the length of sleep episodes (C), number of sleep bouts (D), and sleep latency (E). Statistics was applied as in Figure 3. Asterisks indicate statistical comparisons with sssp1;gabatf,UAS-GABATvh. ***, P<0.001; **, P<0.01; *, P<0.05; ns, not significant. Compared with sssp1;gabatf,UAS-GABATvh controls, overexpression of GABAT in either PL00338 or Repo-GAL4 expressing cells fragments the sleep bout length (MWU test with Bonferroni adjustment, P<0.0001 for both drivers) , but not in Gad1-GAL4 expressing cells (P=0.47). Note, U: UAS-GABATvh; PL: PL00338; Repo: Repo-GAL4; Gad1: Gad1-GAL4. (F-G) Confocal imaging of GAD1-YFP and GABATtvh in Drosophila brains indicates distinct but often proximate staining patterns of GAD1 and GABAT expression. GABATtvh was stained by an Alexa Fluor 647 anti-V5 antibody. The YFP and Alexa Fluor 647 signals were imaged for GAD1-YFP (left panels, green) and GABATtvh (middle panels, magenta) expression patterns with the overlay images shown in the right panels. Representative central brain (F) and medial lateral brain (G) regions are shown.
We next compared the protein expression patterns of GABAT and GAD1, using the tagged GABATtvh transgene and a transposon insertion, carrying a yellow fluorescent protein (YFP) fluorescent marker, in the Gad1 gene, CPTI000977 (31). The CPTI000977 insertion truncates the GAD1 protein and fuses a YFP reporter to its C-terminus (GAD1-YFP). Fluorescent images of brains stained for GABATtvh and GAD1-YFP are largely different from each other (Fig. 6F and 6G). GABATvh is often found neighboring, but not overlapping with the GAD1-YFP signal (Fig. 6G), suggesting that in Drosophila GABAT is largely expressed in non-GABAergic cells. Interestingly, CG1732, the GABA:sodium symporter, required for GABA re-uptake from extra-cellular space in Drosophila, is expressed in a subset of lateral glial cells in the ventral nerve cord at embryonic stage 15-16 and in glia-like cells in adult brains (32, 33). Expression of CG1732 has thus far only been reported in glia, which is consistent with the idea that GABAT acts in glia to break down GABA following re-uptake. Together the expression data support our behavioral analysis that expression of GABAT in non-GABAergic cells, specifically glia, is required for the short sleep phenotype in sleepless mutants.
Discussion
GABA is an inhibitory neurotransmitter that promotes sleep in both mammals and Drosophila. In fact, benzodiazepines, a class of GABAa receptor blockers, are widely prescribed for insomnia (34). However, little is known about the regulation of GABA signaling in the context of sleep:wake cycles. Our data demonstrate that a mitochondrial enzyme, GABA transaminase, expressed in glia regulates GABA levels to affect sleep. GABA transaminase catalyzes GABA turnover to succinic semialdehyde, which in turn is turned over into succinic acid by succinic semialdehyde dehydrogenase, recycling the carbon backbone to the Krebs cycle (35). We note that the role of mitochondria in the control of GABA levels is generally under-appreciated, but our work here indicates that it provides an important link between cellular metabolism and GABA-regulated processes such as sleep.
Notably, it was an unbiased approach that led us to the discovery of the Drosophila CG7433 gene, which turns out to encode a GABA transaminase (GABAT) upregulated in sss brains. We propose that increased GABAT is a mechanism underlying the loss of sleep in sss, rather than a mere consequence of sleep loss, for the following reasons. Firstly, disruption of the GABAT gene results in increased GABA levels, total sleep, and sleep consolidation, indicating that GABAT itself is a bona fide negative regulator of sleep. Secondly, sss mutant brains show increased GABAT and decreased GABA levels, which are predicted to decrease sleep and/or sleep consolidation. Furthermore, increased GABAT as a homeostatic response to the loss of sleep in sss is unlikely as it would only exacerbate the sleep loss. Lastly, disruption of GABAT completely and specifically represses the sleep phenotype of sssp1, confirming both that GABAT has a role in sleep regulation and that GABAT acts downstream of SSS to modulate sleep. Another sleep promoting molecule downstream of GABAT activity is γ-hydroxybutyric acid (GHB) (36). Due to low similarity of the primary sequence of this enzyme among different species, we are uncertain if Drosophila contains succinic semialdehyde reductase, which catalyzes a product of GABAT, SSA, into GHB. However, increased GABAT activity in sss and loss of GABAT in gabat would be predicted to increase and reduce downstream GHB levels, and hence sleep, respectively, contrary to the observed sleep phenotypes of sss and gabat. We therefore exclude GHB as relevant in the mechanism underlying the sleep phenotypes of sss and gabat.
Latency to sleep is used as a clinical measurement of sleepiness for human patients, and was shown by Griffith and colleges to be regulated by the kinetics of GABAa receptor signaling in Drosophila (21). Patients with sleep onset insomnia have prolonged latency to sleep. We show that latency to sleep onset is altered in both sss and gabat mutants, severely lengthened in the sss mutants (Fig. 3E) and reduced in gabat mutants (Fig. 2E), supporting the idea that GABAT affects GABAa signaling to regulate sleep. In addition, although the gabat mutants, like other sleep mutants, accumulate modifiers of the baseline sleep phenotype over time, their reduced latency to sleep remains intact (data not shown). Interestingly, the gabat sleep latency phenotype even dominates in both double mutant backgrounds – sssp1; gabatf as well as datfmn;gabatf, in which loss of gabat is coupled with potentiated dopamine signaling (Fig. 4E). The latter suggests that the sleep promoting effect of gabat is dominant to the wake promoting effect of dopamine at lights-off (ZT12, when sleep latency was measured). The reduced sleep during the later night and the short duration of sleep bouts in datfmn;gabatf animals (Fig. 4A and 4C) indicate that the potentiated dopamine signaling dominates over gabat as the night proceeds. This is vastly different to the increased sleep throughout the night in gabat or sss;gabat mutant flies (Fig. 2A and Fig. 3A).
We showed previously that sss mutants can be rescued by expression of SSS in cholinergic neurons. The present study, indicating a role for GABAergic neurons is not inconsistent with the previous result, as co-expression of GABA and acetylcholine has been reported previously (37). We suggest that loss of sss in cholinergic cells increases cholingeric signaling, which may generally reduce sleep. However, the effect on GABAT reported here likely leads to a lasting and profound sleep loss.
Our data indicate that loss of SSS non-cell-autonomously affects glial GABAT. It is not clear how this cell non-autonomy is achieved, especially given that GABAT is a mitochondrial protein. As loss of SSS causes hyperactivity of neurons that normally express it, this may lead to higher energy demands on surrounding glial cells via a glia-neuron metabolic shuttle, such as the lactate-pyruvate shuttle (38), and affect mitochondrial metabolism in glia. A computational model simulating an astrocyte–GABAergic neuron cellular complex predicts that substantially increasing GABAergic neuron activity would lead to both increased lactate release from glia to neurons and increased GABA turnover to SSA in glia (i.e. increased GABAT activity) (32). We note that increased excitability of GABAergic neurons in sss mutants may not only increase GABAT in glia, but could also ultimately cause depletion of GABA from the GABAergic neurons. Indeed, repetitive stimulation of GABAergic neurons is reported to deplete GABA and reduce the number of recycling vesicles (39). Mammalian data suggest that GABA can also be released from glia in a tonic fashion through an anion channel (40). Should this be the case, modulating GABAT activity would regulate the glial GABA level and therefore its tonic release to the extra-neuronal space, providing a means to regulate sleep through tonic GABA inhibition (41).
An alternative explanation for the increased GABAT is that increased activity of GABAergic neurons triggers a homeostatic response in neighboring glia. As noted earlier, we did not observe increased GABAT in Shaker mutants although these also have high neural activity and display decreased sleep. It is important to note though that the sleep phenotype of Shaker mutants is much weaker than that of sss flies(3, 7). It is not even known if Shaker and SSS act in the same cells to regulate sleep, although SSS affects Shaker activity at the larval neuromuscular junction in a cell-autonomous manner (5). Even if they do act in the same cells, effects of SSS may be greater as it appears to affect channels other than just Shaker (unpublished observations). Regardless of the differences between SSS and Shaker, an effect of neural activity on GABAT in glia is probably not restricted to sss mutants. High neural activity in epilepsy is associated with sleep disruption (42) and, interestingly, it is commonly treated by an inhibitor of GABAT, vigabatrin(43). To date there are only 3 reported cases of patients with GABAT deficiency, compared with hundreds of cases for deficiencies of other enzymes of the GABA shunt such as glutamic acid decarboxylase and succinic semialdehyde dehydrogenase (44, 45), hindering genetic and clinical studies on the human enzyme. These patients exhibited elevated GABA concentrations in the plasma, the cerebrospinal fluid (44) or the basal ganglia (45), and displayed symptoms that include hypotonia and lethargy. Vigabatrin, an irreversible inhibitor of mammalian GABAT used clinically, is known to induce sleepiness in humans (46), and promote non-rapid-eye-movement (NREM) sleep in amygdala-kindled rats (47). Thus, GABAT may have a conserved role in the regulation of sleep. Our data, generated as a result of an unbiased proteomic analysis, provide a mechanistic basis and an animal model to understand the role of GABAT in sleep regulation. Importantly, our studies show that GABAT can be regulated through a cell-non-autonomous and post-transcriptional mechanism.
Supplementary Material
Acknowledgements
The authors thank the Penn Proteomics Core for conducting the 2D-DIGE gel analysis, Erin Forbeck and Xiaobo Wan for help with the HPLC analysis and members of the laboratory for helpful discussions.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Spiegelhalder K, Regen W, Nanovska S, Baglioni C, Riemann D. Comorbid sleep disorders in neuropsychiatric disorders across the life cycle. Curr Psychiatry Rep. 2013 Jun;15(6):364. doi: 10.1007/s11920-013-0364-5. [DOI] [PubMed] [Google Scholar]
- 2.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep state switching. Neuron. 2010 Dec 22;68(6):1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koh K, Joiner WJ, Wu MN, Yue Z, Smith CJ, Sehgal A. Identification of SLEEPLESS, a sleep-promoting factor. Science. 2008 Jul 18;321(5887):372–376. doi: 10.1126/science.1155942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang JW, Humphreys JM, Phillips JP, Hilliker AJ, Wu CF. A novel leg-shaking Drosophila mutant defective in a voltage-gated K(+)current and hypersensitive to reactive oxygen species. J Neurosci. 2000 Aug 15;20(16):5958–5964. doi: 10.1523/JNEUROSCI.20-16-05958.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu MN, Joiner WJ, Dean T, Yue Z, Smith CJ, Chen D, et al. SLEEPLESS, a Ly-6/neurotoxin family member, regulates the levels, localization and activity of Shaker. Nat Neurosci. 2010 Jan;13(1):69–75. doi: 10.1038/nn.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean T, Xu R, Joiner W, Sehgal A, Hoshi T. Drosophila QVR/SSS modulates the activation and C-type inactivation kinetics of Shaker K(+) channels. J Neurosci. 2011 Aug 3;31(31):11387–11395. doi: 10.1523/JNEUROSCI.0502-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, et al. Reduced sleep in Drosophila Shaker mutants. Nature. 2005 Apr 28;434(7037):1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 8.Sherif FM, Ahmed SS. Basic aspects of GABA-transaminase in neuropsychiatric disorders. Clin Biochem. 1995 Apr;28(2):145–154. doi: 10.1016/0009-9120(94)00074-6. [DOI] [PubMed] [Google Scholar]
- 9.Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilestro GF, Cirelli C. pySolo: a complete suite for sleep analysis in Drosophila. Bioinformatics. 2009 Jun 1;25(11):1466–1467. doi: 10.1093/bioinformatics/btp237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marouga R, David S, Hawkins E. The development of the DIGE system: 2D fluorescence difference gel analysis technology. Anal Bioanal Chem. 2005 Jun;382(3):669–678. doi: 10.1007/s00216-005-3126-3. [DOI] [PubMed] [Google Scholar]
- 12.Williams JA, Su HS, Bernards A, Field J, Sehgal A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science. 2001 Sep 21;293(5538):2251–2256. doi: 10.1126/science.1063097. [DOI] [PubMed] [Google Scholar]
- 13.Minden JS, Dowd SR, Meyer HE, Stuhler K. Difference gel electrophoresis. Electrophoresis. 2009 Jun;30(Suppl 1):S156–161. doi: 10.1002/elps.200900098. [DOI] [PubMed] [Google Scholar]
- 14.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007 Jun;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 15.Beckervordersandforth RM, Rickert C, Altenhein B, Technau GM. Subtypes of glial cells in the Drosophila embryonic ventral nerve cord as related to lineage and gene expression. Mech Dev. 2008 May-Jun;125(5-6):542–557. doi: 10.1016/j.mod.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem. 1996 Nov 1;241(3):779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 17.Storici P, De Biase D, Bossa F, Bruno S, Mozzarelli A, Peneff C, et al. Structures of gamma-aminobutyric acid (GABA) aminotransferase, a pyridoxal 5'-phosphate, and [2Fe-2S] cluster-containing enzyme, complexed with gamma-ethynyl-GABA and with the antiepilepsy drug vigabatrin. J Biol Chem. 2004 Jan 2;279(1):363–373. doi: 10.1074/jbc.M305884200. [DOI] [PubMed] [Google Scholar]
- 18.Hacker U, Nystedt S, Barmchi MP, Horn C, Wimmer EA. piggyBac-based insertional mutagenesis in the presence of stably integrated P elements in Drosophila. Proc Natl Acad Sci U S A. 2003 Jun 24;100(13):7720–7725. doi: 10.1073/pnas.1230526100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004 Mar;36(3):283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 20.Parisky KM, Agosto J, Pulver SR, Shang Y, Kuklin E, Hodge JJ, et al. PDF cells are a GABA-responsive wake-promoting component of the Drosophila sleep circuit. Neuron. 2008 Nov 26;60(4):672–682. doi: 10.1016/j.neuron.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agosto J, Choi JC, Parisky KM, Stilwell G, Rosbash M, Griffith LC. Modulation of GABAa receptor desensitization uncouples sleep onset and maintenance in Drosophila. Nat Neurosci. 2008 Mar;11(3):354–359. doi: 10.1038/nn2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R. The GABA(A) receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 2009 Mar 10;19(5):386–390. doi: 10.1016/j.cub.2009.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andretic R, Shaw PJ. Essentials of sleep recordings in Drosophila: moving beyond sleep time. Methods in enzymology. 2005;393:759–772. doi: 10.1016/S0076-6879(05)93040-1. [DOI] [PubMed] [Google Scholar]
- 24.Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002 May 16;417(6886):287–291. doi: 10.1038/417287a. [DOI] [PubMed] [Google Scholar]
- 25.Dahdal D, Reeves DC, Ruben M, Akabas MH, Blau J. Drosophila pacemaker neurons require g protein signaling and GABAergic inputs to generate twenty-four hour behavioral rhythms. Neuron. 2010 Dec 9;68(5):964–977. doi: 10.1016/j.neuron.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fowler LJ, John RA. Active-site-directed irreversible inhibition of rat brain 4-aminobutyrate aminotransferase by ethanolamine O-sulphate in vitro and in vivo. Biochem J. 1972 Nov;130(2):569–573. doi: 10.1042/bj1300569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kume K, Kume S, Park SK, Hirsh J, Jackson FR. Dopamine is a regulator of arousal in the fruit fly. J Neurosci. 2005 Aug 10;25(32):7377–7384. doi: 10.1523/JNEUROSCI.2048-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ge S, Pradhan DA, Ming GL, Song H. GABA sets the tempo for activity-dependent adult neurogenesis. Trends Neurosci. 2007 Jan;30(1):1–8. doi: 10.1016/j.tins.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenbock G. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 2002 Oct 24;36(3):463–474. doi: 10.1016/s0896-6273(02)00975-3. [DOI] [PubMed] [Google Scholar]
- 30.Fei H, Chow DM, Chen A, Romero-Calderon R, Ong WS, Ackerson LC, et al. Mutation of the Drosophila vesicular GABA transporter disrupts visual figure detection. J Exp Biol. 2010 May;213:1717–1730. doi: 10.1242/jeb.036053. Pt 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knowles-Barley S, Longair M, Armstrong JD. BrainTrap: a database of 3D protein expression patterns in the Drosophila brain. Database (Oxford) 2010:baq005. doi: 10.1093/database/baq005. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belelli D, Peden DR, Rosahl TW, Wafford KA, Lambert JJ. Extrasynaptic GABAA receptors of thalamocortical neurons: a molecular target for hypnotics. J Neurosci. 2005 Dec 14;25(50):11513–11520. doi: 10.1523/JNEUROSCI.2679-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thimgan MS, Berg JS, Stuart AE. Comparative sequence analysis and tissue localization of members of the SLC6 family of transporters in adult Drosophila melanogaster. J Exp Biol. 2006 Sep;209:3383–3404. doi: 10.1242/jeb.02328. Pt 17. [DOI] [PubMed] [Google Scholar]
- 34.Crow JM. Insomnia: chasing the dream. Nature. 2013 May 23;497(7450):S16–18. doi: 10.1038/497S16a. [DOI] [PubMed] [Google Scholar]
- 35.Yogeeswari P, Sriram D, Vaigundaragavendran J. The GABA shunt: an attractive and potential therapeutic target in the treatment of epileptic disorders. Curr Drug Metab. 2005 Apr;6(2):127–139. doi: 10.2174/1389200053586073. [DOI] [PubMed] [Google Scholar]
- 36.Wong CG, Bottiglieri T, Snead OC., 3rd GABA, gamma-hydroxybutyric acid, and neurological disease. Ann Neurol. 2003;54;(Suppl 6):S3–12. doi: 10.1002/ana.10696. [DOI] [PubMed] [Google Scholar]
- 37.O'Malley DM, Sandell JH, Masland RH. Co-release of acetylcholine and GABA by the starburst amacrine cells. J Neurosci. 1992 Apr;12(4):1394–1408. doi: 10.1523/JNEUROSCI.12-04-01394.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellerin L. Brain energetics (thought needs food) Curr Opin Clin Nutr Metab Care. 2008 Nov;11(6):701–705. doi: 10.1097/MCO.0b013e328312c368. [DOI] [PubMed] [Google Scholar]
- 39.Wang L, Tu P, Bonet L, Aubrey KR, Supplisson S. Cytosolic transmitter concentration regulates vesicle cycling at hippocampal GABAergic terminals. Neuron. 2013 Oct 2;80(1):143–158. doi: 10.1016/j.neuron.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 40.Lee S, Yoon BE, Berglund K, Oh SJ, Park H, Shin HS, et al. Channel-mediated tonic GABA release from glia. Science. 2010 Nov 5;330(6005):790–796. doi: 10.1126/science.1184334. [DOI] [PubMed] [Google Scholar]
- 41.Occhipinti R, Somersalo E, Calvetti D. Energetics of inhibition: insights with a computational model of the human GABAergic neuron-astrocyte cellular complex. J Cereb Blood Flow Metab. 2010 Nov;30(11):1834–1846. doi: 10.1038/jcbfm.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Derry CP, Duncan S. Sleep and epilepsy. Epilepsy Behav. 2013 Mar;26(3):394–404. doi: 10.1016/j.yebeh.2012.10.033. [DOI] [PubMed] [Google Scholar]
- 43.Hemming K, Maguire MJ, Hutton JL, Marson AG. Vigabatrin for refractory partial epilepsy. Cochrane Database Syst Rev. 2013;(1):CD007302. doi: 10.1002/14651858.CD007302.pub2. [DOI] [PubMed] [Google Scholar]
- 44.Medina-Kauwe LK, Tobin AJ, De Meirleir L, Jaeken J, Jakobs C, Nyhan WL, et al. 4-Aminobutyrate aminotransferase (GABA-transaminase) deficiency. J Inherit Metab Dis. 1999 Jun;22(4):414–427. doi: 10.1023/a:1005500122231. [DOI] [PubMed] [Google Scholar]
- 45.Tsuji M, Aida N, Obata T, Tomiyasu M, Furuya N, Kurosawa K, et al. A new case of GABA transaminase deficiency facilitated by proton MR spectroscopy. J Inherit Metab Dis. 2010 Feb;33(1):85–90. doi: 10.1007/s10545-009-9022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.US-FDA . Sabril Medication Guide. Administration USFaD; Washington, DC: 2012. [Google Scholar]
- 47.Raol YH, Meti BL. Effects of vigabatrin on sleep-wakefulness cycle in amygdala-kindled rats. Epilepsia. 2000 Feb;41(2):128–131. doi: 10.1111/j.1528-1157.2000.tb00131.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.