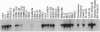Abstract
The ability to carry out high-resolution genetic mapping at high throughput in the mouse is a critical rate-limiting step in the generation of genetically anchored contigs in physical mapping projects and the mapping of genetic loci for complex traits. To address this need, we have developed an efficient, high-resolution, large-scale genome mapping system. This system is based on the identification of polymorphic DNA sites between mouse strains by using interspersed repetitive sequence (IRS) PCR. Individual cloned IRS PCR products are hybridized to a DNA array of IRS PCR products derived from the DNA of individual mice segregating DNA sequences from the two parent strains. Since gel electrophoresis is not required, large numbers of samples can be genotyped in parallel. By using this approach, we have mapped > 450 polymorphic probes with filters containing the DNA of up to 517 backcross mice, potentially allowing resolution of 0.14 centimorgan. This approach also carries the potential for a high degree of efficiency in the integration of physical and genetic maps, since pooled DNAs representing libraries of yeast artificial chromosomes or other physical representations of the mouse genome can be addressed by hybridization of filter representations of the IRS PCR products of such libraries.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cornall R. J., Aitman T. J., Hearne C. M., Todd J. A. The generation of a library of PCR-analyzed microsatellite variants for genetic mapping of the mouse genome. Genomics. 1991 Aug;10(4):874–881. doi: 10.1016/0888-7543(91)90175-e. [DOI] [PubMed] [Google Scholar]
- Cox R. D., Copeland N. G., Jenkins N. A., Lehrach H. Interspersed repetitive element polymerase chain reaction product mapping using a mouse interspecific backcross. Genomics. 1991 Jun;10(2):375–384. doi: 10.1016/0888-7543(91)90322-6. [DOI] [PubMed] [Google Scholar]
- Cox R. D., Lehrach H. Genome mapping: PCR based meiotic and somatic cell hybrid analysis. Bioessays. 1991 Apr;13(4):193–198. doi: 10.1002/bies.950130409. [DOI] [PubMed] [Google Scholar]
- Dietrich W., Katz H., Lincoln S. E., Shin H. S., Friedman J., Dracopoli N. C., Lander E. S. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics. 1992 Jun;131(2):423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein D. J., Vekemans M., Gros P. Splotch (Sp2H), a mutation affecting development of the mouse neural tube, shows a deletion within the paired homeodomain of Pax-3. Cell. 1991 Nov 15;67(4):767–774. doi: 10.1016/0092-8674(91)90071-6. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fleischman R. A., Saltman D. L., Stastny V., Zneimer S. Deletion of the c-kit protooncogene in the human developmental defect piebald trait. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10885–10889. doi: 10.1073/pnas.88.23.10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne C. M., McAleer M. A., Love J. M., Aitman T. J., Cornall R. J., Ghosh S., Knight A. M., Prins J. B., Todd J. A. Additional microsatellite markers for mouse genome mapping. Mamm Genome. 1991;1(4):273–282. doi: 10.1007/BF00352339. [DOI] [PubMed] [Google Scholar]
- Herrmann B. G., Labeit S., Poustka A., King T. R., Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990 Feb 15;343(6259):617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Hunter K. W., Ontiveros S. D., Watson M. L., Stanton V. P., Jr, Gutierrez P., Bhat D., Rochelle J., Graw S., Ton C., Schalling M. Rapid and efficient construction of yeast artificial chromosome contigs in the mouse genome with interspersed repetitive sequence PCR (IRS-PCR): generation of a 5-cM, > 5 megabase contig on mouse chromosome 1. Mamm Genome. 1994 Oct;5(10):597–607. doi: 10.1007/BF00411453. [DOI] [PubMed] [Google Scholar]
- Janocha S., Wolz W., Srsen S., Srsnova K., Montagutelli X., Guénet J. L., Grimm T., Kress W., Müller C. R. The human gene for alkaptonuria (AKU) maps to chromosome 3q. Genomics. 1994 Jan 1;19(1):5–8. doi: 10.1006/geno.1994.1003. [DOI] [PubMed] [Google Scholar]
- Lebeau M. C., Alvarez-Bolado G., Wahli W., Catsicas S. PCR driven DNA-DNA competitive hybridization: a new method for sensitive differential cloning. Nucleic Acids Res. 1991 Sep 11;19(17):4778–4778. doi: 10.1093/nar/19.17.4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledbetter S. A., Nelson D. L., Warren S. T., Ledbetter D. H. Rapid isolation of DNA probes within specific chromosome regions by interspersed repetitive sequence polymerase chain reaction. Genomics. 1990 Mar;6(3):475–481. doi: 10.1016/0888-7543(90)90477-c. [DOI] [PubMed] [Google Scholar]
- Lisitsyn N., Lisitsyn N., Wigler M. Cloning the differences between two complex genomes. Science. 1993 Feb 12;259(5097):946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- Love J. M., Knight A. M., McAleer M. A., Todd J. A. Towards construction of a high resolution map of the mouse genome using PCR-analysed microsatellites. Nucleic Acids Res. 1990 Jul 25;18(14):4123–4130. doi: 10.1093/nar/18.14.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly K. F. A Macintosh program for storage and analysis of experimental genetic mapping data. Mamm Genome. 1993;4(6):303–313. doi: 10.1007/BF00357089. [DOI] [PubMed] [Google Scholar]
- Montagutelli X., Lalouette A., Coudé M., Kamoun P., Forest M., Guénet J. L. aku, a mutation of the mouse homologous to human alkaptonuria, maps to chromosome 16. Genomics. 1994 Jan 1;19(1):9–11. doi: 10.1006/geno.1994.1004. [DOI] [PubMed] [Google Scholar]
- Rowe L. B., Nadeau J. H., Turner R., Frankel W. N., Letts V. A., Eppig J. T., Ko M. S., Thurston S. J., Birkenmeier E. H. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm Genome. 1994 May;5(5):253–274. doi: 10.1007/BF00389540. [DOI] [PubMed] [Google Scholar]
- Simmler M. C., Cox R. D., Avner P. Adaptation of the interspersed repetitive sequence polymerase chain reaction to the isolation of mouse DNA probes from somatic cell hybrids on a hamster background. Genomics. 1991 Jul;10(3):770–778. doi: 10.1016/0888-7543(91)90462-n. [DOI] [PubMed] [Google Scholar]
- Sinnett D., Deragon J. M., Simard L. R., Labuda D. Alumorphs--human DNA polymorphisms detected by polymerase chain reaction using Alu-specific primers. Genomics. 1990 Jul;7(3):331–334. doi: 10.1016/0888-7543(90)90166-r. [DOI] [PubMed] [Google Scholar]
- Tassabehji M., Read A. P., Newton V. E., Harris R., Balling R., Gruss P., Strachan T. Waardenburg's syndrome patients have mutations in the human homologue of the Pax-3 paired box gene. Nature. 1992 Feb 13;355(6361):635–636. doi: 10.1038/355635a0. [DOI] [PubMed] [Google Scholar]
- Ton C. C., Hirvonen H., Miwa H., Weil M. M., Monaghan P., Jordan T., van Heyningen V., Hastie N. D., Meijers-Heijboer H., Drechsler M. Positional cloning and characterization of a paired box- and homeobox-containing gene from the aniridia region. Cell. 1991 Dec 20;67(6):1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- Zehetner G., Lehrach H. The Reference Library System--sharing biological material and experimental data. Nature. 1994 Feb 3;367(6462):489–491. doi: 10.1038/367489a0. [DOI] [PubMed] [Google Scholar]