SUMMARY
The multipass membrane protein ACD6 forms large protein complexes and is subject to constitutive ER-associated degradation. Salicylic acid signaling increases the size of ACD6 complexes and the plasma membrane levels of ACD6 and the ACD6-associated pattern receptor FLS2. ACD6 is necessary for promoting increased levels of FLS2 and its co-receptor BAK1 specifically at the plasma membrane during salicylic acid (SA) signaling, suggesting that ACD6 maturation and plasma membrane levels are tightly controlled as a mechanism for regulating these pattern receptors.
Key words: ACD6, protein trafficking, protein quality control, salicylic acid
Abstract
ACCELERATED CELL DEATH6 (ACD6) is a multipass membrane protein with an ankyrin domain that acts in a positive feedback loop with the defense signal salicylic acid (SA). This study implemented biochemical approaches to infer changes in ACD6 complexes and localization. In addition to forming endoplasmic reticulum (ER)- and plasma membrane (PM)-localized complexes, ACD6 forms soluble complexes, where it is bound to cytosolic HSP70, ubiquitinated, and degraded via the proteasome. Thus, ACD6 constitutively undergoes ER-associated degradation. During SA signaling, the soluble ACD6 pool decreases, whereas the PM pool increases. Similarly, ACD6-1, an activated version of ACD6 that induces SA, is present at low levels in the soluble fraction and high levels in the PM. However, ACD6 variants with amino acid substitutions in the ankyrin domain form aberrant, inactive complexes, are induced by a SA agonist, but show no PM localization. SA signaling also increases the PM pools of FLAGELLIN SENSING2 (FLS2) and BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1). FLS2 forms complexes ACD6; both FLS2 and BAK1 require ACD6 for maximal accumulation at the PM in response to SA signaling. A plausible scenario is that SA increases the efficiency of productive folding and/or complex formation in the ER, such that ACD6, together with FLS2 and BAK1, reaches the cell surface to more effectively promote immune responses.
INTRODUCTION
Important components of the plant defense system include proteins at the cell surface that can perceive microbial patterns or pathogen effectors and amplify defense signaling, respectively (Spoel and Dong, 2012). The latter class of proteins includes ACCELERATED CELL DEATH 6 (ACD6), a positive regulator of cell death and defense responses in Arabidopsis that is present in cells at very low levels (Rate et al., 1999; Lu et al., 2003, 2005, 2009). The dominant gain-of-function mutant acd6-1 shows small stature, spontaneous cell death, and constitutively elevated defenses (Rate et al., 1999). Among the defenses affected in acd6-1 is the regulatory molecule salicylic acid (SA) (Vanacker et al., 2001). SA is required for acd6-1-conferred phenotypes and acd6-1 acts in part via the major SA transducer NON-EXPRESSOR of PR1 (NPR1; Rate et al., 1999). Recent characterization of natural variants of ACD6 suggests that some alleles confer defense-related phenotypes similar to those caused by acd6-1 (Todesco et al., 2010). Analysis of a T-DNA mutant and plants with one extra copy of ACD6, respectively, revealed that ACD6 is a dose-dependent regulator of SA accumulation and signaling that affects susceptibility to Pseudomonas syringae (Lu et al., 2003, 2005). ACD6 transcript levels are also induced by SA signaling (Lu et al., 2003). Hence, ACD6 functions in a positive feedback loop with SA (Lu et al., 2005).
The ACD6 gene encodes a predicted multipass membrane protein with an N-terminal domain harboring nine ankyrin repeat motifs that face the cytosol (Lu et al., 2003, 2005). The acd6-1 mutant has a L591F amino acid substitution in the membrane domain (Lu et al., 2003). Ankyrin-containing proteins have diverse functions, such as transcriptional initiators, cell cycle regulators, cytoskeletal components, ion transporters, and signal transducers (Bork, 1993). An ankyrin repeat typically contains 33 amino acid residues with few conserved residues that are important for maintaining stability of the structure (Gaudet, 2008). The variable residues on the surface of an ankyrin repeat are often involved in mediating protein–protein interactions (Gaudet, 2008). Interestingly, eight intragenic acd6-1 suppressors mapped to the ankyrin repeat domain, with four modeled to affect surface-accessible residues in non-consensus amino acids (Lu et al., 2005). Most suppressor mutants are hypersusceptible to P. syringae, suggesting they are loss-of-function mutants (Lu et al., 2005). These altered residues and the ankyrin repeats affected may be important for protein–protein interactions and the function of ACD6-1 and/or the wild-type ACD6 protein.
The effects of increased SA on plant cells are broad-ranging and include transcriptional reprogramming (Wang et al., 2006) and an increase in the capacity of secretory system to traffic pathogenesis-related proteins such as PR1 to the apoplast (Wang et al., 2005). The secretory pathway is important for the delivery of proteins to the extracellular milieu as well as for the maturation of proteins that target the plasma membrane (Vitale and Denecke, 1999). Since SA affects secretory pathway components (Wang et al., 2005), it is predicted that SA signaling also affects proteins that are trafficked to the plasma membrane, although this has not been carefully examined.
The endoplasmic reticulum (ER) is the initial site where proteins destined for membranes enter into the secretory pathway and the place where membrane proteins complete their folding to achieve their native conformations (Hammond and Helenius, 1995; Vitale and Denecke, 1999). ER quality control (ERQC) monitors this process, exporting only correctly folded and assembled protein complexes to their destination and retaining misfolded and misassembled proteins in the ER (Brodsky and Skach, 2011). Proteins retained in the ER are subject to ER-associated degradation (ERAD), in which proteins are retrotranslocated into cytosol, ubiquitin modified, and eventually broken down via cytoplasmic proteasomes (Meusser et al., 2005; Needham and Brodsky, 2013). Recognition of misfolded proteins is an important step in the ERAD pathway, which involves multiple chaperones (Goeckeler and Brodsky, 2010; Määttänen et al., 2010). These chaperones detect exposed hydrophobic regions and/or incompletely glycan-modified proteins (Römisch, 2005). Depending on which part of a protein is misfolded, cytosolic HSP70 and/or BIP (an HSP70 that resides in the ER lumen) can play important roles in substrate selection and targeting for degradation (Hendershot, 2004; Goeckeler and Brodsky, 2010). They do this by detecting hydrophobic regions of misfolded proteins. In case of glycoproteins, there is also a lectin chaperone calnexin/careticulin (CNX/CRT) system to detect immature glycan within a glycoprotein (Molinari, 2007; Hüttner and Strasser, 2012). Multipass membrane proteins present an added burden in the secretory pathway, since their maturation is more complex and often not efficient (Guerriero and Brodsky, 2012). An extensively studied example of a multipass membrane protein that is constitutively subject to ERAD is the human cystic fibrosis transmembrane conductance regulator (CFTR; Guerriero and Brodsky, 2012). Indeed, a large percentage of wild-type CFTR and certain disease-causing forms never reach maturity (Kopito, 1999).
The ERQC/ERAD system is conserved in eukaryotes, including plants (Guerra and Callis, 2012). Increasing evidence indicates that the ERQC/ERAD system plays important roles in plant immunity (Saijo, 2010; Eichmann and Schäfer, 2012; Tintor and Saijo, 2014). There is a relatively specific requirement for some ER quality-control components for the stability and maturation of the Arabidopsis Ef-Tu receptor important for perceiving an EF-Tu-derived microbial pattern in normal growth conditions and SUPPRESSOR OF BIR1 (SOBIR1) (Li et al., 2009; Nekrasov et al., 2009; Saijo et al., 2009; Sun et al., 2014). However, the underlying mechanism for how ERQC/ERAD is regulated in plants is lacking.
The relationship between SA signaling and the regulation and maturation of multipass plasma membrane proteins important for defense have has to be established. Here we report that, based on biochemical approaches, SA signaling affects the production, localization to the plasma membrane, and complex formation of ACD6. We show that ACD6 forms complexes with chaperones (HSP70 and BIP) and the single pass membrane protein FLAGELLIN SENSING2 (FLS2). FLS2 and its co-receptor BRI1-ASSOCIATED RECEPTOR KINASE 1 (BAK1) are involved in responses to microbial patterns, herbivores, and/or brassinosteroids (Li et al., 2002; Chinchilla et al., 2007; Heese et al., 2007; Yang et al., 2011). SA signaling affects the plasma membrane localization of FLS2 and BAK1 in a manner that depends to varying degrees on ACD6. This suggests that SA may broadly regulate the trafficking of integral membrane protein complexes that act in defense.
RESULTS
ACD6 and ACD6-1 Are Multipass Integral Membrane Proteins
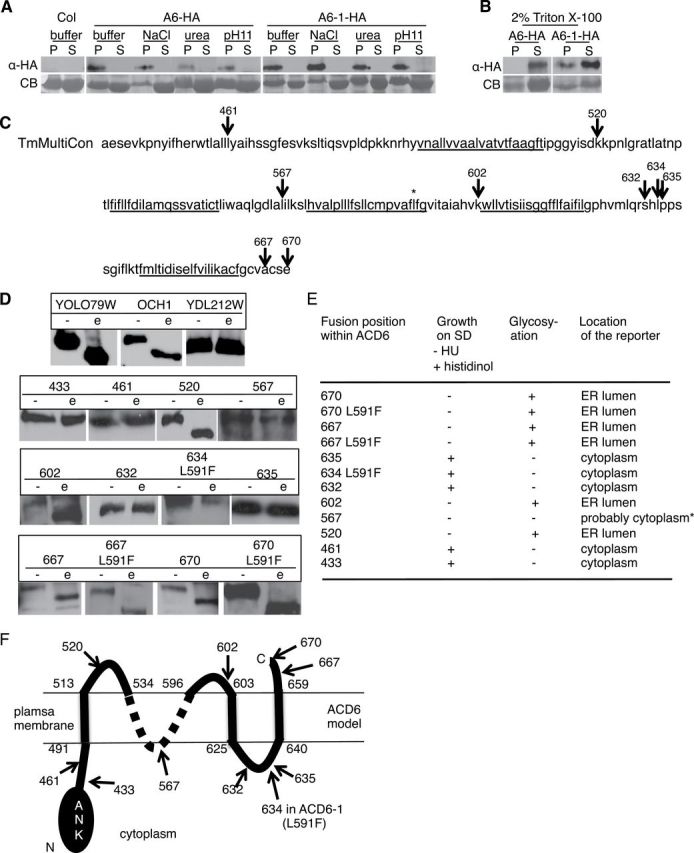
ACD6 and ACD6-1 proteins translated in vitro had the properties of integral membrane proteins (Lu et al., 2005). We confirmed these properties with protein synthesized within plant cells, using previously constructed transgenic Arabidopsis that produces functional HA-tagged versions of ACD6 and ACD6-1, respectively (Lu et al., 2005), and an optimized protocol for preparing plant extracts and isolating microsomal membranes (see the ‘Methods’ section). Under conditions in which peripheral membrane but not integral membrane proteins are solubilized (e.g. 1.5M NaCl, 2M urea, or 100mM Na2CO3 (pH 11); Axtell and Staskawicz, 2003), both ACD6-HA and ACD6-1-HA were retained in the membrane pellet fraction (Figure 1A). However, treatment of the membrane fractions with 2% Triton X-100 resulted in significant solubilization of both ACD6-HA and ACD6-1-HA (Figure 1B). These patterns of fractionation and solubilization were similar to that found with the integral plasma membrane protein H+–ATPase (Supplemental Figure 1). These data indicate that both ACD6-HA and ACD6-1-HA are integral membrane proteins.
Figure 1.
ACD6 and ACD6-1 Are Multipass Integral Membrane Proteins.
(A, B) ACD6-HA and ACD6-1-HA are integral membrane proteins (A) that can be solubilized with detergent (B). Microsomal protein from transgenic plants expressing ACD6-HA (A6-HA)/ACD6-1-HA (A6-1-HA) treated with 1.5M NaCl, 2M urea, 100mM Na2CO3 (pH 11), or 2% Triton X-100, respectively, partitioned into supernatant (S) or membrane pellet (P) fractions and concentrated using trichloroacetic acid were subjected to immunoblotting using HA antibody. CB, Coomassie blue-stained membrane. Experiments in (A) and (B) were repeated twice, with similar results.
(C) A consensus model from the Aramemnon database showing potential membrane spanning regions underlined within the C-terminus of ACD6 (amino acids 441–670). * marks the L591F change in ACD6-1.
(D) ACD6 truncation fusion proteins produced in yeast were treated with endo-glycosidase H (e) or mock-treated (–) and subject to Western blot analysis using HA antibody. Top panel shows yeast positive (YOLO79W and OCH1) and negative (YCL212W) control proteins for deglycosylation.
(E) Summary of the growth of yeast producing the fusion proteins and predicted location of the C-terminus of each fusion protein based on results from (D) and the growth assays. For the growth assay, if the C-terminus of the fusion protein is in the cytosol and folds properly, yeast grows on histidinol. Localization to the ER lumen (evidenced by glycosylation of the reporter fusion partner) predicts an extracellular location in plant cells. The lack of growth of fusion 567 (*) on histidinol was likely due to the lack of productive folding of the fusion partner.
(F) Provisional topology model for ACD6. Numbers demarcating the extent of transmembrane regions are inferred from the ARAMEMNON TmMultiCon analysis and our data. The ambiguity of the structure in the 536–594 region is denoted by a dotted line. Given the predicted topology of ACD6 (panel (C)), its likely that 567 is in a cytosolic loop. We were unable to resolve the ambiguity of the location of the region near 567 with additional fusions. At least two independent yeast transformants per construct were used for each assay. Numbers (C), (D), and (F) mark the junctions of C-terminal reporter.
Since ACD6 is an integral membrane protein, there may be constraints on the efficiency of its maturation if it has multiple membrane spanning regions. Indeed, a query of the Aramemnon plant membrane protein database (Schwacke et al., 2003), which uses multiple algorithms to create consensus topology models, predicted that ACD6 harbors four or more membrane spanning regions (Figure 1C). We used a heterologous yeast dual reporter assay to test for the presence of multiple membrane spanning regions (Kim et al., 2003). The dual reporter was fused to the C-terminus of full-length and truncated versions of ACD6 or ACD6-1. This allowed complementary assays to be used to discern the position of the reporter as being cytoplasmic (assayed by growth on histidinol) or in the ER lumen (equivalent to the extracellular region in a plant cell (Vitale and Denecke, 1999), assayed by an endoglycosidase H-induced band shift due to de-glycosylation of the reporter moiety on a Western blot). Fusions with ACD6-1 gave the same topology results as ACD6, indicating that the two proteins have similar overall topologies, within the resolution of this method. In designing the fusion proteins, we favored the TmMultiCon model provided by Aramenon, because it employed an extended consensus model that included predictions about homologous proteins (Figure 1C). We validated the presence of three transmembrane helices at approximately 491–513, 603–605, and 640–659 (Figure 1D and 1E). Additionally, the data supported the carboxy terminus being extracellular and amino acids 520 and 602 each belonging to extracellular loops. Amino acid 567 was predicted to be in a cytosolic loop, but we did not get growth on histidinol with the strain carrying the fusion at amino acid 567 (Figure 1E). This likely was due to non-productive folding of the reporter moiety in the fusion protein and not due to an extracellular location of amino acid 567, since we did obtain a glycosylated fusion protein (Figure 1D and 1E). Unfortunately, we were unable to resolve the ambiguity of the location of the region near 567 with additional fusions. Figure 1F shows a provisional topology model for ACD6 that integrates the current data with information from previous experiments (Lu et al., 2005) showing that the ankyrin domain faces the cytoplasm. Together, the results show that ACD6 and ACD6-1 are multipass membrane proteins.
ACD6 and ACD6-1 Form Large Complexes
Due to the presence of the ankyrin domain, ACD6 and ACD6-1 are predicted to form multi-protein complexes. Indeed, using the Triton X-100-solubilized microsomal membrane proteins, both ACD6-HA and ACD6-1-HA formed large complexes (560–860kDa) visible on immunoblots of both one- and two-dimensional blue native (2D BN) gels (Figure 2A and 2B). Monomeric ACD6 (or ACD6-1) is 70kDa. Thus, both ACD6-HA and ACD6-1-HA reside in large complexes. However, the abundance of ACD6-1-containing complexes was higher than ACD6-containing complexes in these solubilized membrane extracts.
Figure 2.
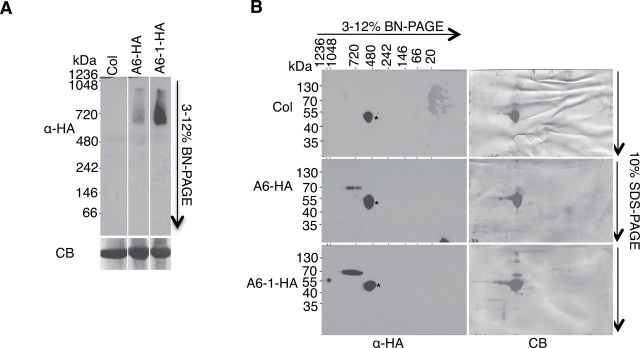
ACD6-HA and ACD6-1-HA Reside In Large Complexes.
Microsomal proteins from the indicated genotypes were solubilized with 0.5% Triton X-100 and separated in the first dimension by BN–PAGE (A) and in the second dimension by SDS–PAGE (B), and analyzed by immunoblotting with HA antibody. CB, Coomassie blue-stained membrane; *, non-specific band. Lanes in (A) were from one continuous membrane. These experiments were repeated three times, with similar results.
To exclude the possibility that Triton X-100 abnormally affected the complexes, we used digitonin, which is a milder detergent than Triton X-100, to characterize ACD6-1-containing complexes. As expected, digitonin treatment solubilized ACD6-1HA, ACD6-HA, and the integral plasma membrane H+–ATPase (Supplemental Figure 2A and 2B). Importantly, digitonin-solubilized ACD6-1-HA complexes were similar in size to the Triton X-100-solubilized complexes in both one and 2D-BN gels and there was no evidence of aggregation as proteins completely entered the gels (Supplemental Figure 2B and 2C). Immunoblots of samples fractionated using Fast Protein Liquid Chromatography (FPLC) confirmed that ACD6-1-HA-containing complexes were large (Supplemental Figure 3). The molecular masses of the ACD6-1-HA complexes were larger than the 669-kDa marker, with the majority of protein eluting at approximately 720–840kDa. This is consistent with results obtained using BN gels. These data indicate that using Triton X-100-treated microsomal membranes and BN gels, which have the advantage of requiring smaller volumes and providing higher resolution than FPLC, is suitable for studying ACD6- and ACD6-1-containing complexes.
ACD6-Containing Membrane Complexes Increase in Abundance, Size, and Localization to the Plasma Membrane during SA Signaling
ACD6 functions in a positive feedback loop with SA (Lu et al., 2003). Therefore, it seemed possible that ACD6-HA- and/or ACD6-1-HA-containing complexes might be regulated by SA. After treatment of plants with the SA agonist benzo (1, 2, and 3) thiadiazole-7-carbothioic acid (BTH), the level of ACD6-HA in membrane fraction was strongly increased (Figure 3A). The abundance and size of the ACD6-HA complexes as assayed by 2D BNG was also increased after BTH treatment, with complexes up to 1000kDa appearing (Figure 3B). In contrast, there was a more modest increase in ACD6-1-HA content in the membrane fraction after BTH treatment compared with mock-treated plants and plants without any treatment (Figure 3A). However, there were also larger ACD6-1-HA-containing complexes visible after BTH treatment, similar to the ACD6-HA-containing complexes (Figure 3B). FPLC analysis confirmed that larger complexes of ACD6-1-HA were detectable after BTH treatment, with the new peak eluting at approximately 1250kDa (Supplemental Figure 3). Thus, SA signaling significantly boosts both the level of ACD6-HA in the membrane fraction and the size of ACD6-HA- and ACD6-1-HA-containing complexes.
Figure 3.
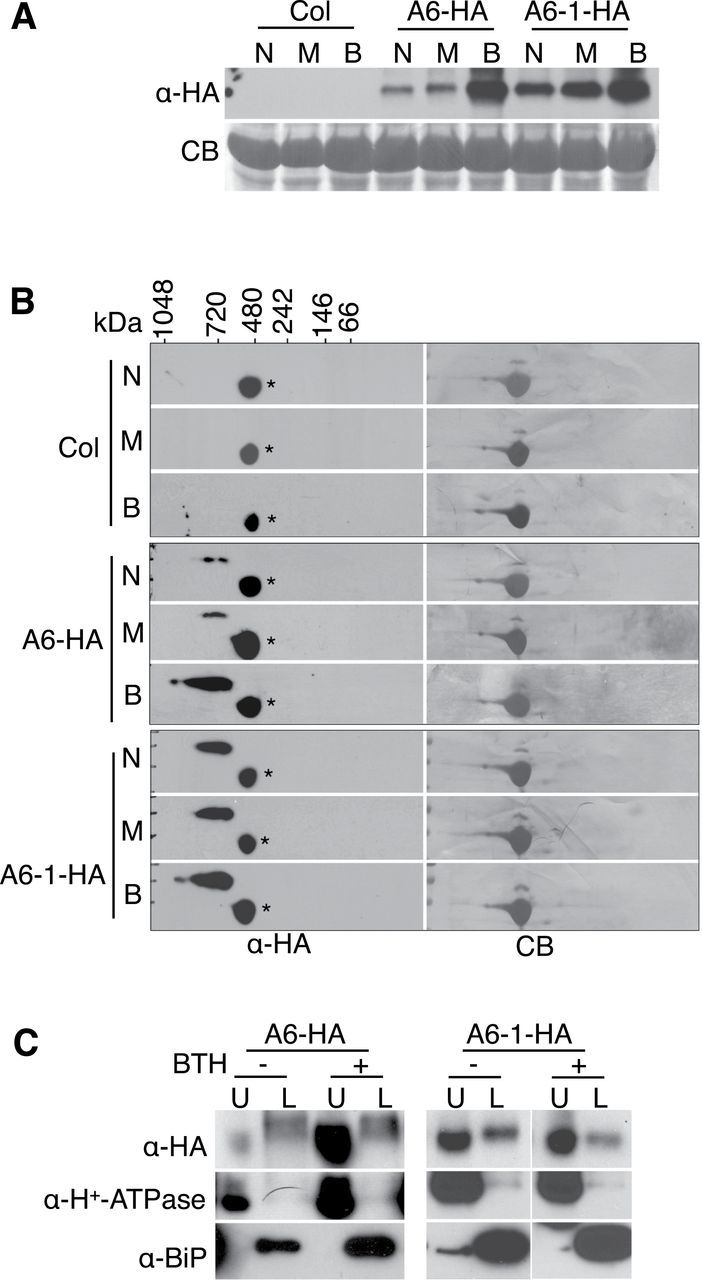
The SA Agonist BTH Causes Increases in the Levels and Sizes of ACD6 and ACD6-1 Complexes and the Fraction of Each Protein Localized to the Plasma Membrane.
Microsomal proteins from Col, ACD6-HA (A6-HA), and ACD6-1-HA (A6-1-HA) untreated leaves (N) or leaves treated for 48 h with 150 μM BTH (B) or water with 0.005% Silwet-77 (M, mock control) were separated by SDS–PAGE, 2D BNG, or SDS–PAGE following aqueous two-phase partitioning and immunoblotted with antibodies against HA, BAK1, H+–ATPase (plasma membrane marker), and/or BIP (ER marker), respectively. CB, Coomassie blue-stained membrane.
(A) ACD6-HA and ACD6-1-HA levels in microsomal membranes are increased after stimulation of the SA pathway using BTH.
(B) ACD6-HA and ACD6-1-HA microsomal membrane complexes analyzed by 2D BNG are large and increase in size after BTH treatment. *, non-specific band.
(C) The fraction of ACD6-HA at the plasma membrane (U, upper phase) versus ER-enriched membranes (L, lower phase), assayed by aqueous two-phase partitioning, increases after BTH treatment. A large amount of ACD6-1-HA is in the upper phase even without BTH treatment. These experiments were repeated three times, with similar results. For the right two panels in (C) (A6-1-HA), blots on the left (– BTH) and right (+ BTH) sides were from the same experiment, but separate membranes were simultaneously processed.
ACD6-HA and ACD6-1-HA were previously detected in plasma membrane-enriched and non-plasma membrane fractions of untreated plants (Lu et al., 2005). To determine whether an increase in SA signaling affects the membrane localization of these proteins, we treated plants with BTH and used aqueous two-phase partitioning. We used H+-ATPase as a marker for plasma membrane-enriched (upper phase) fractions. BIP was used to assess the degree to which ER proteins contaminated the upper phase. The levels of both marker proteins increased after BTH treatment (Supplemental Figure 4). BIP induction by BTH treatment was shown previously (Wang et al., 2005). Additionally, plasma membrane H+-ATPase activity and levels were shown to increase in response to SA in pea leaves (Liu et al., 2009). In agreement with our prior results (Lu et al., 2005), ACD6-HA and ACD6-1-HA were in both the upper phase (plasma membrane-enriched) and lower phase (non-plasma membrane) after partitioning (Figure 3C). Supplemental Figure 5 shows that total protein levels in the respective upper and lower phases with or without BTH treatment were similar. After BTH treatment, most of the ACD6-HA proteins were in the upper phase, consistently with SA signaling increasing the fraction of ACD6 at the plasma membrane (Figure 3C). A large amount of ACD6-1-HA was at the plasma membrane even without BTH treatment (Figure 3C), consistent with plants already having a constitutive defense response. As expected, H+-ATPase was mainly in the upper phase under all conditions tested. These data show that SA signaling affects the complexes and subcellular localization of ACD6, which likely are important for its function at the plasma membrane.
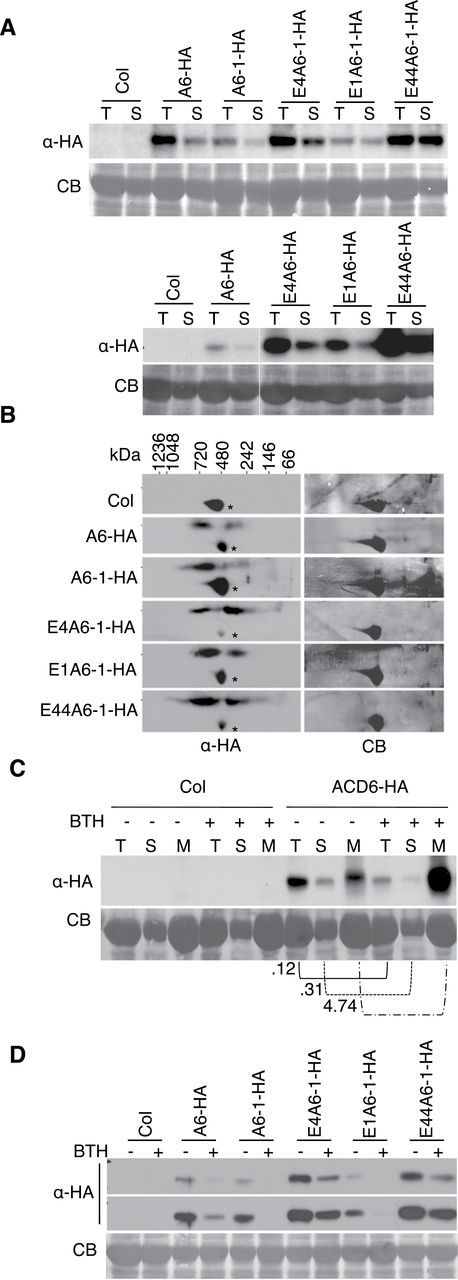
Signaling-Inactive Ankyrin Domain Variants of ACD6-1 and ACD6 Form Altered Complexes that Fail to Reach the Plasma Membrane
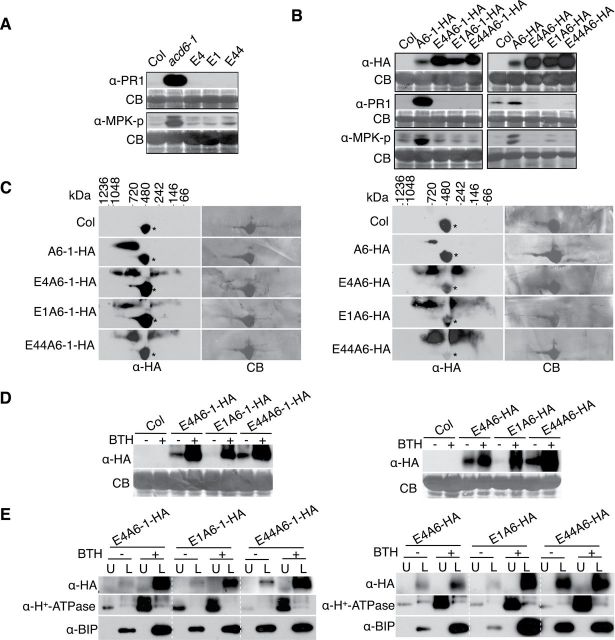
Previous structural modeling of acd6-1 intragenic suppressor mutants implicated predicted non-conserved, surface-exposed residues within the ankyrin repeat of ACD6-1 as important for function (Lu et al., 2005). We hypothesized that such mutations may cause disruptions in ACD6-1- or ACD6-containing complexes formed through ankyrin domain interactions. Three ankyrin repeat variants, E1 (G307E), E4 (G303E), and E44 (E348K), in the context of the original acd6-1 mutant suppressed the accumulation of PR1 protein and the activation of Mitogen Activated Protein Kinases 3 and 6 (MPK3 and MPK6), assessed by auto-phosphorylation. These defense markers were elevated in acd6-1 plants (Figure 4A). Unlike plants that produced ACD6-1-HA, plants that produced ankyrin variants of ACD6-1-HA failed to show increased PR1 accumulation or MPK3/MPK6 activation (Figure 4B) and appeared morphologically similar to wild-type plants (Supplemental Figure 6). Plants that have an extra copy of ACD6 or ACD6-HA are morphologically similar to wild-type (Lu et al., 2005; Supplemental Figure 6), but have slightly elevated PR1 transcript levels (Lu et al., 2005). As expected, plants carrying ACD6-HA showed modestly elevated levels of PR1 and phosphorylated MPK3 and MPK6 proteins relative to wild-type (Figure 4B). However, these phenotypes were not conferred by the ankyrin variants in the context of ACD6-HA (Figure 4B). Thus, ankyrin mutations cause reduced function in the absence of the original ACD6-1 (L591F) amino acid substitution.
Figure 4.
Mutations That Alter the Ankyrin Domain of ACD6-1 or ACD6 Cause the Formation of Aberrant Complexes That Fail to Reach the Plasma Membrane.
(A) acd6-1 ankyrin suppressors lack activation of defense markers. Total proteins isolated from leaves of the indicated plants and subjected to SDS–PAGE were analyzed by immunoblotting with antibodies against PR1, phospho-p44/42 MPK (to detect MPK-p), and/or HA, respectively. E4, E1, and E44 are variants of ACD6-1 with G303E, G307E, E348K amino acid changes in the anykrin domain, respectively.
(B) Transgenic plants expressing ACD6-HA or ACD6-1-HA containing the indicated ankyrin mutations fail to show activation of defense markers. A6-HA and A6-1-HA: transgene-encoded ACD6-HA and ACD6-1-HA, respectively. E4A6-HA, E1A6-HA, E44A6-HA: transgene-encoded versions of ACD6-HA with the indicated amino acid substitutions. E4A6-1-HA, E1A6-1-HA, E44A6-1-HA: transgene-encoded versions of ACD6-1-HA with the indicated amino acid substitutions. Defenses were measured in total extracts as in (A). Antibodies against HA were used to detect ACD6-HA, ACD6-1-HA, and their variant proteins. For each panel shown, lanes were from one continuous membrane.
(C–E) Microsomal membrane protein extracts from plants carrying transgenes with ankyrin mutations harbor the indicated ACD6-HA and ACD6-1-HA ankyrin variants, which (C) form aberrant complexes as assayed by 2D BNG, (D) are still induced by BTH, and (E) do not accumulate at the plasma membrane as assayed by aqueous two-phase partition analysis. Complexes, protein levels, and localization were detected by immunoblotting with HA antibody. U, upper; L, lower; CB, Coomassie blue staining; *, non-specific band. Experiments in the left panel of (B) were done twice, with similar results. All other experiments were repeated three times, with similar results.
Membrane-associated protein complexes that contained ankyrin variants in the context of ACD6-1-HA or ACD6-HA ranged in size: some were smaller than expected and some appeared to be similar in size to complexes that contained ACD6-1-HA or ACD6-HA (Figure 4C). Ankyrin variant protein levels in the microsomal membrane fraction were still induced by BTH (Figure 4D). However, aqueous two-phase partitioning indicated the proteins were retained in the ER-enriched fractions, even after BTH treatment (Figure 4E). Such localization was probably due to the action of the ER quality-control system that retains misfolded proteins or misassembled membrane protein complexes in the ER (Ellgaard and Helenius, 2003). Together, these data indicate that the ankyrin domain mutations cause aberrant complex formation and a failure of the variant ACD6 and/or ACD6-1 proteins to accumulate at the plasma membrane.
A Fraction of the ACD6, ACD6-1, and Ankyrin Variant Proteins Is Soluble and Shows Reduced Abundance after BTH Treatment
The retention of ACD6-1-HA ankyrin variant proteins in the ER-enriched fraction and the presence of a fraction of ACD6-HA and ACD6-1-HA proteins in the ER-enriched fraction indicated that these proteins might be subject to ERQC and ERAD. If this occurred, proteins would be retrotranslocated from the ER to the cytosol prior to degradation by proteasomes (Nakatsukasa et al., 2008; Vembar and Brodsky, 2008). Such cytosolic proteins should be soluble after fractionation. Indeed, ACD6-HA, ACD6-1-HA, and variants of ACD6-1-HA and ACD6-HA were all present in the soluble fraction, albeit less ACD6-1-HA was detected in this fraction relative to ACD6-HA (Figure 5A).
Figure 5.
Characterization of ACD6, ACD6-1, and Ankyrin Variant Proteins Present in the Soluble Fraction.
The indicated protein extracts were separated by SDS–PAGE and analyzed by immunoblotting with HA antibody.
(A) ACD6-HA (A6-HA), ACD6-1-HA (A6-1-HA), and the ankyrin variants E4A6-1-HA, E1A6-1-HA, and E44A6-1-HA (see legend of Figure 4) proteins are present in the soluble fraction. Total (T) proteins were isolated from leaves of the indicated plants. Soluble (S) proteins are the supernatant fraction from total proteins after ultracentrifugation (see the ‘Methods’ section). The same volume of T and S fractions were loaded.
(B) Soluble complexes containing ACD6-HA, ACD6-1-HA, or variant ACD6-1-HA are similar in size. Soluble proteins from the indicated plants were separated by 2D BNG and immunoblotted with anti-HA antibody. *, non-specific band.
(C) Comparison of the level ACD6-HA in the total extract and soluble and membrane fractions isolated from leaf tissue treated for 48 h with 150 μM BTH (+) or water with 0.005% Silwet-77 (–). The same volume of T and S fractions, respectively, were loaded for mock and BTH treatments. Membrane fractions (M) were 50-fold concentrated relative to the T fractions. Numbers under the blot indicate the fraction of ACD6-HA in the BTH samples relative to the corresponding value in the mock-treated samples. All samples of the same type (T, S, or M, respectively) had the same amount of protein loaded, as validated by densitometry of the total Coomassie gel lanes).
(D) ACD6-HA, ACD6-1-HA, and ankyrin variant ACD6-1-HA protein levels in the soluble fraction are reduced after treatment with BTH. Soluble proteins from the indicated plants treated for 48 h with 150 μM BTH (+) or water with 0.005% Silwet-77 (–). Two exposures of the same Western blot are shown. CB, Coomassie blue staining. These experiments were repeated three times, with similar results. Lanes in the bottom panel of (A) were from one continuous membrane.
Soluble ACD6-HA, ACD6-1-HA, and the ankyrin variant proteins formed similarly sized complexes, as detected by 2D BNG analysis (Figure 5B). These complexes formed larger (520–740kDa) and smaller (260–390kDa) sized complexes, suggesting variant and wild-type proteins may follow the same path for degradation (Figure 5B). Some of the soluble complexes of ACD6-HA and ACD6-1-HA were smaller than those found in the respective membrane fractions (compare Figures 5B and 2B).
After BTH treatment, ACD6-HA showed decreased abundance in the soluble pool as well as in the total extract, which contrasted with the greatly increased ACD6-HA level in the membrane fraction (Figure 5C). As with ACD6-HA, the levels of ACD6-1-HA and the ankyrin variant proteins in the soluble fractions were reduced after BTH treatment (Figure 5D). We validated that the soluble fractions were free of membrane protein by probing all the fractions from Figure 5 with antibodies to membrane and cytosolic marker proteins (Supplemental Figure 7). Thus, all versions of ACD6 analyzed were present in distinct soluble complexes and showed decreased abundance in response to SA signaling.
Degradation of ACD6, ACD6-1, and Ankyrin Variant Proteins Is Proteasome Dependent
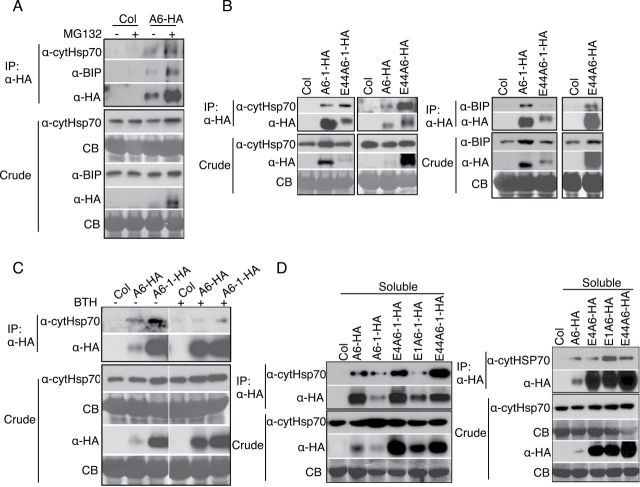
Misfolded and misassembled protein complexes retained in the ER are eventually degraded by ERAD, a multistep process that utilizes the ubiquitin-proteasome system (Vembar and Brodsky, 2008; Guerra and Callis, 2012). To test whether ACD6-HA, ACD6-1-HA, and ACD6-1-HA variant proteins were degraded by proteasomes, we treated leaves from plants expressing each protein with MG132, a proteasome inhibitor. Figure 6A shows that MG132 treatment caused significant accumulation of ACD6-HA, ACD6-1-HA, and ankyrin variant proteins in both the membrane and the soluble fractions (see Supplemental Figure 8 for fractionation controls for Figure 6), suggesting that these proteins are degraded by proteasomes.
Figure 6.
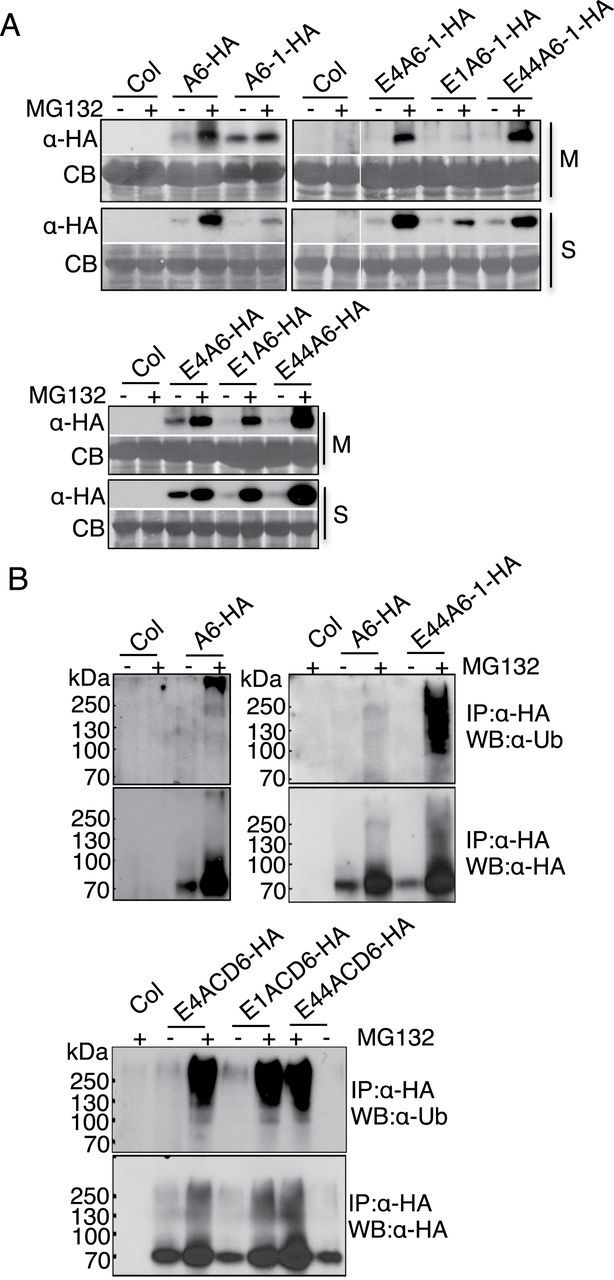
Degradation of ACD6, ACD6-1, and Ankyrin Variant Proteins Depends on the Proteasome.
(A) ACD6-HA, ACD6-1-HA, and ankyrin variant protein (see legend to Figure 4) degradation is inhibited by the proteasome inhibitor MG132. Microsomal (M) proteins and supernatant/soluble (S) proteins isolated from indicated plants treated with 0.1% DMSO (–) or 50 μm MG132 (+) for 6h were separated by SDS–PAGE and analyzed by immunoblotting with HA antibody. Lanes in the right panel were from one continuous blot.
(B) ACD6-HA and the indicated ankyrin variant proteins (see legend to Figure 4) proteins are polyubiquitinated in planta. Proteins from the soluble fraction of indicated plants were immunoprecipitated (IP) using anti-HA high-affinity matrix were separated by SDS–PAGE and immunoblotted with ub (upper panel) and HA antibodies (lower panels), respectively. These experiments were repeated three times, with similar results.
Proteins destined for proteasome-mediated degradation are expected to be ubiquitinated prior to their degradation (Vembar and Brodsky, 2008; Guerra and Callis, 2012). Indeed, immunoprecipitation of ACD6-HA and ankyrin variants from the soluble fraction allowed detection of varying amounts of higher-molecular-weight species that were ubiquitinated (Figure 6B). Although the amount of ubiquitinated ACD6-HA was low, it was reproducibly detected.
The Molecular Chaperone Hsp70 Family Members Are Associated with ACD6, ACD6-1, and Ankyrin Variants during ERAD
Molecular chaperones are central mediators during ERAD. ER luminal HSP70, BIP, and cytosolic HSP70 are major chaperones for non-glycosylated proteins during ERAD; they facilitate substrate selection and targeting (Meacham et al., 2001; Okuda-Shimizu and Hendershot, 2007; Otero et al., 2010; Matsumura et al., 2011). As ACD6 is not a glycosylated protein (Lu et al., 2005), BIP and cytosolic HSP70 might mediate ERAD of ACD6. Consistent with this idea, ACD6-HA formed membrane complexes with both HSP70 and BIP, and levels of these complexes increased after blocking proteasome-mediated degradation with MG132 (Figure 7A). BIP and HSP70 were also detected in membrane complexes with ankyrin variant proteins. HSP70 in other systems can contribute to ERAD by binding portions of misfolded membrane proteins that reside in the cytoplasm (Needham et al., 2011; Donnelly et al., 2013). Consistent with this possibility, the abundance of HSP70-ankyrin variant protein complexes was increased relative to HSP70-ACD6- and HSP70-ACD6-1 complexes (Figure 7B). Together, these results suggest that BIP and HSP70 regulate ACD6 for ERAD. The finding that ACD6 and ACD6-1 (Figure 7A and 7B) were found in complexes with BIP, an ER protein, shows that a fraction of these proteins found in the lower phase after two-phase partitioning resides in the ER (Figure 3C).
Figure 7.
ER-Resident BIP and Cytosolic HSP70 Are Involved in ERAD of ACD6.
(A) BIP and cytosolic HSP70 form complexes with ACD6-HA (A6-HA) that increase after 6 h of MG132 treatment (50 μM).
(B) Cytosolic HSP70 and BIP from complexes with ACD6-1-HA and ankyrin variants.
(C) Less cytosolic HSP70 forms complexes with ACD6-HA and ACD6-1-HA (A6-1-HA) after 48 h of BTH treatment (150 μM) compared with mock controls (–).
(D) ACD6, ACD6-1, and ankyrin variant proteins form soluble complexes cytosolic HSP70. Solubilized microsomal proteins (A–C) and soluble proteins (D) were isolated from the indicated plants. Immunoprecipitations (IPs) were performed as in Figure 6B and analyzed by Western blot using BIP and cytHSP70 antibodies. Crude represents 1% of the input for the IPs. There experiments were repeated three times, with similar results.
We also investigated the effect of BTH treatment on HSP70-ACD6/ACD6-1 membrane complexes. Less cytosolic HSP70 was associated with the membrane fractions of both ACD6-HA and ACD6-1-HA after BTH treatment (Figure 7C). This suggests that the fraction of ACD6 and ACD6-1 that was misfolded was reduced due to SA signaling. This result is also consistent with our finding that SA increases the efficiency of ACD6 maturation and trafficking.
The cytoplasmic chaperone HSP70 (cytHSP70) can bind to the hydrophobic regions of proteins and prevent their aggregation in the cytoplasm (Schmid et al., 1994; Hartl, 1996; Young et al., 2004; Kampinga and Craig, 2010). Therefore, cytHSP70 might associate with ACD6-HA, ACD6-1-HA, and ankyrin variants of ACD6-HA/ACD6-1-HA proteins to help maintain their solubility in the cytoplasm and facilitate turnover of proteins that are not productively folded or assembled into complexes. ACD6-HA, ACD6-1-HA, and variant ACD6-HA/ACD6-1-HA proteins immunoprecipitated from the soluble fraction were all associated with cytHSP70 (Figure 7D). Validation that the soluble fraction was not contaminated with membrane proteins is shown in Supplemental Figure 9.
FLS2 Forms Complexes with ACD6 and ACD6-1
To find additional components of ACD6/ACD6-1-containing complexes, we performed LC–MS/MS analysis after immunoprecipitation of ACD6-1-HA and identified peptides of the flagellin receptor FLS2 (Supplemental Table 1). Other peptides identified will be presented elsewhere. We confirmed that FLS2 formed complexes with both ACD6-HA and ACD6-1-HA using immunoprecipitation and immunoblotting (Figure 8A). Additionally, FLS2 was present in large complexes in plants that express ACD6-1-HA, consistently with its presence in ACD6-/ACD6-1-containing complexes (compare Figure 8B and Supplemental Figure 10 with Figure 3B and Supplemental Figure 3, respectively). A role for ACD6 in FLS2-mediated signaling is indicated by the reduced transcriptional response of the acd6-2 loss-of-function mutant to the FLS2 ligand flg22 (Supplemental Figure 11).
Figure 8.
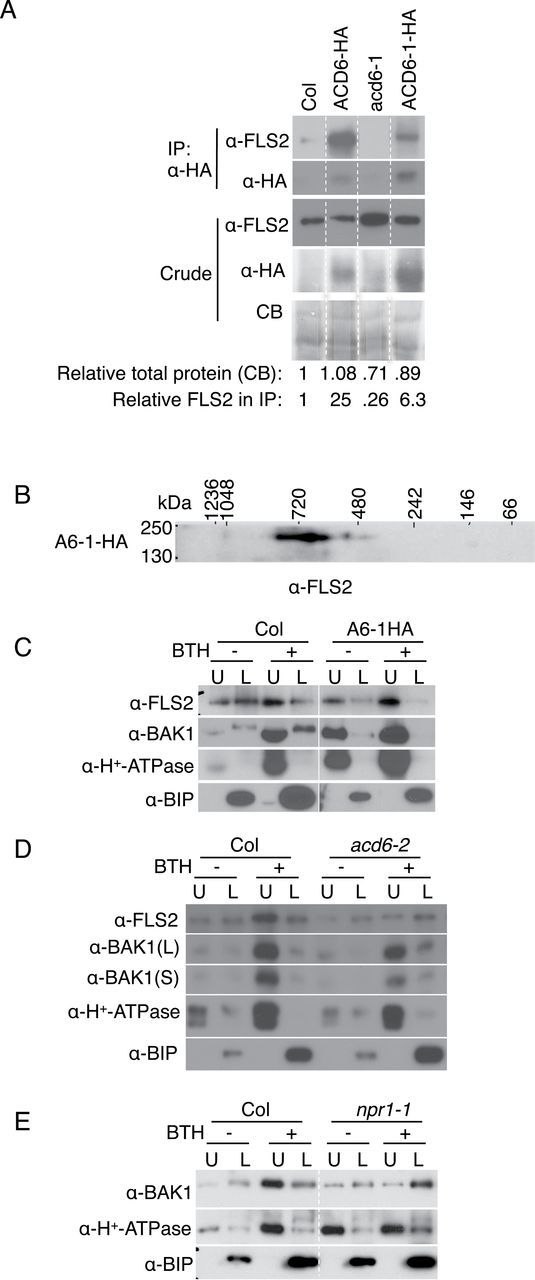
ACD6/ACD6-1 Form Complexes with FLS2 and Affect the Trafficking of FLS2 and BAK1 to Plasma Membrane after BTH Treatment.
(A) Co-immunoprecipitation of FLS2 with ACD6-HA and ACD6-1-HA, respectively. Numbers below the blots show the relative amount of total protein quantified from the Coomassie blots and the relative amount of FLS2 after immunoprecipitation, normalized to total protein quantified by densitometry.
(B) Two-dimensional blue native gel analysis shows that FLS2 forms large complexes in ACD6-1-HA (A6-1-HA).
(C) The proportion of FLS2 and BAK1 in the plasma membrane (upper phase) is increased after BTH treatment (48 h, 150 μM) in Col. Additionally, the fraction of FLS2 and BAK1, respectively, at the plasma membrane was higher in ACD6-1-HA plants versus Col even without BTH.
(D) Loss of ACD6 (acd6-2) affects the plasma membrane levels (and fractions of proteins at the plasma membrane) of FLS2 and, to a lesser extent, BAK1 after BTH treatment.
(E) In the npr1 mutant plants, trafficking to the plasma membrane of BAK1 is severely compromised after BTH treatment. Microsomal proteins were isolated from leaves of the indicated plants and solubilized with 0.5% Triton X-100. Solubilized microsomal proteins were obtained for immunoprecipitations, separated by two-dimensional BNG/SDS–PAGE or two phase, and analyzed by immunoblotting with the indicated antibodies. All the lanes in panels (A), (C), and (E), respectively, were from one continuous membrane. These experiments were repeated three times, with similar results.
Like ACD6, FLS2 and BAK1 were present in both the plasma membrane and non-plasma membrane fractions (Figure 8C). The localization pattern for FLS2 based on fractionation is in agreement with a previous study, albeit Lee and colleagues reported greater enrichment of FLS2 in the upper phase under basal conditions (Lee et al., 2011). Differences in plant growth conditions probably account for the differences in the precise percentage of FLS2 in different fractions. Other receptors, such as EFR and Xa21, also have significant non-plasma membrane pools under basal conditions (Saijo et al., 2009; Park et al., 2010). Importantly, 48h after BTH treatment, FLS2 and BAK1 were predominantly at the plasma membrane (Figure 8C). In plants that expressed ACD6-1-HA, both FLS2 and BAK1 were predominantly at the plasma membrane (Figure 8C). In BTH-treated plants that lacked ACD6 (acd6-2 mutants), FLS2 failed to show increased levels at the plasma membrane relative to the non-plasma membrane fraction (Figure 8D). BAK1 showed reduced plasma membrane levels in acd6-2 relative to wild-type after BTH treatment, albeit the effect was only partial (Figure 8D). However, in BTH-treated plants that were compromised for SA signal transduction due to an npr1-1 mutation, there was a severe defect in the trafficking of BAK1 to the plasma membrane (Figure 8E). Together, these data show that a major function for ACD6 is to regulate the plasma membrane pool of FLS2 (and to a lesser extent BAK1), especially in response to SA signaling.
DISCUSSION
Plasma membrane proteins represent an important class of proteins, some of which are involved in immune signaling and are regulated during pathogen attack (Elmore et al., 2012). This study focused on the regulation and maturation of ACD6, which we found to be a multipass membrane protein. We focused on using biochemical approaches, because ACD6 is a very low abundance protein for which cell biological approaches (e.g. imaging of fluorescent fusion proteins) have not been possible. Like many other multipass membrane proteins, events at the ER regulate the fate of ACD6. In untreated plants, ACD6 resides in soluble, ER-enriched, and plasma membrane pools, and is constitutively degraded with hallmarks of ERAD (Vembar and Brodsky, 2008; Guerra and Callis, 2012). Specifically, soluble ACD6 binds cytosolic HSP70 and is ubiquitinated and degraded by the proteasome. ACD6 and ACD6-1 each reside in large complexes that include BIP, presumably to help with folding in the ER, and the membrane proteins FLS2 and BAK1, which are important for responses to infection. ACD6/ACD6-1-containing complexes increase in abundance and size during SA signaling. In the presence of ankyrin variants with amino acid substitutions in surface-accessible non-conserved residues, complexes fail to form normally and thus do not reach the cell surface. This suggests that the ankyrin domain may act as a platform for assembling protein complexes, similarly to other ankyrin-containing proteins (Sedgwick and Smerdon, 1999).
In response to SA signaling, the fraction of membrane-associated ACD6 at the plasma membrane increases within 48h. The same is true for FLS2 and BAK1, which also require ACD6 to different degrees for their elevated levels at the plasma membrane. SA-regulated increases in proteins at the plasma membrane could be due to increased stability of ACD6, FLS2, and BAK1 specifically at this subcellular site. However, we favor a model in which there is increased trafficking of these proteins to the plasma membrane in response to SA signaling. Plasma membrane proteins are trafficked by the secretory pathway (Vitale and Denecke, 1999; Popescu, 2012), which is up-regulated by SA (Wang et al., 2005). Therefore, it is likely that SA signaling causes increased maturation and trafficking of ACD6, FLS2, and BAK1 to the plasma membrane. The idea that a higher level at the plasma membrane of ACD6 occurs due to increased trafficking from the ER to the plasma membrane is consistent with the behavior of the ankyrin variants. The membrane-associated pools of these proteins are induced by BTH. However, their levels at the plasma membrane are low, probably due to the ER quality-control system that retains them in the ER and thus stops their trafficking to the plasma membrane. The formation of aberrant ER-associated complexes by these variants supports this idea. In the case of BAK1, the increased fraction of protein at the plasma membrane during SA signaling requires NPR1, which controls up-regulation of secretory pathway components (including a transmembrane trafficking protein) in response to BTH (Wang et al., 2005). The behavior of BAK1 is consistent with the observation that potentiation of receptor-mediated pattern responses after SA treatment fails to occur in npr1 plants (Xu et al., 2014). Figure 9 summarizes the major findings about the fate of ACD6 before and during SA signaling. Note that SA signaling increases the level of H+-ATPase, but this plasma membrane protein efficiently traffics to the plasma membrane under all conditions used in this study. This indicates that there are differences in how SA signaling affects different membrane proteins.
Figure 9.
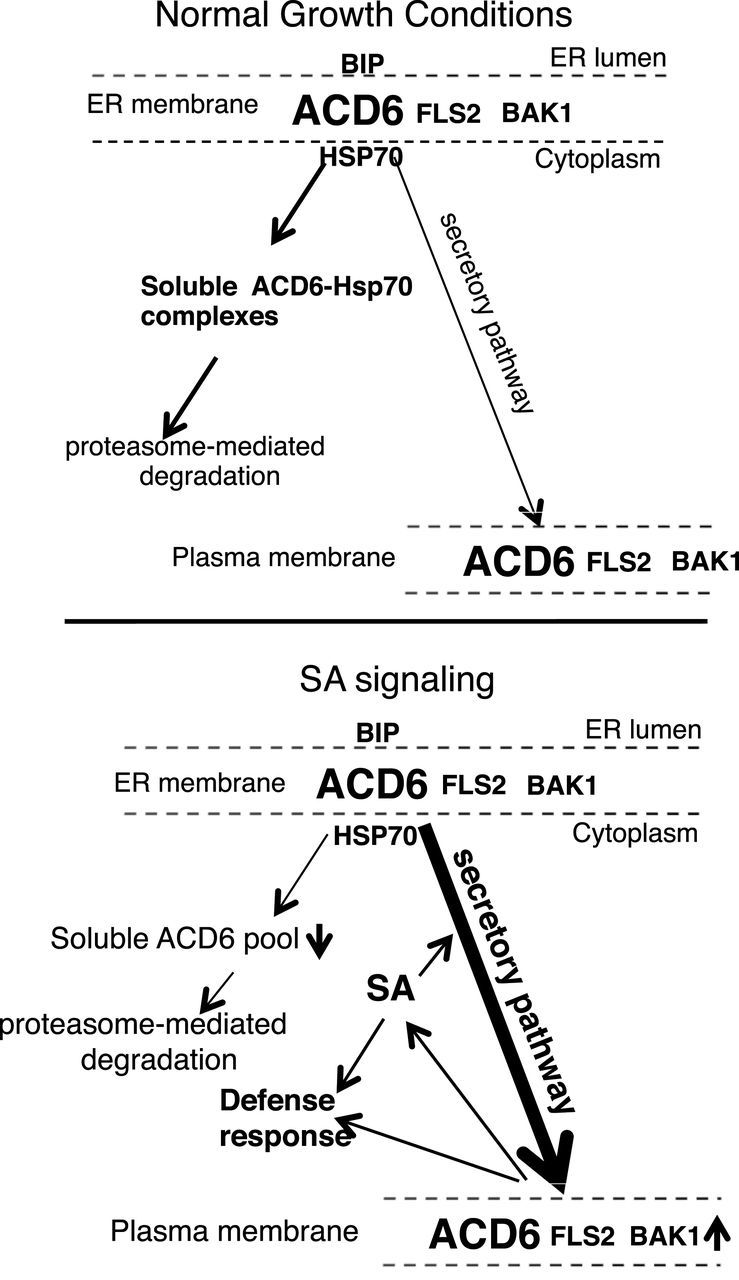
Working Model for the Regulation by SA of the Maturation and Localization/Transport of the ACD6 Protein and ACD6’s Co-Trafficking with FLS2 and BAK1.
Under normal growth conditions, plants have a low level of SA and ACD6 resides in large complexes (top panel). Maturation of ACD6 proteins and/or formation of productive complexes are not efficient and, as a consequence, a significant pool of ACD6 does not reach the plasma membrane. Instead, misfolded ACD6 and/or misassembled ACD6 complexes are constitutively retained in the ER and retrotranslocated into the cytosol (contained in the soluble fraction), where they form complexes with HSP70 and are degraded by the proteasome at a certain rate. This prevents the untimely activation of ACD6. Upon infection by some pathogens, plants accumulate a relatively high level of SA (lower panel), which regulates the efficiency of the maturation and transport of ACD6 proteins. As a result, the levels and sizes of ACD6 complexes and the amount of ACD6 transport to the plasma membrane are increased. SA signaling is also correlated with a reduction in the soluble pool of ACD6, which may be an indirect consequence of the increased efficiency of maturation. The increased plasma membrane pool of ACD6 ensures that cells with elevated SA can activate stronger defenses when exposed to a pathogen. FLS2 forms complexes with ACD6/ACD6-1. Additionally, FLS2 and its co-receptor BAK1 show increased abundance at the plasma membrane in response to SA, a process that depends strongly (for FLS2) or moderately (for BAK1) on ACD6. The model of regulation of ACD6 signaling by SA allows plants to finely tune its defense response to the invaders with minimal impact on plant growth.
Limiting the amount of ACD6 protein that resides at the plasma membrane under normal growth conditions may have the advantage of reducing the impact that ACD6 has on plant growth. The correlation between the higher level of the gain-of-function ACD6-1 protein at the plasma membrane and the small stature of acd6-1 plants supports this idea. Some natural alleles of ACD6 also have reduced stature (Todesco et al., 2010), although the levels and localization of these natural ACD6 variant proteins have not been assessed. Limiting the fraction of additional defense proteins such FLS2, BAK1 (this work) and Ef-Tu receptor (Saijo et al., 2009) that localize to the plasma membrane could be a way to decrease the chance that their ectopic activation will create a growth trade-off when no pathogen is present. Since SA signaling increases the levels of these proteins in the plasma membrane (this work), we suggest that the plasma membrane is an important site of action for ACD6, FLS2, and BAK1. Additionally, the induction of SA during infection may allow plants to have stronger receptor-mediated responses.
The ACD6, ACD6-1, and their variant proteins found in the soluble fraction are presumed to be intermediates of the ERAD process, which is largely conserved between plants and other systems (Muller et al., 2005; Su et al., 2011; Guerra and Callis, 2012; Su et al., 2012). A certain amount of the unubiquitinated form of proteins in cytosol may be due to deubiquitination by a deubiquitinating enzyme after dislocation from ER (Ernst et al., 2009; Claessen et al., 2012). An example of an intact membrane protein found in the cytosol of mammalian cells is class I MHC (Wiertz et al., 1996). The exact steps that occur between protein extraction from the ER to its degradation by the proteasome are not clear (Smith et al., 2011). The sizes of soluble complexes of ACD6, ACD6-1, and their variants are similar, suggesting that there is a common way to degrade them. It is largely unknown how cells maintain the solubility of retrotranslocated membrane proteins before they are degraded by cytoplasmic proteasomes. However, chaperones such as HSP70 can bind to hydrophobic regions of proteins to prevent their aggregation (Buchberger et al., 2010). Our results showing the association of HSP70 with ACD6, ACD6-1, and the ankyrin variant proteins in the soluble complexes supports such a role for cytoplasmic HSP70 in Arabidopsis. For human CFTR, Hsc70 plays an important role in its degradation in the cytosol during ERAD (Matsumura et al., 2011); plant HSP70 probably also has a similar role.
Plants that produce ACD6-1 have very different phenotypes than those with the wild-type version of ACD6, showing much more elevated defenses and small stature (Rate et al., 1999; Lu et al., 2003, 2005). Nevertheless, the sizes of complexes in which ACD6 and ACD6-1 reside appear to be similar. The level of ACD6 complexes in the membrane is much lower than that of ACD6-1 complexes, suggesting that the abundance of complexes at the membrane is an important way to regulate ACD6 function. In contrast, soluble but presumably inactive complexes of ACD6 are more abundant than those of ACD6-1. Interestingly, stimulating the SA pathway using BTH causes a dramatic increase of ACD6 in the plasma membrane, reduced levels of ACD6 (and ACD6-1) in the soluble fraction, and the appearance of new larger membrane complexes. Although plants with ACD6-1 already have elevated SA (Vanacker et al., 2001; Lu et al., 2005), they lack detectable levels of the largest complexes until further stimulation with the SA agonist BTH. One possibility is that in ACD6-1-containing plants, the level of ACD6-1 is still below a critical threshold for forming the largest complex. For ACD6, this largest complex appears only when most of the protein is localized to the plasma membrane; therefore, it seems to be a plasma membrane-specific complex, which may have a distinct function. Given the similarity of the sizes of ACD6- and ACD6-1-containing complexes, it may be that a primary effect of the acd6-1 mutation is to allow the ACD6-1 protein to fold more efficiently compared to ACD6. The consequence of this is could be to enable more protein to productively mature and become trafficked to the plasma membrane together with FLS2 and BAK1. However, it is also possible that the ACD6-1 protein is more active as a signaling protein apart from its level at the membrane or ability to fold more efficiently. More work is necessary to resolve this question.
METHODS
Plant Materials and Growth Conditions
All plants used in this study are in the Arabidopsis thaliana Columbia (Col-0) ecotype background. The npr1-1 mutant, the loss-of-function mutant acd6-2, gain-of-function mutant acd6-1, acd6-1 intragenic suppressors (E4, E1, and E44), and transgenic plants with hemagglutinin (HA)-tagged genomic ACD6 or ACD6-1 (encoding ACD6-HA and ACD6-1-HA, respectively) driven by the native promoter at single insertion sites and were previously described (Cao et al., 1994; Rate et al., 1999; Lu et al., 2005; Todesco et al., 2010). Plants were grown in a growth chamber with 16-h light/8-h dark photoperiod as described (Lu et al., 2005). For all experiments, 19–20-day-old plants were used.
Generation of Transgenic Plants
The genomic constructs used for generating transgenic plants with functional ACD6-HA and ACD6-1-HA were described previously (Lu et al., 2005). Three suppressor mutations (E4: 2503 g>a, G303E; E1: 2515 g>a, G307E; and E44: 2637 g>a, E348K) were introduced into both of the ACD6-HA and ACD6-1-HA constructs, respectively, by site-directed mutagenesis using the GeneTailor™ Site-Directed Mutagenesis System (Invitrogen). Since there was no available Col mutant completely lacking ACD6 expression, all six constructs were sequence-validated and individually transformed into flowering Col Arabidopsis (Clough and Bent, 1998). Independent transgenic progeny (T0) was selected on soil using BASTA (AgrEvo) treatment as described (Lu et al., 2005) and single insertion lines from the T2 generation were identified using growth on ½ MS plates with BASTA. At least 10 independent transformed lines with single insertions were identified and analyzed for gene expression by RT–PCR and protein production by Western blot analysis. At least two lines with reproducible protein levels from each construct were used for further experiments and similar results were obtained.
Chemical Treatments
For benzo (1, 2, 3) thiadiazole-7-carbothioic acid (BTH, from Robert Dietrich, Syngenta, Research Triangle Park, NC) treatments, plants were sprayed with 150 μM BTH in water with 0.005% Silwet-77 or water with 0.005% Silwet-77 as mock treatment until all the leaves were wet. Leaves were collected 48h after treatment. For MG132 treatments, the procedure of Qin et al. (2008) was followed except that the treatment time was shortened to 6h and leaf pieces were used instead of seedlings. Briefly, 2-mm leaf pieces were floated in 50 μM MG132 (Sigma) in 0.1% DMSO or 0.1% DMSO for mock control with gentle shaking in a growth chamber for 6h.
Subcellular Fractionation and Immunoblotting
Protein extracts were obtained as described (Lu et al., 2005) with some modifications. Leaves were frozen and ground in liquid nitrogen to a fine powder and then thawed in 2 volumes ice-cold extraction buffer (50mM Tris–HCl pH 7.5; 0.33M Sucrose; 5mM EDTA; 150mM NaCl, and 1 complete protease inhibitor cocktail from Roche). The crude extracts were filtered through two layers of miracloth and centrifuged at 10000 g for 10min to get total protein extract (supernatant), which was further ultracentrifuged at 100000 g for 60min to get the microsomal membrane and soluble fractions, respectively. Membrane pellets were re-suspended in re-suspension buffer 1 (50mM Tris–HCl pH 7.5, 10% glycerol, 0.5% Triton X-100, protease inhibitor cocktail), re-suspension buffer 2 (0.33M sucrose, 5mM potassium phosphate, pH 7.8, 3mM KCl, 0.1mM EDTA, 1 complete protease inhibitor cocktail), or re-suspension buffer 3 (50mM Tris-HCL pH 8.0, 10% glycerol, 0.5% sodium deoxycholate, 1% Igepal CA-630 from SIGMA, and complete protease inhibitor cocktail from Roche; Chinchilla et al., 2007). Aliquots of fractionated extracts were stored at –80ºC until use.
For testing whether ACD6/ACD6-1 are integral membrane proteins, microsomal proteins were treated with 1.5M NaCl, 2M urea, 100mM Na2CO3 (pH 11), or 2% Triton X-100, respectively, at 4ºC for 2h with gentle shaking and fractionated into supernatant (S) or membrane pellet (P) fractions by ultracentrifugation. Protein precipitated by trichloroacetic acid from supernatant (S) and protein from membrane pellet (P) fractions were then subjected to immunoblotting using an antibody against HA.
Concentrations of plant protein extracts were measured by Bradford assay (Bradford, 1976) and 15–20 μg proteins were used for Western blot analysis. Proteins were separated using sodium dodecyl sulfate (SDS)–PAGE using 10% gels and analyzed by immunoblotting using antibodies against HA (16B12; Covance), phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (Cell Signalling Technologies), PR1 (from Dr. Xinnian Dong, Duke University), BAK1 (from Dr. Delphine Chinchilla, University of Basel), and FLS2 (from Dr. Delphine Chinchilla, University of Basel and from Dr. Antje Heese, University of Missouri). H+–ATPase (AS07260, Agrisera), cytosolic HSP70 (SPA817, Stressgen), or BIP (SPA-818, Stressgen), respectively. Yeast protein extracts were analyzed using 6% gels. Horseradish peroxidase conjugated anti-mouse (Thermo Scientific, Santa Cruz Biotechnology) and anti-rabbit antibodies (Thermo Scientific) were used as secondary antibodies. SuperSignal West Pico Stable Peroxidase (Thermo Scientific) and SuperSignal West Femto Stable Peroxidase (Thermo Scientific) were used to detect the signals.
Blue Native Gel Electrophoresis
Microsomes in re-suspension buffer 1 were gently shaken at 4ºC for 1h and centrifuged at 100000 g for 30min to get the supernatant (solubilized membrane proteins). 30–40 μg of solubilized membrane proteins and the soluble fraction after 100000 g ultracentrifugation were used for two-dimensional blue native gels (BNGs). Strips of BN lanes run according to Invitrogen’s NativePAGE Novex Bis-Tris Gel System on a 3%–12% Bis-Tris gradient gel (Invitrogen, Cat. BN1001box) were mounted and analyzed by SDS–PAGE. ACD6-HA and ACD6-1-HA proteins were detected by immunoblotting.
FPLC Gel Filtration
Solubilized membrane protein used for FPLC were prepared in the same way as for BNG and applied to a Pharmacia FPLC System AKTA FPLC U PC900 at 4ºC. A superpose 610/300 GL column was used and calibrated using a Gel Filtration HMW Calibration Kit (GE Healthcare). Fractions of 0.5ml each were collected at a flow rate of 0.3 ml min–1, concentrated using 10 μl StrataClean resin (Stratagene), and solubilized in 40 μl SDS sample buffer (125mM Tris, pH 6.8; 20% glycerol; 4% SDS; 5M urea; 0.01% bromophenol blue; 166mM DTT). Equal volumes of each fraction were analyzed by SDS–PAGE and immunoblotting.
Aqueous Two-Phase Partitioning
Microsomal membrane proteins re-suspended in re-suspension buffer 2 were subject to two-phase aqueous partitioning to separate PM and ER membrane as described (Larsson et al., 1987). Briefly, 1ml membrane fraction from 5 g leaves was loaded to 3 g aqueous two-phase solution with a final composition of 6.2% dextran T-500 (Sigma), 6.2% polyethylene glycol 3350 (Sigma), 0.33M sucrose, 5mM potassium phosphate pH 7.8, and 3mM KCl. The mixtures were inverted 100 times and separated by centrifugation at 1500 g for 5min. Upper phase and lower phase were then diluted with re-suspension buffer 2 and centrifuged at 100000 g for 1 h to get the pellet. The pellets containing enriched PM from upper phase and non-PM proteins (ER-enriched) from lower phase were solubilized in SDS sample buffer and 3–4 μg of proteins were analyzed by immunoblotting.
Immunoprecipitation of Different Complexes from Soluble or Membrane Fractions
For co-immunoprecipitation experiments from soluble fraction extracts, samples were incubated with anti-HA-matrix (Roche) overnight at 4°C with gentle shaking. The resulting immunoprecipitates were washed five times with the washing buffer (50mM Tris–HCl pH7.5, 150mM NaCl, 0.5% Triton X-100, and protease inhibitor cocktail). The eluted proteins were analyzed by SDS–PAGE and immunoblotting. For identification of FLS2 in immunoprecipitated ACD6-1-HA complexes by mass spectrometry, solubilized membrane proteins were prepared from 20-day-old leaves of transgenic plants expressing HA-tagged ACD6-1 and wild-type as for BNGs. Possible interacting proteins were purified with anti-HA high-affinity matrix, separated by SDS–PAGE and stained by Coomassie blue. Differential bands were cut, digested with trypsin, and analyzed by LC/MS/MS. For validating FLS2–ACD6 complexes by immunoprecipitation, membrane proteins solubilized in re-suspension buffer 3 were incubated with anti-HA high-affinity matrix (Roche).
ACD6 Topology Analysis
The ARAMEMNON database (Schwacke et al., 2003; see http://aramemnon.botanik.uni-koeln.de/) was queried to predict the number of ACD6 transmembrane-spanning domains, the location of their hydrophilic loops, and the C-terminus. The sites with the highest probability of lying within a loop region were chosen for fusions with a dual topology reporter for use in yeast (Kim et al., 2003). Topology reporter plasmid pJK90 was used to create constructs with ACD6/ACD6-1 fusions integrated into yeast by homologous recombination as described (Kim et al., 2003). pJK90 was treated with SmaI restriction enzyme to linearize the vector between the end of the Triosephosphate Isomerase promoter and the start codon of OST4. The 5’-end homologous recombination region matched the 3’-end of SmaI-digested pJK90, and the 3’-end homologous region matched the linker between the end of OST4 and the start of the HA sequence. ACD6, ACD6-1, and various truncations were amplified by PCR from cDNAs (Lu et al., 2005), using primers that also included the homologous sequence. For forward primer, the homologous region (5’-AGGTGGTTTGTTACGCATGCAAGCTTGATATCGAA-3’) was immediately followed by the start codon of ACD6. The 3’ reverse primer was chosen so as to maintain the C-terminal fusion reporter in-frame with the ACD6/ACD6-1 gene. The reverse primers were designed to include the 3’ homologous sequence (5’-GATGGTCT AGAGGTGTAACCACTTGAGTTCTTAGG-3’) plus 14–22 nucleotides of the ACD6 or ACD6-1 cDNA. Yeast strain STY 50 (MATa, his4-401, leu2, –3, and –112, trp1-1, ura3-52, HOL1-1, SUC2::LEU2) was transformed with the linearized pJK90 and purified PCR product carrying ACD6 flanked by the homologous sequences. Transformation was carried out using the Frozen-EZ Yeast Transformation II Kit (Zymo Research). Successful transformants were selected by plating on synthetic media lacking Uracil (SD-Ura). Plasmids from successful transformants were isolated and verified by PCR and sequencing. Yeast carrying ACD6–His3–Suc2–His4c fusion protein were streaked onto SD-Ura plates lacking histidine but containing 6mM histidinol (Sigma) and grown at 30°C for 3–4 d. The same yeast strains were streaked onto SD-Ura and SD-Ura-His plates as positive and negative controls for assessing yeast growth. Yeast was grown in SD-Ura liquid media overnight. Total cellular protein was extracted by re-suspending the cell pellet in 10% trichloroacetic acid, incubating at 60°C for 5min, chilling on ice for 5min, and vortexing with 425–600 micron glass beads (Sigma) for 1min. The supernatant was transferred to a new tube and centrifuged at 3000 g for 5min. The precipitated protein was re-suspended in SDS–PAGE sample buffer supplemented with 50mM Na+-PIPES (piperazine-N,N′-bis (2-ethanesulfonic acid); pH 7.5) and with 8M urea. The mixture was kept at 60°C for 5min and then purified using Micro Bio-Spin Columns with Bio-Gel P-6 in Tris buffer (Bio-Rad Laboratories). The eluate was aliquoted into two tubes and 800mM sodium acetate (pH 5.6) buffer stock was added to give a final buffer concentration of 80mM sodium acetate. For deglycosylation, the samples were treated with 10 mU of endoglycosidase H (Roche) and incubated at 37°C overnight. The untreated control samples were also incubated at 37°C without the addition of endoglycosidase H enzyme. After completion of endoglycosidase H reaction, the samples were treated with SDS–PAGE sample buffer and heated at 60°C for 10 min before SDS–PAGE analysis.
Quantitation of Immunoblots
Immunoblots were quantified using Gel-Pro analyzerTM software for densitometry. Relative values were normalized against the total protein contents on the Coomassie-stained membranes, except that the Rubisco bands were excluded.
Real-Time RT–PCR Analyses
Total RNA was isolated using Trizol Reagent (Invitrogen) 1 h after water or 1 μM flg22 infiltration of leaves of 21-day-old plants. cDNAs were synthesized using Prime Masterscript (Takara). qRT–PCR were performed as described as SYBR Green Master ROX reagent (Roche) using the Applied Biosystem 7900HT Fast Real-Time PCR system and SDS2.3 software. Primer sets were previously described (FRK1 and EF1a) (Lu et al., 2003; He et al., 2006). FRK1 transcript levels were normalized to the levels of EF1a.
Accession Numbers
Sequence data from this article can be found in the EMBL/GenBank data libraries under accession number(s) At5g46330 (FLS2), At4g33430 (BAK1), At4g14400 (ACD6), At3g45640 (MPK3), At2g43790 (MPK6), At1g64280 (NPR1), At2g14610 (PR1), and At2g19190 (FRK1). Accession numbers for mutant lines are SALK_045869 (acd6-2), SALK_141277 (fls2), and CS3726 (npr1-1).
SUPPLEMENTARY DATA
Supplementary Data are available at Molecular Plant Online.
FUNDING
This work was supported by grants to J.T.G. from the National Institutes of Health (R01 GM54292) and the National Science Foundation (IOS 0822393). C.T. was supported by a JSPS Postdoctoral Fellowship for Research Abroad.
Supplementary Material
ACKNOWLEDGMENTS
We thank Xinnian Dong, Delphine Chinchilla, Antje Heese, Benjamin Glick, Robert Dietrich, Hyun Kim, and Gunnar von Heijne for sharing reagents and strains, and Milan Mrksich for use of his FLPC. We thank members of the Greenberg lab, Delphine Chinchilla, Gayle Lamppa, Uddhav Shigdel, Yang Liu, Chuan He, and Benjamin Glick for helpful discussions. No conflict of interest declared.
REFERENCES
- Axtell M.J., Staskawicz B.J. (2003). Initation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 112, 369–377 [DOI] [PubMed] [Google Scholar]
- Bork P. (1993). Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins. 17, 363–374 [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., Skach W.R. (2011). Protein folding and quality control in the endoplasmic reticulum: recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23, 464–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A., Bukau B., Sommer T. (2010). Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol. Cell. 40, 238–252 [DOI] [PubMed] [Google Scholar]
- Cao H., Bowling S.A., Gordon A.S., Dong X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell. 6, 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Zipfel C., Robatzek S., Kemmerling B., Nurnberger T., Jones J.D., Felix G., Boller T. (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 448, 497–500 [DOI] [PubMed] [Google Scholar]
- Claessen J.H., Kundrat L., Ploegh H.L. (2012). Protein quality control in the ER: balancing the ubiquitin checkbook. Trends Cell Biol. 22, 22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Donnelly B.F., Needham P.G., Snyder A.C., Roy A., Khadem S., Brodsky J.L., Subramanya A.R. (2013). Hsp70 and Hsp90 multichaperone complexes sequentially regulate thiazide-sensitive cotransporter endoplasmic reticulum-associated degradation and biogenesis. J. Biol. Chem. 288, 13124–13135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann R., Schäfer P. (2012). The endoplasmic reticulum in plant immunity and cell death. Front Plant Sci. 3, 200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L., Helenius A. (2003). Quality control in the endoplasmic reticulum. Nat. Rev. Mol. Cell Biol. 4, 181–191 [DOI] [PubMed] [Google Scholar]
- Elmore J.M., Liu J., Smith B., Phinney B., Coaker G. (2012). Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Prot. 11, M111 014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst R., Mueller B., Ploegh H.L., Schlieker C. (2009). The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell. 36, 28–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudet R. (2008). A primer on ankyrin repeat function in TRP channels and beyond. Mol. BioSys. 4, 372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeckeler J.L., Brodsky J.L. (2010). Molecular chaperones and substrate ubiquitination control the efficiency of endoplasmic reticulum-associated degradation. Diabetes Obes. Metab. 2, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra D.D., Callis J. (2012). Ubiquitin on the move: the ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol. 160, 56–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerriero C.J., Brodsky J.L. (2012). The delicate balance between secreted protein folding and endoplasmic reticulum-associated degradation in human physiology. Physiol. Rev. 92, 537–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond C., Helenius A. (1995). Quality control in the secretory pathway. Curr. Opin. Cell Biol. 7, 523–529 [DOI] [PubMed] [Google Scholar]
- Hartl F.U. (1996). Molecular chaperones in cellular protein folding. Nature. 381, 571–579 [DOI] [PubMed] [Google Scholar]
- He P., Shan L., Lin N.C., Martin G.B., Kemmerling B., Nürnberger T., Sheen J. (2006). Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell. 125, 563–575 [DOI] [PubMed] [Google Scholar]
- Heese A., Hann D.R., Gimenez-Ibanez S., Jones A.M., He K., Li J., Schroeder J.I., Peck S.C., Rathjen J.P. (2007). The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc. Natl Acad. Sci. U S A. 104, 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendershot L.M. (2004). The ER function BiP is a master regulator of ER function. Mt. Sinai. J. Med. 71, 289–297 [PubMed] [Google Scholar]
- Hüttner S., Strasser R. (2012). Endoplasmic reticulum-associated degradation of glycoproteins in plants. Front Plant Sci. 3, 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., Craig E.A. (2010). The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Melen K., von Heijne G. (2003). Topology models for 37 Saccharomyces cerevisiae membrane proteins based on C-terminal reporter fusions and predictions. J. Biol. Chem. 278, 10208–10213 [DOI] [PubMed] [Google Scholar]
- Kopito R.R. (1999). Biosynthesis and degradation of CFTR. Physiol. Rev. 79, S167–S173 [DOI] [PubMed] [Google Scholar]
- Larsson C., Widell S., Kjellbom P. (1987). Preparation of high-purity plasma membranes. Methods Enzymol. 148, 558–568 [Google Scholar]
- Lee H.Y., Bowen C.H., Popescu G.V., Kang H.-G., Kato N., Ma S., Dinesh-Kumar S., Snyder M., Popescu S.C. (2011). Arabidopsis RTNLB1 and RTNLB2 reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell. 23, 3374–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wen J., Lease K.A., Doke J.T., Tax F.E., Walker J.C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 110, 213–222 [DOI] [PubMed] [Google Scholar]
- Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J.D. (2009). Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl Acad. Sci. U S A. 106, 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu H., Pan Q., Yang H., Zhan J., Huang W. (2009). The plasma membrane H+-ATPase is related to the development of salicylic acid-induced thermotolerance in pea leaves. Planta. 229, 1087–1098 [DOI] [PubMed] [Google Scholar]
- Lu H., Liu Y., Greenberg J.T. (2005). Structure–function analysis of the plasma membrane- localized Arabidopsis defense component ACD6. Plant J. 44, 798–809 [DOI] [PubMed] [Google Scholar]
- Lu H., Rate D.N., Song J.T., Greenberg J.T. (2003). ACD6, a novel ankyrin protein, is a regulator and an effector of salicylic acid signaling in the Arabidopsis defense response. Plant Cell. 15, 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H., Salimian S., Gamelin E., Wang G., Fedorowski J., LaCourse W., Greenberg J.T. (2009). Genetic analysis of acd6-1 reveals complex defense networks and leads to identification of novel defense genes in Arabidopsis . Plant J. 58, 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Määttänen P., Gehring K., Bergeron J.J., Thomas D.Y. (2010). Protein quality control in the ER: the recognition of misfolded proteins. Semin Cell Dev. Biol. 21, 500–511 [DOI] [PubMed] [Google Scholar]
- Matsumura Y., David L.L., Skach W.R. (2011). Role of Hsc70 binding cycle in CFTR folding and endoplasmic reticulum-associated degradation. Mol. Biol. Cell. 22, 2797–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham G.C., Patterson C., Zhang W., Younger J.M., Cyr D.M. (2001). The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nature Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- Meusser B., Hirsch C., Jarosch E., Sommer T. (2005). ERAD: the long road to destruction. Nature Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- Molinari M. (2007). N-glycan structure dictates extension of protein folding or onset of disposal. Nat. Chem. Biol. 3, 313–320 [DOI] [PubMed] [Google Scholar]
- Muller J., Piffanelli P., Devoto A., Miklis M., Elliott C., Ortmann B., Schulze-Lefert P., Panstruga R. (2005). Conserved ERAD-like quality control of a plant polytopic membrane protein. Plant Cell. 17, 149–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Huyer G., Michaelis S., Brodsky J.L. (2008). Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell. 132, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham P.G., Brodsky J.L. (2013). How early studies on secreted and membrane protein quality control gave rise to the ER associated degradation (ERAD) pathway: the early history of ERAD. Biochim. Biophys. Acta. 1833, 2447–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham P.G., Mikoluk K., Dhakarwal P., Khadem S., Snyder A.C., Subramanya A.R., Brodsky J.L. (2011). The thiazide-sensitive NaCl cotransporter is targeted for chaperone-dependent endoplasmic reticulum-associated degradation. J. Biol. Chem. 286, 43611–43621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V., Li J., Batoux M., Roux M., Chu Z.H., Lacombe S., Rougon A., Bittel P., Kiss-Papp M., Chinchilla D, et al. (2009). Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438; Sinai J. Med. 71, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda-Shimizu Y., Hendershot L.M. (2007). Characterization of an ERAD pathway for nonglycosylated BiP substrates, which require Herp. Mol. Cell. 28, 544–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero J.H., Lizák B., Hendershot L.M. (2010). Life and death of a BiP substrate. Semin. Cell Dev. Biol. 21, 472–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.-J., Bart R., Chern M., Canlas P.E., Bai W., Ronald P.C. (2010). Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One. 5, e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C. (2012). A model for the biosynthesis and transport of plasma membrane-associated signaling receptors to the cell surface. Front. Plant Sci. 3, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F., Sakuma Y., Tran L.S., Maruyama K., Kidokoro S., Fujita Y., Fujita M., Umezawa T., Sawano Y., Miyazono K, et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 20, 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rate D.N., Cuenca J.V., Bowman G.R., Guttman D.S., Greenberg J.T. (1999). The gain-of-function Arabidopsis acd6 mutant reveals novel regulation and function of the salicylic acid signaling pathway in controlling cell death, defenses, and cell growth. Plant Cell. 11, 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römisch K. (2005). Endoplasmic reticulum-associated degradation. Annu. Rev. Cell Dev. Biol. 21, 435–456 [DOI] [PubMed] [Google Scholar]
- Saijo Y. (2010). ER quality control of immune receptors and regulators in plants. Cellular Microbiol. 12, 716–724 [DOI] [PubMed] [Google Scholar]
- Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Haweker H., Dong X., Robatzek S., Schulze-Lefert P. (2009). Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28, 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid D., Baici A., Gehring H., Christen P. (1994). Kinetics of molecular chaperone action. Science. 263, 971–973 [DOI] [PubMed] [Google Scholar]
- Schwacke R., Schneider A., van der Graaff E., Fischer K., Catoni E., Desimone M., Frommer W.B., Flugge U.I., Kunze R. (2003). ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol. 131, 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S.G., Smerdon S.J. (1999). The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci. 24, 311–316 [DOI] [PubMed] [Google Scholar]
- Smith M.H., Ploegh H.L., Weissman J.S. (2011). Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 334, 1086–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel S.H., Dong X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nature Reviews. Immunology. 12, 89–100 [DOI] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2011). Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis . Proc. Natl Acad. Sci. U S A. 108, 870–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su W., Liu Y., Xia Y., Hong Z., Li J. (2012). The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant. 5, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Zhang Q., Gao M., Zhang Y. (2014). Regulation of SOBIR1 accumulation and activation of defense responses in bir1-1 by specific components of ER quality control. Plant J. 77, 748–756 [DOI] [PubMed] [Google Scholar]
- Tintor N., Saijo Y. (2014). ER-mediated control for abundance, quality, and signaling of transmembrane immune receptors in plants. Front Plant Sci. 5, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todesco M., Balasubramanian S., Hu T.T., Traw M.B., Horton M., Epple P., Kuhns C., Sureshkumar S., Schwartz C., Lanz C, et al. (2010). Natural allelic variation underlying a major fitness trade-off in Arabidopsis thaliana . Nature. 465, 632–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacker H., Lu H., Rate D.N., Greenberg J.T. (2001). A role for salicylic acid and NPR1 in regulating cell growth in Arabidopsis . Plant J. 28, 209–216 [DOI] [PubMed] [Google Scholar]
- Vembar S.S., Brodsky J.L. (2008). One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Denecke J. (1999). The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell. 11, 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Amornsiripanitch N., Dong X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Path. 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Weaver N.D., Kesarwani M., Dong X. (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science. 308, 1036–1040 [DOI] [PubMed] [Google Scholar]
- Wiertz E.J., Jones T.R., Sun L., Bogyo M., Geuze H.J., Ploegh H.L. (1996). The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 84, 769–779 [DOI] [PubMed] [Google Scholar]
- Xu J., Xie J., Yan C., Zou X., Ren D., Zhang S. (2014). A chemical genetic approach demonstates that MPK3/MPK6 activation and NADPH oxidase-mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D.H., Hettenhausen C., Baldwin I.T., Wu J. (2011). BAK1 regulates the accumulation of jasmonic acid and the levels of trypsin proteinase inhibitors in Nicotiana attenuata’s responses to herbivory. J. Exp. Bot. 62, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.C., Agashe V.R., Siegers K., Hartl F.U. (2004). Pathways of chaperone-mediated protein folding in the cytosol. Nat. Rev. Mol. Cell Biol. 5, 781–791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.