Abstract
Recent advances in digital imaging is impacting the practice of pathology. One of the key enabling technologies that is leading the way towards this transformation is the use of whole slide imaging (WSI) which allows glass slides to be converted into large image files that can be shared, stored, and analyzed rapidly. Many applications around this novel technology have evolved in the last decade including education, research and clinical applications. This publication highlights a collection of abstracts, each corresponding to a talk given at Carnegie Mellon University's (CMU) Bioimaging Day 2014 co-sponsored by the Biomedical Engineering and Lane Center for Computational Biology Departments at CMU. Topics related specifically to digital pathology are presented in this collection of abstracts. These include topics related to digital workflow implementation, imaging and artifacts, storage demands, and automated image analysis algorithms.
Keywords: Challenges, digital pathology, image analysis, opportunities
INTRODUCTION
Advances in digital imaging promises to revolutionize the workflow of Pathology Departments in Health Care Institutions. Much like Radiology Departments over the past decade, the advent of more affordable and accurate computing and imaging systems is currently facilitating the conversion of typical manual and analog pathology workflows toward digital implementation. The cornerstone of this digital transition is the widespread use of whole slide imaging (WSI). WSI allows glass slides to be converted into large image files that can be shared, stored, and analyzed, thereby extending the possibilities of using these digital files beyond just morphological assessment. The transition to a digital workflow and adoption of digital imaging brings with it key opportunities in information management, image sharing (telepathology), and image analysis. This, in turn, could open the door to numerous new diagnostic technologies based on increasing throughput and employing concomitant automation, digital image analysis, and better accuracy through the ability of modern computer systems and algorithms to quantitatively “mine” large quantities of data.
This collection of abstracts, each corresponding to a talk given at Carnegie Mellon University's (CMU) Bioimaging Day 2014, illustrates several of the challenges and opportunities that lie ahead. The CMU Bioimaging day is an annual event, co-sponsored by the Biomedical Engineering and Lane Center for Computational Biology Departments at CMU, where the main goal is to bring together researchers working in quantitative biomedical imaging in the Pittsburgh Community. This particular event was held on February 26, 2014 and included several invited talks and posters. Topics related specifically to digital pathology are presented in this collection of abstracts. These include topics related to digital workflow implementation, imaging and artifacts, storage demands, and automated image analysis algorithms. The discussion reveals that despite promising preliminary results, much work is still needed if the promise of digital imaging in pathology is to be fulfilled.
Digital Imaging Tools for Translational and Personalized Medicine: The Critical Role of Pathologists
Anil V Parwani
Department of Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, USA
Digital imaging tools are widely being used in pathology today ranging from simple static images to more complex digital whole slide images and three-dimensional images in multiple planes. WSI uses computerized technology to scan and convert entire pathology glass slides into digital images at high resolution, which are then made available to pathologists and investigators on a network or locally. Unlike glass slides, which can be lost, fade or damaged with time, images can be archived and retrieved easily. The quality of the images produced is of diagnostic quality, and with viewing software, it is possible to have annotations and clinical metadata presented with the image, potentially resulting in a virtual microscope with all the clinical and prognostic information needed to correlate morphology with genomic or proteomic or immunohistochemical data.[1,2,3,4] WSI images, when packaged with the relevant clinical information, provides the pathologist with the ability to manage all the information and creating opportunities for increased efficiency. Other benefits are to be able to share and discuss cases remotely with colleagues and experts.
The third and one of the most important aspects of digitization of slides is the ability to perform image analysis and use computer-aided diagnostic tools on WSI.[1,5] Quantification of immunohistochemistry using image analysis improves the accuracy and reproducibility of pathologists’ interpretations by eliminating inter/intraobserver variability, leading to better information for clinicians in treatment decisions for patients. Standardization and automation are essential to foster translational research efforts in the study of new biomarkers for cancer care. Automated image analysis provides a standard, reproducible, sensitive and specific method of biomarker quantitation, unlike semi-quantitative (manual) scoring which is inherently subjective and laborious.[5]
Organizations such as the College of American Pathologists (CAP) and Digital Pathology Association are advocating the use of this promising technology to advance patient care and provide rapid access to expert opinion. The CAP convened an expert panel, which was tasked to develop guidelines for the validation of WSI for diagnostic purposes. These guidelines, which are based on scientific evidence and expert opinion, will allow pathologists to now move closer toward actually using validated WSI technology in a safe manner to improve patient care.[6,7]
In summary, digital imaging tools have the potential to provide new insights into translational and personalized medicine, and pathologists and researchers will continue to play a pivotal role in applying these tools to their practice to advance the care of patients.
Detecting Cancer Using Quantitative Analysis of Nuclear Morphology
Gustavo K. Rohde
Department of Biomedical Engineering, Lane Center for Computational Biology, Carnegie Mellon University, Pittsburgh, PA, USA
The advent of digital imaging systems and their eventual adoption in the pathologist's workflow will create new opportunities for the utilization of quantitative image analysis methods in diagnostic and prognostic pathology and cytology. Given the strong association of nuclei with cancerous processes, the quantification of nuclear structure and morphology quantification has long been a target of study in pathology. The precise quantification of nuclear structure differences between two populations of nuclei (e.g., benign vs. malignant) is still an open and critical question, given that the choice of different features and classification methods can dramatically impact the outcome. Recently developed algorithms for comparing the intensities in digital images have the potential for rendering the process of cancer detection from nuclear morphology highly accurate, specific, and interpretable.
Nuclear Structure in Pathology
It is well-known that nuclear shape and chromatin distribution are intricately linked to biological processes related to cancers. These cellular changes are spurred by aberrations in the genetic code and the transcription of different messenger RNAs compared to their normal tissue of origin. These changes occur in the nucleus and are accompanied by the unfolding and repackaging of chromatin that in part or in whole produces changes in nuclear size (pleomorphism), shape, membrane contours, chromatin structure, and emergence of a nucleolus.[8] Numerous staining techniques (e.g., H and E) commonly used in standard laboratories are able to highlight nuclear features, including nuclear chromatin distribution patterns. It is therefore no surprise that there have been numerous attempts at quantifying nuclei in a wide variety of cancers with different degrees of success.[9,10,11] Notably, Mohler et al.[12] have shown that quantitative analysis of nuclear morphology can be superior to the Gleason score often used for grading prostate cancer, which has prognostic significance.
Nuclear Segmentation
The first step in a nuclear information processing “pipeline” is the detailed and accurate extraction of nuclei from histopathology or cytology images. A plethora of approaches for this task have been previously described, given the wide variety of appearance nuclei can have in different organs as well as the different staining techniques (e.g. H and E, Feulgen, Diff Quik, etc.,) that can be used. A variety of software (e.g., ImageJ, CellProfilert, etc., are freely available to the academic community for performing this task. Recent developments utilizing supervised learning of nuclear shape and texture models have the potential for rendering this task more reliable and easy to use.[13]
Numerical Features for Quantifying Nuclei
Once extracted from digital images, nuclei are most commonly quantified with the so-called numerical feature approach, a long standing and generic approach in image analysis[14] that has found applications in numerous areas of science and technology. The goal in this step is to measure certain qualities of nuclei by specific computations based on their intensity values. Features that aim to characterize shape and size normally include the area (for two-dimensional measurements), perimeter, form factor, and others. Features aimed at extracting texture and chromatin placement information include the variance and other moments of the intensity values, as well as features based on the pixel intensity co-occurrence matrix and filter (e.g. Gabor) outputs. In modern studies,[15] several hundred features are used to characterize each nucleus. This high-dimensional feature space is then “mined” using pattern recognition approaches in the hope of finding differences that can be used to reliably tell apart two or more cell populations (e.g., benign vs. malignant).
Transport-Based Morphometry for Quantifying Nuclear Structure Differences
We recently described a novel image analysis approach, termed transport-based morphometry,[16] based on the Mathematics of optimal transport[17] that is especially well suited for quantifying mass distributions within a confine, such as chromatin placement within a nuclear envelope. The idea is to measure how close or far nuclei are situated from each other, by measuring the minimum total effort (in terms of mass times distance that it must be transported) necessary to arrange the chromatin distribution from one nucleus to another. The idea has recently been used to detect and differentiate cancers in the liver,[17] thyroid,[18] and lung,[19] using both tissue and cytology specimens. In total these tests utilized nearly 140 patients. The accuracy of detection in blind predictive studies (using “held out” data) was nearly 100% (near perfect sensitivity and specificity), while the accuracy obtained using modern numerical feature approaches was only in the 80 percentile.[18,19] Moreover, as Figure 1 indicates, the approach can be used to also visualize differences in chromatin distribution between different classes (e.g., cancer types).
Figure 1.
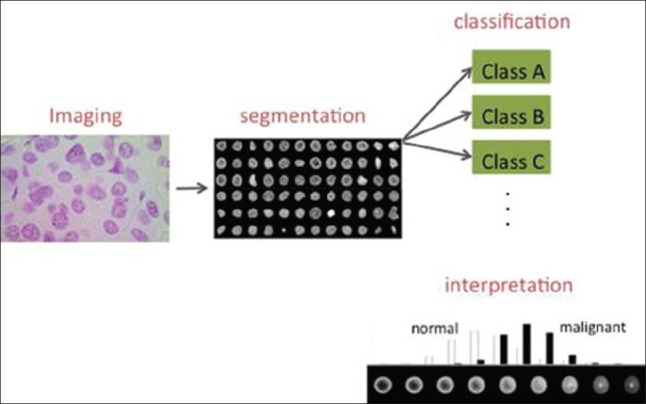
Nuclear structure extraction and quantification process. A Feulgen stained tissue section from a patient suspected of having fetal-type hepatoblastoma. Nuclei are first automatically segmented, and then utilized for cancer detection and subtyping using classification approaches. Modern mathematical algorithms for image analysis are also able to display intuitive visualizations depicting differences between nuclear classes. In this case, malignant cell distributions, on average, tend to have their chromatin more evenly distributed throughout the nuclear envelope
Conclusion
Although numerous barriers still remain for widespread adoption of digital imaging in pathology, there is little doubt that digital imaging is set to play an increasing role in the pathologist's workflow. This in turn will further facilitate the utilization of quantitative image analysis to enable higher precision in diagnosis and prognosis. The quantification of nuclear structure will continue to play an important, and perhaps increasing role in the diagnosis of numerous pathologies. Recent studies have shown that, if mined properly, the quantitative analysis of large numbers of nuclei can be highly accurate and thus “clear up” certain diagnostic dilemmas. New mathematical algorithms can play a crucial role in ensuring high accuracy as well as allowing for an intuitive understanding of morphology differences between cancer types.
Automated Tissue Screening: A Challenging Prospect for Digital Image Analysis in Pathology
Michael T. McCann1, Ramamurthy Bhagavatula2, Matthew C. Fickus3, John A. Ozolek4, and Jelena Kovacevic1,5
1Department of Biomedical Engineering and Center for Bioimage Informatics, 5Department of Electrical and Computer Engineering Carnegie Mellon University, 2Massachusetts Institute of Technology Lincoln Laboratory, 3Department of Mathematics and Statistics, Air Force Institute of Technology Wright-Patterson AFB, 4Department of Pathology, Children's Hospital of Pittsburgh University of Pittsburgh School of Medicine, Pittsburgh, PA, USA
In most surgical pathology practices, certain specimens recur at relatively high frequency. In our practice at Children's Hospital of Pittsburgh, gastrointestinal (GI) biopsies are by far the most common specimen. In the last calendar year, we accessioned approximately 2500 GI biopsies. This roughly translates to 337,500 pieces of tissues that are reviewed by the pathologist of which an estimated 202,500 will be diagnosed as “unremarkable”. Having an automated tissue screening tool could potentially save time and money, improve diagnoses and promote investigative efforts. The latter three elements stem from saving time. Cutting a pathologist's time that is spent diagnosing normal tissue can translate to saving money (perhaps by having fewer pathologists). Pathologists would have more time to think about difficult cases and more time for pursuing an investigation. A precedent for automated screening of patient pathology samples is found in gynecologic cytopathology where automated systems are used to screen Pap smears for abnormal cells. Slides with abnormal cells are then forwarded to the pathologist for final diagnosis.
Developing a tissue screening tool is challenging because unlike cytology, tissue has architectural context, stroma, glands, inflammation, and biopsy artifacts that must be taken into account. Pathology in the GI tract can be patchy and therefore a system must scan all tissue fragments on all levels and be evaluated at different magnifications. Criteria used for detection of pathological processes may need to be individualized for the specific pathology and take into account the location of the biopsy since histologies vary within the GI tract. Furthermore, algorithms designed for detection of colitis may differ from algorithms for other diseases. If a robust screening system is developed, just how independent could a system be allowed to operate? Tissue evaluation lacks the intermediary examiners (cytotechnologist) to review slides flagged by their automated systems. Giving an automated system total authority for diagnosis (even for normal tissue) could be problematic since at least in the pediatric realm certain serious GI conditions can have relatively normal appearing biopsies yet have subtle pathology that can be recognized and diagnosed (e.g. tufting enteropathy).
In collaboration with the laboratory of Professor Jelena Kovačević, preliminary work on the development of an automated image analysis platform to distinguish colitis from normal colon in biopsies has yielded promising results. In brief, the methods involve using a pixel-level classifier derived from a set of features described by a “histopathology vocabulary”.[20] The histopathology vocabulary takes prioritized descriptive terms to describe key pathological findings and translates them to mathematical expressions. This classifier was able to accurately distinguish biopsies with colitis from normal with 90% or greater accuracy [Figure 2]. This classifier performed similarly or better compared to other available classifiers.[21] Developing a tissue recognition system that can work in the diagnostic/clinical setting may have value in the current health care climate.
Figure 2.
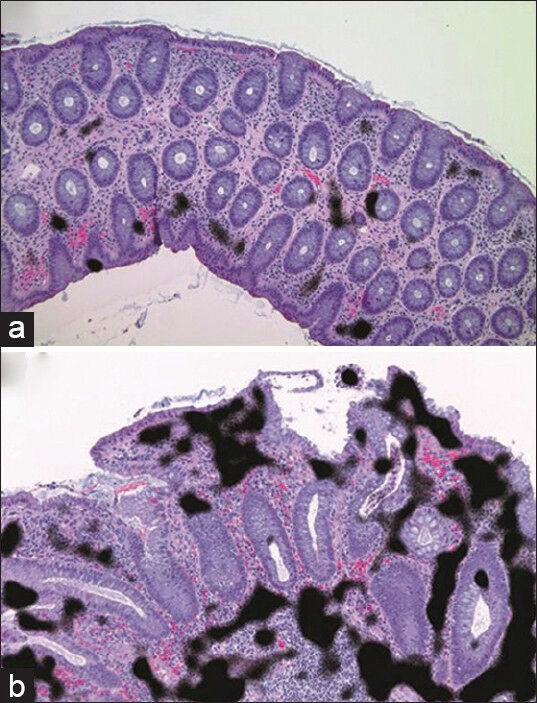
Medium magnification view of normal colon (a) and colitis (b). The pixel level classifier detected areas of high nuclear density within the stroma (black regions) and classified this biopsy image as having colitis
Barriers to the Adoption of Digital Pathology in Clinical Practice
Liron Pantanowitz
Department of Pathology, University of Pittsburgh Medical Center, Pittsburgh, PA, USA
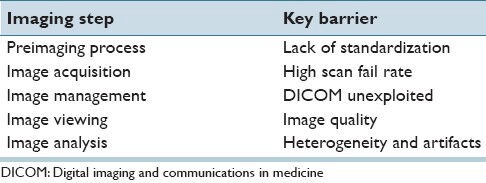
Although digital pathology is being utilized for a plethora of reasons, there are several barriers responsible for its slow adoption in current clinical practice. One of the key barriers remains the pathologist's mindset. Besides those pathologists who are technophobic, there are many who feel that the available technology is not mature enough to replace their microscopes. Perhaps others are concerned that digital pathology may ultimately replace pathologists. Despite the fact it is logistically easier to move an image around than a patient or pathologist, even across geographic boundaries, physician licensure in countries like the United States is limited by state borders. This problem represents a major federal barrier to the practice of telepathology.[22] Limitations by the United States Food and Drug Administration on making primary diagnoses using digital pathology has many pathologists anxious about using this technology in their clinical practice. In order to address many of the other key barriers, it is best to approach them systematically according to the steps involved in the digital imaging process [Table 1].
Table 1.
Principal barriers to the adoption of digital pathology
Preimaging Problems
The garbage in, garbage out principle readily applies to preimaging barriers in digital pathology. The quality of a digital image is dependent on the quality of the fixed/frozen, stained and sometimes sectioned cellular material to be imaged. Studies have shown that thinner, more consistent tissue sectioning results in faster WSI capture times and better image quality.[23] Better efforts are needed to standardize fixation, staining and slide preparation, akin to those that were applied to liquid-based cytology in order to successfully perform automated digital screening of Pap tests. Slides without air bubbles [Figure 3a], dirt and tissue section folds will need to be prepared. Even pen marks may have to be removed, as is commonly performed when dotting screened glass cytology slides, because they may cause digital image artifacts [Figure 3b].[24]
Figure 3.
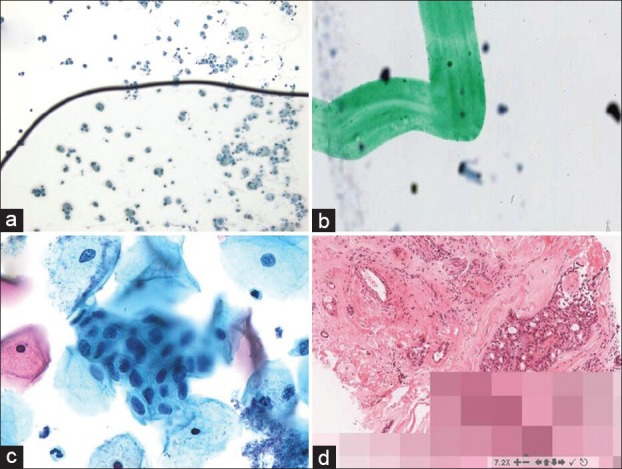
Digital pathology image aberrations. (a) An air bubble on this slide has caused many of the cells to be out of focus. (b) The green dotting pen mark on this slide is in focus whereas the cells are not. (c) Not all of the endocervical cells in this cell group are in focus because this Pap test slide was scanned with a single z plane. (d) Pixilated image due to slow internet connectivity
Image Acquisition Issues
Recent evaluations of WSI scanners show that, while these devices have improved considerably in the last 5 years, they still have a way to go. Indeed, some instruments can scan glass slides <60 s at ×40. However, when you factor in the time for slide preparation and image interpretation, which may amount to 9-35 min for a frozen section case,[25] this is slower than what pathologists are used to when using glass slides. Moreover, WSI scanners may fail to scan certain glass slides, albeit that the scan failure rate is reported to be low by vendors. Pathologists should be aware that glass slides that are too thick, broken or have material on them that is scant and very pale very often may not be digitized. Computerized workflow simulations indicate that if a WSI robot is introduced into the current workflow of a high-volume histology laboratory, without making significant changes to the current workflow of that laboratory, it would be very disruptive and costly.[26] In addition, not all WSI scanners offer z-staking to view the multiple focal planes sometimes needed for interpreting cytology slides with thick smears or that contain three-dimensional cell groups [Figure 3c]. Unfortunately, those scanners that do provide the functionality to acquire multiplane images take a long time to scan slides and produce large digital files.
Image Management Dilemmas
The compliance by picture archiving and communication system vendors with the digital imaging and communications in medicine (DICOM) standard plays an important role in the success of digital radiology. Although supplement 145 of DICOM promotes some standardization in digital pathology,[27] most digital pathology and laboratory information system (LIS) vendors have not yet adopted the DICOM standard for handling digital images. Therefore, images integrated into a LIS currently inhibit their portability. In order to reliably evaluate a digital image it is important that it be associated with relevant metadata. In order for this to happen, more vendors will need to offer LIS integration with digital imaging systems. Given the large size of many WSI files, particularly when compared to radiology images, the magnitude of WSI datasets has demanding storage needs, which are an impediment for many laboratories. It is unclear if commercial cloud services will solve this problem in the near future.
Image Viewing Woes
Despite technical advances with digital cameras, for many pathologists image resolution is still not ideal. Pathologists complain that images are never quite in focus at low magnification and appear too pixilated when they zoom in for higher magnification views. Image “quality” is dependent on the digital camera's sensor used to acquire images, the microscope optics (where objectives with higher numerical aperture provide better resolution), and the display on which images are viewed. Medical grade monitors have been shown to be superior to commercial computer displays.[28] However, given the expense of some medical grade monitors many pathologists currently do not use these displays to view digital images. Individual display parameters such as color are also important, and only recently is attention being devoted to this level of detail when interpreting digital pathology images.[29] Guidelines advising pathologists of the ideal display parameters are currently lacking. Perhaps nothing is more frustrating for a pathologist than waiting for pixels to slowly fill in or freeze on their monitor when viewing a digital image [Figure 3d]. Reliable network connectivity with ample bandwidth is imperative when viewing digital pathology images. Such problems have tainted many pathologists’ experience with digital pathology.
Image Analysis Quandaries
Image analysis indisputably has many benefits such as computer-aided diagnosis. However, in order for many image algorithms to become a part of daily pathology practice several concerns need to addressed. For example, it is unclear if whole slides or just “hot spots” of images should be analyzed. Many tumors contain tumor infiltrating lymphocytes. Hence, algorithms that include such nonneoplastic cells may overestimate scores. Furthermore, tissues often exhibit artifacts such as crushed regions of tumor. Algorithms that exclude crushed nuclei may accordingly underestimate scores. Finally, although image analysis has been shown to produce more accurate and reproducible quantification than pathologists are able to manually determine, only a few studies have shown that this really makes a difference clinically.[30]
Clearly, there are scores of barriers that explain why digital pathology is not currently being widely used by pathologists. In order for this to happen, the following are needed to overcome these barriers:
Standardization of the entire imaging process is needed
Technology needs to be better, faster and cheaper
Digital pathology applications need to be cost-effective
Rules and regulations need to facilitate digital practice
Algorithms need to better mimic real pathology practice
More pathologists need to start using digital pathology.
CONCLUSION
The 2014 CMU Bioimaging day provided the audience with an opportunity to interact with experts in the field of digital pathology and quantitative biomedical imaging. Digital imaging in pathology has evolved significantly over the last decade and has transformed modern pathology practice to the extent that very few practices in pathology today can function without relying in part on digital images and the information gleaned from certain imaging modalities. The future of digital pathology is promising. New technologies are constantly being developed and tested to observe and record the structural, functional and molecular characteristics of cells and tissues with finer detail. The CMU Bioimaging day provided key insights into some of the current applications of digital pathology in clinical practice, use of novel imaging modalities, and provided an overview of current and potential limitations and barriers. This symposium promises to be an attractive forum to bring individuals with varying interest and expertise in digital imaging together to discuss important aspects of bio-imaging as applied to the life sciences and healthcare.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2014/5/1/32/139712
REFERENCES
- 1.Pantanowitz L, Valenstein PN, Evans AJ, Kaplan KJ, Pfeifer JD, Wilbur DC, et al. Review of the current state of whole slide imaging in pathology. J Pathol Inform. 2011;2:36. doi: 10.4103/2153-3539.83746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho J, Parwani AV, Jukic DM, Yagi Y, Anthony L, Gilbertson JR. Use of whole slide imaging in surgical pathology quality assurance: Design and pilot validation studies. Hum Pathol. 2006;37:322–31. doi: 10.1016/j.humpath.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH. Digital pathology: Exploring its applications in diagnostic surgical pathology practice. Pathology. 2010;42:512–8. doi: 10.3109/00313025.2010.508787. [DOI] [PubMed] [Google Scholar]
- 4.Wilbur DC, Madi K, Colvin RB, Duncan LM, Faquin WC, Ferry JA, et al. Whole-slide imaging digital pathology as a platform for teleconsultation: A pilot study using paired subspecialist correlations. Arch Pathol Lab Med. 2009;133:1949–53. doi: 10.1043/1543-2165-133.12.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornish TC, Swapp RE, Kaplan KJ. Whole-slide Imaging: Routine Pathologic Diagnosis. Adv Anat Pathol. 2012;19:152–9. doi: 10.1097/PAP.0b013e318253459e. [DOI] [PubMed] [Google Scholar]
- 6.Isaacs M, Lennerz JK, Yates S, Clermont W, Rossi J, Pfeifer JD. Implementation of whole slide imaging in surgical pathology: A value added approach. J Pathol Inform. 2011;2:39. doi: 10.4103/2153-3539.84232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pantanowitz L, Sinard JH, Fatheree LA, Henricks WH, Carter AB, Contis L, et al. Validating whole slide imaging for diagnostic purposes in pathology: Guideline from the College of American Pathologists Pathology and Laboratory Quality Center. Arch Pathol Lab Med. 2013;137:1710–122. doi: 10.5858/arpa.2013-0093-CP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. 5th ed. Garland Science; 2008. Molecular Biology of the Cell. [Google Scholar]
- 9.Karakitsos P, Cochand-Priollet B, Guillausseau PJ, Pouliakis A. Potential of the back propagation neural network in the morphologic examination of thyroid lesions. Anal Quant Cytol Histol. 1996;18:494–500. [PubMed] [Google Scholar]
- 10.Mohler JL, Partin AW, Lohr WD, Coffey DS. Nuclear roundness factor measurement for assessment of prognosis of patients with prostatic carcinoma. I. Testing of a digitization system. J Urol. 1988;139:1080–4. doi: 10.1016/s0022-5347(17)42791-1. [DOI] [PubMed] [Google Scholar]
- 11.Deacu M, Aşchie M, Boşoteanu M, Petcu L. Nuclear comparative morphometric study between DCIS and normal resting mammary gland tissue. Rom J Morphol Embryol. 2011;52:303–8. [PubMed] [Google Scholar]
- 12.Partin AW, Walsh AC, Pitcock RV, Mohler JL, Epstein JI, Coffey DS. A comparison of nuclear morphometry and Gleason grade as a predictor of prognosis in stage A2 prostate cancer: A critical analysis. J Urol. 1989;142:1254–8. doi: 10.1016/s0022-5347(17)39049-3. [DOI] [PubMed] [Google Scholar]
- 13.Cheng C, Wang W, Ozolek JA, Rohde GK. A flexible and robust approach for segmenting cell nuclei from 2D microscopy images using supervised learning and template matching. Cytometry A. 2013;83:495–507. doi: 10.1002/cyto.a.22280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castleman KR. Upper Saddle River. New Jersey: Prentice Hall; 1995. Digital Image Processing. [Google Scholar]
- 15.Wang W, Ozolek JA, Rohde GK. Detection and classification of thyroid follicular lesions based on nuclear structure from histopathology images. Cytometry A. 2010;77:485–494. doi: 10.1002/cyto.a.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu S, Kolouri S, Rohde GK. Detecting and visualizing cell phenotype differences from microscopy images using transport-based morphometry. Proc Natl Acad Sci U S A. 2014;111:3448–53. doi: 10.1073/pnas.1319779111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W, Slepčev D, Basu S, Ozolek JA, Rohde GK. A linear optimal transportation framework for quantifying and visualizing variations in sets of images. Int J Comput Vis. 2013;101:254–269. doi: 10.1007/s11263-012-0566-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozolek JA, Tosun AB, Wang W, Chen C, Kolouri S, Basu S, et al. Accurate diagnosis of thyroid follicular lesions from nuclear morphology using supervised learning. Med Image Anal. 2014;18:772–80. doi: 10.1016/j.media.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tosun AB, Yergiyev O, Kolouri S, Silverman JF, Rohde GK. Novel computeraided diagnosis of mesothelioma using nuclear structure of mesothelial cells in effusion cytology specimens. SPIE Medical Imaging. 2014:90410Z–90410Z-6. [Google Scholar]
- 20.Bhagavatula R, Fickus MC, Kelly JW, Guo C, Ozolek JA, Castro CA, et al. Automatic identification and delineation of germ layer components in H&E stained images of teratomas derived from human and nonhuman primate embryonic stem cells. Proc. IEEE Int Symp Biomed Imaging, Rotterdam, Netherlands. 2010 Apr;:1041–4. doi: 10.1109/ISBI.2010.5490168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCann MT, Bhagavatula R, Fickus MC, Ozolek JA, Kovacevic J. Orlando, FL: Processing IEEE International Conference Image Process; 2012. Automated colitis detection from endoscopic biopsies as a tissue screening tool in diagnostic pathology; pp. 2809–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allen TC. Digital pathology and federalism. Arch Pathol Lab Med. 2014;138:162–5. doi: 10.5858/arpa.2013-0258-ED. [DOI] [PubMed] [Google Scholar]
- 23.Yagi Y, Gilbertson JR. A relationship between slide quality and image quality in whole slide imaging (WSI) Diagn Pathol. 2008;(3 Suppl 1):S12. doi: 10.1186/1746-1596-3-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collins R, Parwani AV, Khalbuss WE, Duboy J, Pantanowitz L. Effect of pen marking on glass slides for whole slide image scanning. J Pathol Inform. 2012;3:S3. [Google Scholar]
- 25.Evans AJ, Chetty R, Clarke BA, Croul S, Ghazarian DM, Kiehl TR, et al. Primary frozen section diagnosis by robotic microscopy and virtual slide telepathology: The University Health Network experience. Hum Pathol. 2009;40:1070–81. doi: 10.1016/j.humpath.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 26.McClintock DS, Lee RE, Gilbertson JR. Using computerized workflow simulations to assess the feasibility of whole slide imaging full adoption in a high-volume histology laboratory. Anal Cell Pathol (Amst) 2012;35:57–64. doi: 10.3233/ACP-2011-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R, Chubb L, Pantanowitz L, Parwani A. Standardization in digital pathology: Supplement 145 of the DICOM standards. J Pathol Inform. 2011;2:23. doi: 10.4103/2153-3539.80719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma G, Sharma G, Shah A, Duboy J, Parwani AV, Khalbuss WE, et al. Evaluation of different display modalities for whole slide images in pathology. J Pathol Inform. 2011;2:43. S4-S5. [Google Scholar]
- 29.Bautista PA, Hashimoto N, Yagi Y. Color standardization in whole slide imaging using a color calibration slide. J Pathol Inform. 2014;5:4. doi: 10.4103/2153-3539.126153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown JR, DiGiovanna MP, Killelea B, Lannin DR, Rimm DL. Quantitative assessment Ki-67 score for prediction of response to neoadjuvant chemotherapy in breast cancer. Lab Invest. 2014;94:98–106. doi: 10.1038/labinvest.2013.128. [DOI] [PubMed] [Google Scholar]


