Abstract
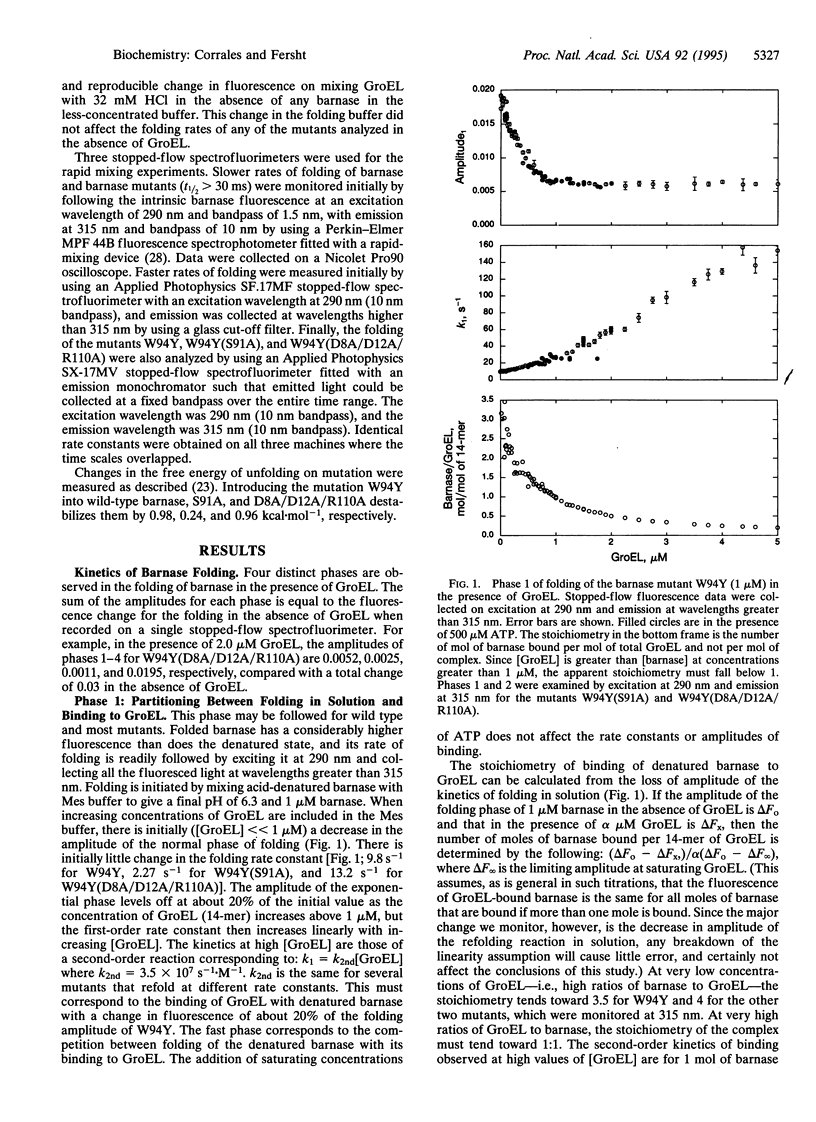
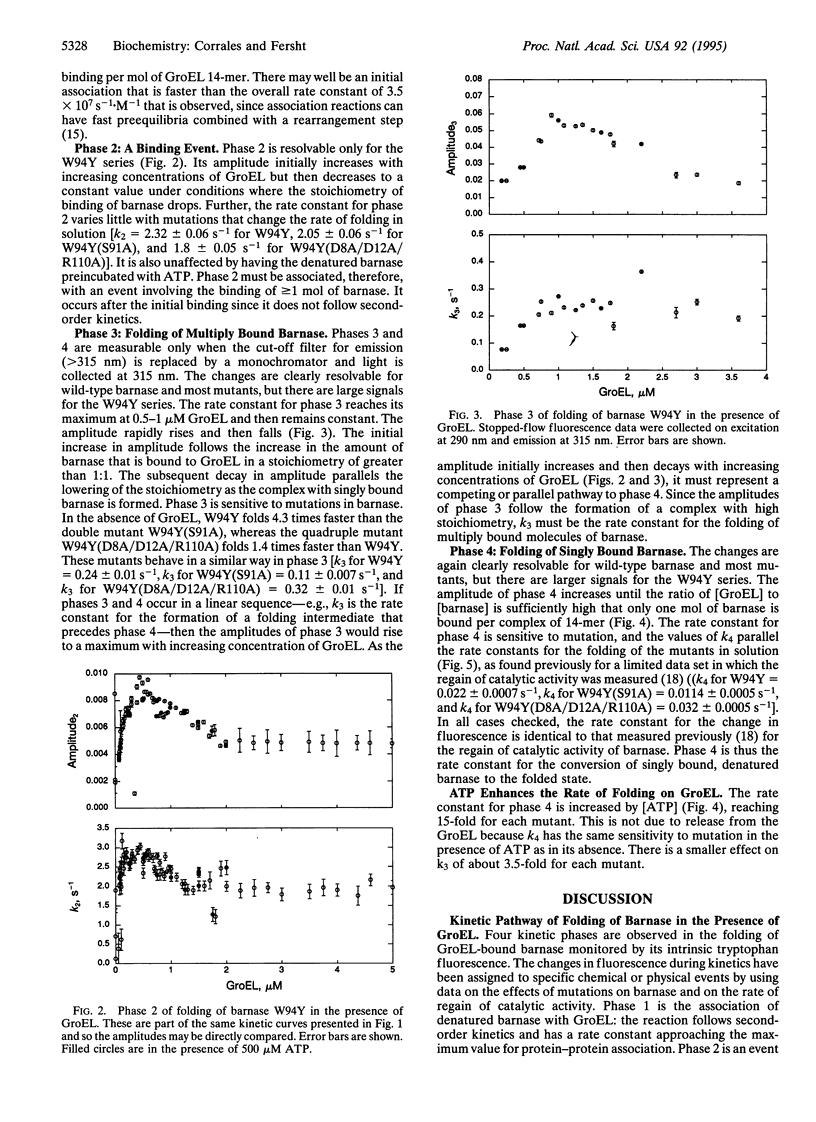
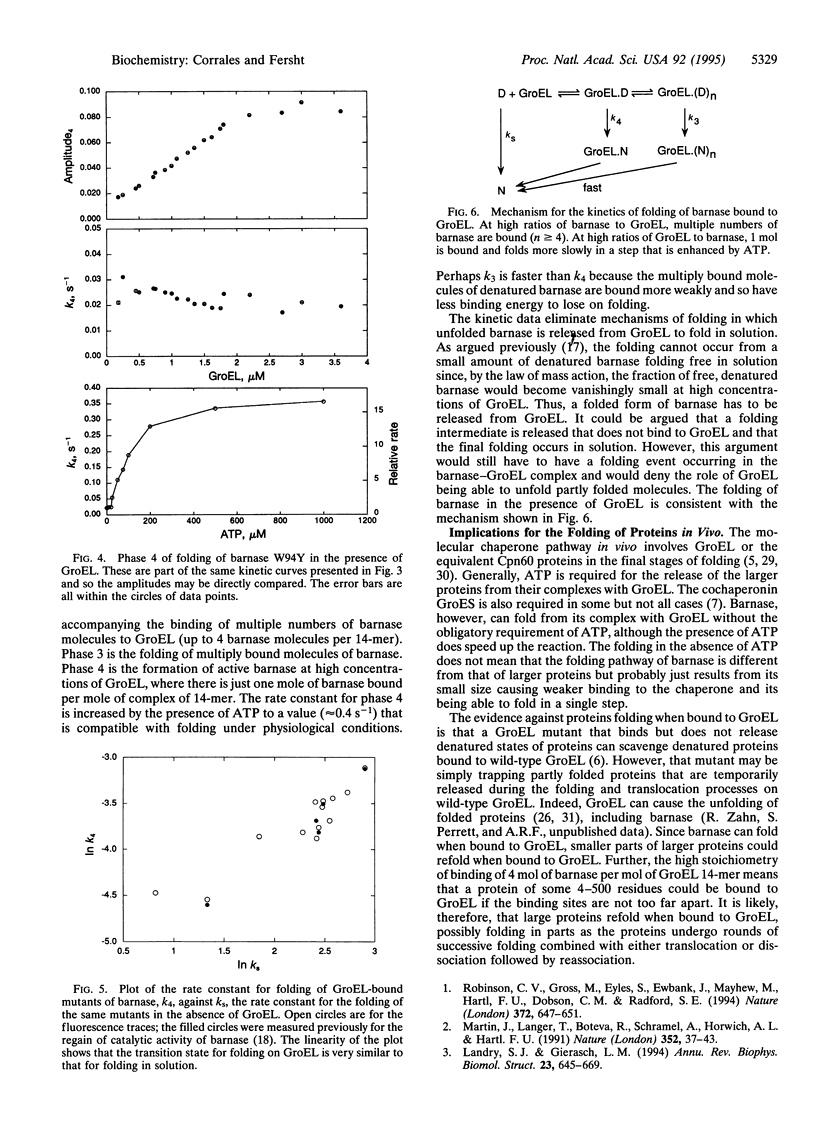
We have analyzed the pathway of folding of barnase bound to GroEL to resolve the controversy of whether proteins can fold while bound to chaperonins (GroEL or Cpn60) or fold only after their release into solution. Four phases in the folding were detected by rapid-reaction kinetic measurements of the intrinsic fluorescence of both wild type and barnase mutants. The phases were assigned from their rate laws, sensitivity to mutations, and correspondence to regain of catalytic activity. At high ratios of denatured barnase to GroEL, 4 mol of barnase rapidly bind per 14-mer of GroEL. At high ratios of GroEL to barnase, 1 mol of barnase binds with a rate constant of 3.5 x 10(7) s-1.M-1. This molecule then refolds with a low rate constant that changes on mutation in parallel with the rate constant for the folding in solution. This rate constant corresponds to the regain of the overall catalytic activity of barnase and increases 15-fold on the addition of ATP to a physiologically relevant value of approximately 0.4 s-1. The multiply bound molecules of barnase that are present at high ratios of GroEL to barnase fold with a rate constant that is also sensitive to mutation but is 10 times higher. If the 110-residue barnase can fold when bound to GroEL and many moles can bind simultaneously, then smaller parts of large proteins should be able to fold while bound.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen S., Roseman A. M., Hunter A. S., Wood S. P., Burston S. G., Ranson N. A., Clarke A. R., Saibil H. R. Location of a folding protein and shape changes in GroEL-GroES complexes imaged by cryo-electron microscopy. Nature. 1994 Sep 15;371(6494):261–264. doi: 10.1038/371261a0. [DOI] [PubMed] [Google Scholar]
- Frydman J., Nimmesgern E., Ohtsuka K., Hartl F. U. Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature. 1994 Jul 14;370(6485):111–117. doi: 10.1038/370111a0. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Gray T. E., Eder J., Bycroft M., Day A. G., Fersht A. R. Refolding of barnase mutants and pro-barnase in the presence and absence of GroEL. EMBO J. 1993 Nov;12(11):4145–4150. doi: 10.1002/j.1460-2075.1993.tb06098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. E., Fersht A. R. Refolding of barnase in the presence of GroE. J Mol Biol. 1993 Aug 20;232(4):1197–1207. doi: 10.1006/jmbi.1993.1471. [DOI] [PubMed] [Google Scholar]
- Hartl F. U., Hlodan R., Langer T. Molecular chaperones in protein folding: the art of avoiding sticky situations. Trends Biochem Sci. 1994 Jan;19(1):20–25. doi: 10.1016/0968-0004(94)90169-4. [DOI] [PubMed] [Google Scholar]
- Hendrick J. P., Hartl F. U. Molecular chaperone functions of heat-shock proteins. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- Horovitz A., Fersht A. R. Co-operative interactions during protein folding. J Mol Biol. 1992 Apr 5;224(3):733–740. doi: 10.1016/0022-2836(92)90557-z. [DOI] [PubMed] [Google Scholar]
- Horovitz A., Serrano L., Avron B., Bycroft M., Fersht A. R. Strength and co-operativity of contributions of surface salt bridges to protein stability. J Mol Biol. 1990 Dec 20;216(4):1031–1044. doi: 10.1016/S0022-2836(99)80018-7. [DOI] [PubMed] [Google Scholar]
- Landry S. J., Gierasch L. M. Polypeptide interactions with molecular chaperones and their relationship to in vivo protein folding. Annu Rev Biophys Biomol Struct. 1994;23:645–669. doi: 10.1146/annurev.bb.23.060194.003241. [DOI] [PubMed] [Google Scholar]
- Loewenthal R., Sancho J., Fersht A. R. Fluorescence spectrum of barnase: contributions of three tryptophan residues and a histidine-related pH dependence. Biochemistry. 1991 Jul 9;30(27):6775–6779. doi: 10.1021/bi00241a021. [DOI] [PubMed] [Google Scholar]
- Martin J., Mayhew M., Langer T., Hartl F. U. The reaction cycle of GroEL and GroES in chaperonin-assisted protein folding. Nature. 1993 Nov 18;366(6452):228–233. doi: 10.1038/366228a0. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Kellis J. T., Jr, Serrano L., Fersht A. R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989 Jul 13;340(6229):122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- Robinson C. V., Gross M., Eyles S. J., Ewbank J. J., Mayhew M., Hartl F. U., Dobson C. M., Radford S. E. Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature. 1994 Dec 15;372(6507):646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Buchner J., Todd M. J., Lorimer G. H., Viitanen P. V. On the role of groES in the chaperonin-assisted folding reaction. Three case studies. J Biol Chem. 1994 Apr 8;269(14):10304–10311. [PubMed] [Google Scholar]
- Serrano L., Horovitz A., Avron B., Bycroft M., Fersht A. R. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry. 1990 Oct 9;29(40):9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- Serrano L., Kellis J. T., Jr, Cann P., Matouschek A., Fersht A. R. The folding of an enzyme. II. Substructure of barnase and the contribution of different interactions to protein stability. J Mol Biol. 1992 Apr 5;224(3):783–804. doi: 10.1016/0022-2836(92)90562-x. [DOI] [PubMed] [Google Scholar]
- Staniforth R. A., Burston S. G., Atkinson T., Clarke A. R. Affinity of chaperonin-60 for a protein substrate and its modulation by nucleotides and chaperonin-10. Biochem J. 1994 Jun 15;300(Pt 3):651–658. doi: 10.1042/bj3000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd M. J., Viitanen P. V., Lorimer G. H. Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science. 1994 Jul 29;265(5172):659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Donaldson G. K., Lorimer G. H., Lubben T. H., Gatenby A. A. Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry. 1991 Oct 8;30(40):9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- Weissman J. S., Kashi Y., Fenton W. A., Horwich A. L. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994 Aug 26;78(4):693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- Zahn R., Spitzfaden C., Ottiger M., Wüthrich K., Plückthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994 Mar 17;368(6468):261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]



