Abstract

Parkinson’s disease (PD) is a multifactorial disorder with a complex etiology including genetic risk factors, environmental exposures, and aging. While energy failure and oxidative stress have largely been associated with the loss of dopaminergic cells in PD and the toxicity induced by mitochondrial/environmental toxins, very little is known regarding the alterations in energy metabolism associated with mitochondrial dysfunction and their causative role in cell death progression. In this study, we investigated the alterations in the energy/redox-metabolome in dopaminergic cells exposed to environmental/mitochondrial toxins (paraquat, rotenone, 1-methyl-4-phenylpyridinium [MPP+], and 6-hydroxydopamine [6-OHDA]) in order to identify common and/or different mechanisms of toxicity. A combined metabolomics approach using nuclear magnetic resonance (NMR) and direct-infusion electrospray ionization mass spectrometry (DI-ESI-MS) was used to identify unique metabolic profile changes in response to these neurotoxins. Paraquat exposure induced the most profound alterations in the pentose phosphate pathway (PPP) metabolome. 13C-glucose flux analysis corroborated that PPP metabolites such as glucose-6-phosphate, fructose-6-phosphate, glucono-1,5-lactone, and erythrose-4-phosphate were increased by paraquat treatment, which was paralleled by inhibition of glycolysis and the TCA cycle. Proteomic analysis also found an increase in the expression of glucose-6-phosphate dehydrogenase (G6PD), which supplies reducing equivalents by regenerating nicotinamide adenine dinucleotide phosphate (NADPH) levels. Overexpression of G6PD selectively increased paraquat toxicity, while its inhibition with 6-aminonicotinamide inhibited paraquat-induced oxidative stress and cell death. These results suggest that paraquat “hijacks” the PPP to increase NADPH reducing equivalents and stimulate paraquat redox cycling, oxidative stress, and cell death. Our study clearly demonstrates that alterations in energy metabolism, which are specific for distinct mitochondiral/environmental toxins, are not bystanders to energy failure but also contribute significant to cell death progression.
Parkinson’s disease (PD) has been presented as a complex and heterogeneous disease with unclear pathological and etiological mechanisms. Since epidemiological data suggest an association between PD and environmental toxicant exposure, the multifactorial etiology of PD has been now indicated to include environmental toxicity in addition to mutations and aging as major risk factors.1 To date, there is no experimental model that recapitulates all biochemical, pathological, or symptomatic aspects of PD. A number of toxicological models have been established to study dopaminergic cell death, which address the role of oxidative stress, mitochondrial dysfunction, and dopamine metabolism. Recent studies have demonstrated that environmental exposure to the pesticides paraquat or rotenone could increase the risk of developing PD.2 In addition, a dysfunction in the electron transport chain (ETC) has been found in PD brains. Thus, inhibitors of complex I activity such as methyl-4-phenylpyridinium (MPP+)/1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) and rotenone are used to induce mitochondrial dysfunction in dopaminergic cells.3 Oxidative stress in PD is also associated with the pro-oxidant metabolism of dopamine. When injected into the SNpc, the hydroxylated analogue of dopamine, 6-hydroxydopamine (6-OHDA), induces degeneration of the nigrostriatal dopaminergic system by oxidative damage generated via its auto-oxidation.4
Exposures to paraquat, rotenone, MPP+/MPTP or 6-OHDA have been largely used in vitro and in vivo as experimental PD models.3 However, distinct mechanisms are known to mediate their toxic effects. For example, even though paraquat and 6-OHDA are known to induce oxidative stress, the former is known to act as a generator of mitochondrial superoxide anion,5 while 6-OHDA’s auto-oxidation triggers the formation of reactive quinones.6 Thus, both similar and different signal transduction pathways have been described to regulate the toxicity of both neurotoxins.7−9 Similarly, while the complex I inhibitors rotenone and MPP+ are thought to exert their toxic effects by similar mechanisms, other studies have shown that MPP+/MPTP and rotenone toxicity is mediated by mechanisms independent from complex I inhibition10 and the generation of ROS.8,11 Furthermore, recent reports have demonstrated that rotenone and MPP+ actually exert distinct alterations in cellular metabolism and activation of signaling cascades, supporting the idea that their toxicity is mediated by distinct mechanisms.12 Because these different toxicological models address a specific hallmark of PD, that is, mitochondrial dysfunction, oxidative stress, and dopamine toxic metabolism, understanding the molecular mechanisms that mediate their toxicity is of great importance.
In the brain, both energy metabolism and bioenergetics are tightly coupled. Glucose is the obligatory energy substrate of the adult brain. Neurons primarily metabolize glucose via the pentose phosphate pathway (PPP) to provide reducing equivalents required to maintain antioxidant defenses via the production of nicotinamide adenine dinucleotide phosphate (NADPH).13 Dopaminergic neurons in the substantia nigra consume a significant amount of energy during their pacemaking activity, which leads to increased levels of basal oxidative stress.14 Energy failure associated with mitochondrial dysfunction is the hallmark of PD. Dysfunction of the electron transport chain (ETC), tricarboxylic acid cycle (TCA or Krebs cycle), and oxidative phosphorylation (OXPHOS) has been reported in PD brains.15,16 A decrease in glucose metabolism and abnormally elevated lactate levels has also been reported in PD patients.17−19 In addition, down-regulation of PPP enzymes and a failure to increase the antioxidant reserve is an early event in the pathogenesis of sporadic PD.20 While energy failure has been largely associated with the loss of dopaminergic cells in PD and the toxicity induced by mitochondrial/environmental toxins, very little is known regarding the alterations in energy metabolism associated with mitochondrial dysfunction and their causative role in cell death progression.
Biochemical biomarkers represent changes which can be indicative of disease mechanisms.21 Most of the studies so far regarding the identification of biochemical biomarkers for PD have been focused primarily on proteomic studies.22 While the identification of biomarkers from biofluids or neuroimaging are invaluable for diagnosing PD, metabolomics can also provide insights into the molecular mechanisms of disease development and progression for the development of effective and personalized treatments of PD. In this study, we aimed first to identify the specific alterations in the metabolome of dopaminergic cells induced by exposure to environmental/mitochondrial toxins to reveal novel molecular mechanism involved in dopaminergic cell death, and second, to establish a causative role for changes in energy/redox metabolism in dopaminergic cell death. Our data shows that paraquat, rotenone, MPP+, and 6-OHDA elicit major and distinct metabolic alterations with significant differences between them. Paraquat selectively up-regulates the pentose phosphate pathway (PPP) and glucose-6-phosphate dehydrogenase (G6PD) levels, the rate-limiting enzyme of the PPP, which was paralleled by a concomitant down-regulation of glycolysis and the TCA cycle. G6PD was shown to selectively regulate paraquat-induced oxidative stress and apoptotic cell death. These findings provide a valuable insight into the neurotoxicity mechanism of paraquat and demonstrate the importance of alterations in energy/redox metabolism in environmental toxicity. Our results reveal that alterations induced by environmental/mitochondrial toxins are not bystanders to energy failure, but instead, contribute significantly to cell death progression.
Results and Discusion
Cell Death Is Irreversible after 24 h of Exposure to Toxic Paraquat Concentrations
Exposures to environmental/mitochondrial toxins are widely used to study dopaminergic cell death associated with PD. We observed that the exposure of dopaminergic cells to a toxic dose of paraquat (>0.5 mM) for 24 h, followed by a 48 h incubation period in media without paraquat, induced a similar degree of cell death relative to a continuous 72 h exposure to this toxin (Figure 1A–C). When the medium was exchanged after 24 h of treatment with paraquat with fresh medium also containing paraquat, no additional toxicity was observed compared to either a 24 h exposure and medium removal (wash-out) or a continuous 72 h treatment with paraquat (Figure 1B). Interestingly, MPP+- (Figure 1A–C), 6-OHDA- and rotenone-induced cell death (Figure 1A) required their continuous presence for 72 h in order to induce a comparable degree of cell death to that induced by paraquat treatment for only 24 h followed by a 48 h incubation period with fresh medium (Figure 2C). These results suggested that a significant biochemical alteration occurs in cells treated with paraquat for 24 h that irreversibly commits the cells to undergo cell death.
Figure 1.
Cell death is triggered irreversibly after 24 h of exposure to a toxic paraquat (PQ) concentration. (A) Human dopaminergic neuroblastoma cells (SK-N-SH) were exposed to paraquat (0.5 mM, PQ), MPP+ (2.5 mM), rotenone (4 μM) or 6-OHDA (50 μM). Phase contrast (20×) images were taken at the time indicated. Insets represent a 2.3× magnification from the area indicated (broken line squares). (B–C) Cells were exposed to paraquat (0.5 mM) or MPP+ (2.5 mM) for 24 h, and then, (a) cells were incubated with fresh medium for 48 h; (b) cells were kept with the neurotoxin for additional 48 h (72 h total); or (bi) media was exchanged with fresh medium + neurotoxin for additional 48 h. Cell death was quantified after 72 h using PI uptake as a marker for plasma membrane integrity. Data in C represent means ± SE of 3 independent experiments. *p < 0.05, 72 h vs 24 h treatments. (D) Cell death induced by different periods of incubation with paraquat evaluated at 72 h after treatment.
Figure 2.
Alterations in the metabolome induced by exposure to neurotoxins. Cells were treated with paraquat (0.5 mM), rotenone (4 μM), MPP+ (2.5 mM), or 6-OHDA (50 μM) for 24 h. LDA plots were generated from 1D 1H NMR spectra (A), DI-ESI-MS spectra (B), or the combined 1D 1H NMR and DI-ESI-MS data sets (C). The group separation in a LDA plot identifies the similarity or difference between the cellular metabolomes of cells treated with the different toxins. The ellipsoids correspond to the 95% confidence limits from a normal distribution for each cluster. The associated dendrograms were generated based on the 3D MB-PCA scores and were used to further visualize the class separation in the LDA plots. The statistical significance of the class separation is indicated by the p-value listed at each node. (D) Cell death was evaluated at 48 h after exposure to the indicated neurotoxin using PI. Data in A–C represent means of 6 independent samples. Data in D represent means ± SE of 3 independent experiments.
Toxicity of Environmental/Mitochondrial Toxins Is Associated with Changes in Their Metabolome Prior to Cell Death
A combination of analytical techniques provides an enhanced view of the metabolome since each individual method is typically limited to detecting only a subset of metabolites.23 For the first time, we integrated positive-ion direct infusion electrospray ionization mass spectrometry (DI-ESI-MS) and (one-dimensional) 1D 1H NMR techniques along with an integrated chemometrics approach to characterize the alterations in the metabolome induced by neurotoxins. The Linear Discriminant Analysis (LDA) plots and the three-dimensional (3D) Multiblock Principle Component Analysis (MB-PCA) dendrograms for the 1D 1H NMR spectra, DI-ESI-MS spectra, and the combined NMR and DI-ESI-MS data set are shown in Figures 2A, B, and C, respectively. The LDA plot is used to project a 3D MB-PCA scores plot in two-dimensions with an orientation that captures the maximal between class separations. The relative clustering of each group in the LDA plot is an indicator of the similarity in the spectral data, and, correspondingly, the cellular metabolome. The 3D MB-PCA dendrograms provides an alternative approach for quantifying group similarities by determining the statistical significance of the group separation in the MB-PCA scores plots. The fact that the clustering patterns in the LDA plots differ slightly between the 1D 1H NMR (Figure 2A) and DI-ESI-MS (Figures 2B) data sets is not unexpected since the two methods highlight different subregions of the metabolome. More importantly, the LDA plot of the combined NMR and DI-ESI-MS data set (Figures 2C) generated the best separation between the four treatment groups and the control, demonstrating that our integrated NMR/DI-ESI-MS approach enhances our ability to distinguish subtle changes in the metabolome of dopaminergic cells upon treatment with environmental/mitochondrial toxins.
The pairwise p-values from the dendrogram calculation for the combined NMR and DI-ESI-MS data set are listed in Table 1. The p-value represents a relative distance between each pair, where a lower p-value corresponds to a larger pairwise separation. Correspondingly, all groups were found to be significantly separated between each other when using the combined NMR and DI-ESI-MS data sets (Figure 2C and Table 1). Thus, all treatments (paraquat, rotenone, MPP+, and 6-OHDA) induced not only a significant metabolic change when compared to control conditions but also specific and distinct metabolic changes between them. The different metabolic shifts between experimental treatments cannot be attributed to differences in the degree of toxicity induced, as evidenced by the similar degree of cell death induced at 48 h post-treatment by the concentrations tested (Figure 2D).
Table 1. Pairwise Matrix of p Values from 3D MB-PCA Dendograma.
| C | P | M | R | O | |
|---|---|---|---|---|---|
| C | 5.50 × 10–6 | 4.67 × 10–8 | 4.72 × 10–7 | 2.99 × 10–5 | |
| P | 5.50 × 10–6 | 6.25 × 10–8 | 7.70 × 10–6 | 5.91 × 10–4 | |
| M | 4.67 × 10–8 | 6.25 × 10–8 | 1.06 × 10–5 | 2.40 × 10–6 | |
| R | 4.72 × 10–7 | 7.70 × 10–6 | 1.06 × 10–5 | 1.67 × 10–3 | |
| O | 2.99 × 10–5 | 5.91 × 10–4 | 2.40 × 10–6 | 1.67 × 10–3 |
3D MB-PCA scores plot was generated from the integrated 1D 1H NMR and DI-ESI-MS data set (Figure 2C). Pairwise p values were calculated from the 3D MB-PCA scores using our PCA/PLS-DA utilities (http://bionmr.unl.edu/pca-utils.php). The p value represents a relative distance between each pair, where a lower p value indicates a larger separation.
C, untreated control; P, paraquat; M, MPP+; R, rotenone; O, 6-OHDA.
We further aimed to identify the metabolites that significantly contributed to the class separations in the LDA plot between the paraquat treated cells vs the control and vs other treatment groups. The Multiblock Partial Least Squares Discriminant Analysis (MB-PLS-DA) S-plot generated from the combined 1D 1H NMR and DI-ESI-MS spectra data sets (Figure 3A) identifies the metabolites that were significantly increased (upper right corner) or decreased (lower left corner) after paraquat exposure when compared to control samples. An increase in citrate, glucose-6-phosphate/fructose-6-phoshate, heptose (sedoheptulose), and hexose (glucose or myoinositol), and a decrease in lactate, glutamate, dopamine, and phospho-aspartate were clearly observed in the MB-PLS-DA S-plot (Figure 3A) and in the original 1D 1H NMR (Figure 3C) and DI-ESI-MS spectra (Figure 3D). To determine if these metabolic changes were unique for paraquat, a similar comparison was made between paraquat and the other toxins. The resulting MB-PLS-DA Shared and Unique Structures (SUS) plot is shown in Figure 3B. The SUS plot is a union of two S-plots (paraquat vs controls and paraquat vs MPP+, rotenone, and 6-OHDA [other drugs or toxins]), where features shared by the two S-plots are plotted along the diagonal and features unique to either of the S-plots are plotted off-diagonally. The SUS plot shows that changes in the concentrations of citrate, glucose-6-phosphate/fructose 6-phoshate, hexose, lactate, and dopamine are a unique result of paraquat treatment (Figure 3B).
Figure 3.
Alterations in citrate, glucose-6-phosphate/fructose-6-phosphate, lactate, and glucose content are specific for paraquat treatment. (A) S-plot was generated from the combined MB-PLS-DA of 1D 1H NMR spectra and DI-ESI-MS spectra. The S-plot was used to identify metabolites that significantly contribute to the class separation between untreated controls and paraquat treatment. The metabolites located in the upper right quadrant significantly increased while those located in the lower left quadrant significantly decreased after paraquat exposure. (B) Combined shared and unique structure (SUS)-plots of 1D 1H NMR and DI-ESI-MS data set highlights the correlation between metabolites significantly altered in response to paraquat treatment (control vs paraquat) with the alterations induced by the other neurotoxins (paraquat vs MPP+, rotenone, and 6-OHDA). The positively shared changes (upregulated metabolites) from the both models are located on the upper right corner, while the negatively shared changes (downregulated metabolites) are presented on the lower left corner. The metabolites falling into the blue boxes are unique to the model of other drugs vs paraquat. The red boxes are the boundaries for the metabolites unique to the model of control vs paraquat. The model validation parameters for the S-plot are R2 = 0.999; Q2 = 0.970; CV-ANOVA p value = 1.21 × 10–4. Selected regions of representative 1D 1H NMR (C) and DI-ESI-MS (D) spectra obtained from the metabolome of cells treated with paraquat identifies peaks corresponding to metabolites whose concentrations changed after paraquat treatment (red) when compared to untreated controls (black).
Paraquat Induces an Increase in Metabolites within the Pentose Phosphate Pathway (PPP)
Glucose is the obligatory energy substrate of the adult brain. To further our understanding of the changes in the metabolome resulting from the treatment with the distinct toxins, media was supplemented with 13C glucose, and the distribution of 13C-carbons throughout the metabolome was monitored by two-dimensional (2D) 1H–13C HSQC NMR. Metabolite identification was accomplished by comparing the experimental 1H and 13C chemical shifts to standard values in NMR metabolomics databases and concentration changes were inferred based on changes in peak intensities relative to untreated controls. Consistent with the observed changes in the 1D 1H NMR and DI-ESI-MS data sets, paraquat treatment resulted in an increase in glucose, glucose-6-phosphate, fructose-6-phosphate, glucose-1-phosphate, and glucono-1,5-lactone, which are associated with the pentose phosphate pathway (PPP) (Figure 4A and 7). Similar to MPP+, paraquat decreased purine levels (ATP, ADP, and AMP) (Figure 4B). Paraquat also induced a decrease in metabolites associated with the glycolytic pathway as evidenced by a decrease in 3-phospho glycerate, dihydroxyacetone phosphate (DHAP or glycerone phosphate), lactate, and alanine (Figure 4C and 7). Extracellular metabolite analysis (Figure 4E) was consistent with the intracellular metabolomics data. A decrease in extracellular glucose (Figure 4E) in conjunction with an increase in intracellular glucose (Figure 4A) is indicative of an increase in glucose uptake (Figure 7). A decrease in extracellular lactate and alanine (Figure 4E) correlated with their decreased intracellular levels (Figure 4A). These results demonstrate that paraquat increases PPP metabolite accumulation while decreasing glycolysis (Figure 7).
Figure 4.
Paraquat induces selective changes in glucose metabolism, TCA cycle and the PPP pathway. Cells were treated with paraquat (0.5 mM), rotenone (4 μM), MPP+ (2.5 mM), or 6-OHDA (50 μM) for 24 h in glucose free media supplemented with 13C-glucose (3.5 g/L). Analysis of 2D 1H–13C HSQC NMR spectra was used to evaluate changes in glucose-derived metabolites. Bar graphs indicate the relative changes in peak intensity (concentration) for metabolites associated with (A) the pentose phosphate pathway (PPP), (B) nucleotide biosynthesis, (C) glycolysis, (D) the TCA cycle, and (E) metabolites derived from glucose metabolism and found accumulated in the extracellular media. Data represent means ± SD of 3 independent experiments. *p < 0.05, control vs neurotoxin treatments. ATP/ADP/AMP, ATP, ADP or AMP; DHAP, dihydroxyacetone phosphate; NADP/NADPH, NADP, or NADPH; UDP/UMP, UDP, or UMP.
Figure 7.
Paraquat hijacks the PPP to induce oxidative stress and cell death. Our results demonstrate that paraquat induces an increase in the PPP (highlighted in green), which is reflected by an increase in glucose uptake, and in glucose-6-phosphate, glucono-1,5-lactone, erythrose-4-phosphate, and fructose-6-phosphate content (red arrows). In addition, paraquat decreases glycolysis as demonstrated by a decrease in 3-phosphoglycerate, alanine, and lactate levels (green arrows). These metabolic changes were also paralleled by (1) an increase in G6PD (the rate-limiting enzyme in the PPP), and the expression levels of citrate synthase and pyruvate kinases M1/M2 and (2) a decrease in lactate dehydrogenase A/B chains, which participate in glycolysis and the TCA cycle (highlighted in orange). Paraquat also induced an increase in citrate accumulation, which is associated with the well-known inhibitory effect on aconitase (highlighted in orange). An abnormal increase in citrate levels has been reported to exert an inhibitory effect on glycolysis by allosteric inhibition of PFK (broken red line), which explains why an increase glucose uptake and impaired TCA cycle is not translated to an upregulation in glycolysis. Paraquat also induced a decrease in total GSH and glutamate content. Modulation of G6PD levels and activity was directly linked to paraquat toxicity and oxidative stress (highlighted in green). These results demonstrate a role for PPP and G6PD in paraquat induced toxicity. 6-AN, 6-aminonicotinamide, ACO, aconitase, or aconitate hydratase [EC:4.2.1.3]; ADC, aspartate 4-decarboxylase [EC:4.1.1.12]; ALDO, fructose-bisphosphate aldolase [EC:4.1.2.13]; ALT, alanine transaminase [EC:2.6.1.2]; CS, citrate synthase [EC:2.3.3.1]; FBP, fructose-1,6-bisphosphatase I [EC:3.1.3.11]; FUM, fumarate hydratase [EC:4.2.1.2]; G6PD, glucose-6-phosphate 1-dehydrogenase [EC:1.1.1.49]; GAPDH, glyceraldehyde-3-phosphate dehydrogenase [EC:1.2.1.12]; GLDH, glutamate dehydrogenase, [EC: 1.4.1.2]; GLS, glutaminase [EC: 3.5.1.2]; GOT1, aspartate aminotransferase, cytoplasmic [EC:2.6.1.1]; GPI, glucose-6-phosphate isomerase [EC:5.3.1.9]; HK, hexokinase [EC:2.7.1.1]; IDH, isocitrate dehydrogenase [EC:1.1.1.42]; LDH, l-lactate dehydrogenase [EC:1.1.1.27]; MDH, malate dehydrogenase [EC:1.1.1.37]; OGDH, 2-oxoglutarate dehydrogenase, [EC:1.2.4.2]; LSC, succinyl-CoA synthetase [EC:6.2.1.4 6.2.1.5]; PC, pyruvate carboxylase [EC:6.4.1.1]; PGD, 6-phosphogluconate dehydrogenase [EC:1.1.1.44]; PDH, pyruvate dehydrogenase [EC:1.2.4.1]; PGK1, phosphoglycerate kinase [EC:2.7.2.3]; PGM, phosphoglucomutase [EC:5.4.2.2]; PFK, 6-phosphofructokinase 1 [EC:2.7.1.11]; RPI, ribose-5-phosphate isomerase A [EC:5.3.1.6]; SDH, succinate dehydrogenase [EC:1.3.5.1]; TPI, triosephosphate isomerase [EC:5.3.1.1]; TAL, transaldolase [EC:2.2.1.2]; TKT, transketolase [EC:2.2.1.1].
Both the 1D 1H NMR and DI-ESI-MS data sets and the 2D 1H–13C HSQC NMR analysis of 13C-carbon flux also identified a large increase in citrate and a decrease in aspartate resulting from paraquat treatment (Figure 4D and 7). Iron–sulfur cluster containing proteins, such as aconitase, are important targets for ROS. Aconitase catalyzes the stereospecific isomerization of citrate to isocitrate in the tricarboxylic acid cycle (TCA). Thus, the inhibition of aconitase activity by ROS would be expected to increase the cellular pool of citrate. The observed decrease in aspartate, a product of the TCA cycle generated from the addition of an amino group to oxaloacetate by aspartate aminotransferase (GOT1) (Figure 7), also indirectly supports the inactivation of aconitase by paraquat. Overall, our results are consistent with prior observations demonstrating an increase in citrate accumulation via inhibition of aconitase by paraquat-induced superoxide anion formation,24 as well as an increase in the PPP upon paraquat exposure25 (Figure 7).
Interestingly, the increase in glucose uptake and impairment in the TCA cycle induced by paraquat were not translated in an increase in glycolytic rate (measured by the production of lactate), but rather a decrease in lactate content (Figure 4C and 7). This might be explained by the increased accumulation of citrate, a well-known allosteric inhibitor of phosphofructokinase 1 (PFK, Figure 7, dotted red line),26 which catalyzes the phosphorylation of fructose-6-phosphate to fructose-1,6-bisphosphate, a key regulatory step in the glycolytic pathway. Another possibility is that the increase in acetyl-glucosamine induced by paraquat (Supporting Information Figure 2A) could also inhibit PFK by glycosylation as reported elsewhere.27 In either case, paraquat activity appears to result in a decrease in glycolysis activity through an indirect inhibition of PFK, as evidenced by the decrease in the content of metabolites associated with glycolysis downstream of PFK (Figure 7).
MB-PLS-DA S-plot and SUS plot analyses were also generated from the 1D 1H NMR and DI-ESI-MS data sets for the other neurotoxin treatments (Supporting Information Figure 1) and compared to the 2D 1H–13C HSQC NMR results (Figure 4). Strikingly different shifts in the metabolome were observed for cells treated with rotenone, MPP+ or 6-OHDA compared to paraquat. MPP+, rotenone, and 6-OHDA were shown to increase extracellular lactate accumulation (Figure 4E), which is likely associated with an increase in glycolysis, as previously reported.8,12 Additionally, MPP+, but not rotenone, increased the accumulation of choline-containing metabolites (Supporting Information Figure 2).28 Accordingly, previous findings have revealed abnormally elevated lactate and choline metabolite levels in PD subjects.29,30 MPP+ and rotenone decreased purine (ATP, ADP AMP, and GMP) levels, while pyrimidine content (CMP, UDP, and UMP) were only affected by MPP+.
While 1D 1H NMR analysis identified a decrease in citrate and alanine upon MPP+ treatment, only a slight but nonsignificant decrease was found by 2D 1H–13C HSQC NMR, which might be ascribed to the low basal levels of these metabolites. Similar to paraquat, 6-OHDA and rotenone were found to induce a decrease in alanine and aspartate. In contrast to paraquat, and as reported elsewhere,31 a decrease in citrate content by rotenone was consistently detected by both 1D 1H NMR (Supporting Information Figure 1) and 2D 1H–13C HSQC NMR experiments (Figure 4D), which was similarly reported in PD plasma samples.32 All neurotoxins where shown to significantly reduce glutamate content, while intracellular glutamine levels remained unaltered (Supplementary Figure 2). However, extracellular glutamine accumulation was reduced (Figure 4E). Contradictory findings have been reported regarding the changes in levels of glutamate and glutamine in response to these neurotoxins33,34 or in the serum and cerebrospinal fluid of PD patients.35,36 Interestingly, despite TCA cycle blockage by paraquat, both TCA cycle metabolites oxoglutarate and succinate remained unchanged. We can hypothesize that both glucose and aspartate transamination via glutamate dehydrogrenase (GLDH) and aspartate aminotransferase (GOT1), respectively, can compensate for the blockage of the TCA cycle via inhibition of aconitase by paraquat (Figure 7). It is important to highlight that the observed alterations in the metabolomes due to neurotoxin treatments occurred prior to cell death, since samples were harvested after only a 24 h treatment (Figure 1A). Conversely, in vivo studies and clinical sample analysis cannot distinguish between metabolic alterations that occur before cell death (alterations in cell metabolism per se), or metabolic changes associated with cell death (lysis). This uncertainty might explain the observed discrepancies.
Paraquat Induces an Increase in Glucose-6-Phosphate Dehydrogenase Levels
A proteomic analysis was performed to determine whether the alterations in energy/redox metabolic pathways correlated to some extent with changes in protein levels. A 24 h of paraquat exposure induced a significant upregulation or downregulation (>25%) in the expression levels of a number of proteins (Figure 5A). Conversely, paraquat induced the expression of very few de novo proteins as most of the proteins identified were already present in control cells (Figure 5D). The proteins with alterations in their expression levels were classified by their biological function and were shown to be involved in a number of processes including cytoskeleton organization, redox signaling, and mitochondrial function (Figure 5B). Particularly noteworthy was the observed increase in glucose-6-phosphate dehydrogenase (G6PD), mitochondrial malate dehydrogenase (MDH), phosphoglycerate kinase 1 (PGK1), ATP-citrate synthase (CS), and pyruvate kinases isozymes M1/M2, as well as a decrease in lactate dehydrogenase A/B chains (Figure 5C and 7), which correlated with the alterations in the PPP, TCA cycle, and glycolysis pathway identified from the metabolomics analysis (Figure 4 and 7). G6PD is the rate-limiting enzyme of the PPP, and a major source of NADPH required by antioxidant pathways.37 The proteomics result was confirmed by western-blot where a clear increase in G6PD levels was induced by increasing doses of paraquat (Figure 5E). Thus, the increase in G6PD expression correlates with the increase in PPP metabolites induced by paraquat (Figure 7).
Figure 5.
Paraquat induces an increase in G6PD and alterations in proteins involved in apoptosis and redox signaling. (A) Proteomic analysis of cells treated for 24 h with paraquat (0.5 mM). (B–C) Biological function and identification of proteins whose expression levels were found to be significantly changed (>25% increase or decrease, p < 0.05) by paraquat exposure (see squares in A). An increase in the expression levels of glucose-6-phosphate dehydrogenase (G6PD), mitochondrial malate dehydrogenase, phosphoglycerate kinase, ATP-citrate synthase, and pyruvate kinases M1/M2 as well as a reduction in lactate dehydrogenases A/B chain are highlighted by asterisks as they relate to the metabolic alterations observed by NMR/DI-ESI-MS metabolomics (see Figure 7). (D) Changes in the overall expression levels of proteins demonstrate that only a small subset of proteins was identified in either control or paraquat-treated cells. (E) Western blot analysis of changes in G6PD expression induced by paraquat. Numbers below (italics) represent the densitometry analysis with respect to the loading control (GAPDH). Data in A–D were generated from 4 independent samples.
G6PD Regulates Paraquat Toxicity
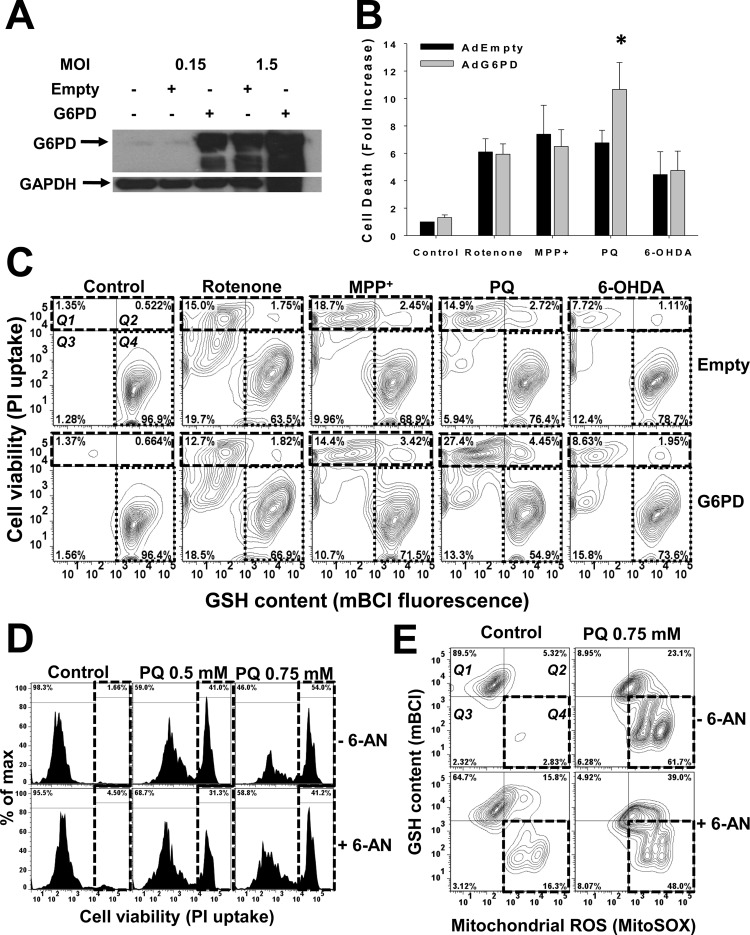
The metabolomic and proteomic data suggested that alterations in the PPP and G6PD expression levels might be involved in the regulation of paraquat toxicity. To further investigate this relationship, cells were transduced with adenovirus encoding for human G6PD (Ad-G6PD) or empty adenovirus (Ad-Empty) (Figure 6A). Ad-G6PD induced a robust expression of G6PD at low titers. Because high titers of Ad-Empty also increased G6PD expression, we used low titers (0.15 MOI) to induce G6PD overexpression and compare it to empty (control) virus infection. G6PD overexpression increased cell death (Figures 6B and C [Q1and 2 quadrants in contour plots, broken lines]) and oxidative stress (measured as GSH loss) (Figure 6C, Q4 regions in contour plots, dotted lines]) induced by paraquat, but not by exposure to the other toxins (Figure 6B–C). Accordingly, inhibition of G6PD with 6-AN selectively reduced paraquat-induced toxicity (Figure 6D), mitochondrial ROS formation, and GSH depletion (Figure 6E, Q4 quadrants in contour plots, dotted lines). 6-AN had no effect on cell death and GSH depletion induced by either rotenone, MPP+, or 6-OHDA (Supporting Information Figure 3A and B). In most circumstances, increased G6PD activity has been reported to protect against oxidative stress-induced cell death.38 However, the increased paraquat toxicity induced by G6PD overexpression and the protective effects of 6-AN can be explained by the requirement of reducing equivalents for paraquat to redox cycle (Figure 7).39 Accordingly, G6PD inhibition has been shown to reduce paraquat-induced cell death,40 while G6PD overexpression was demonstrated to increase it.41 These results also suggest that by “hijacking” the PPP, paraquat outcompetes the GSH recycling by glutathione reductase (GR), which also requires NADPH, thus, arguing against the protective role of this antioxidant system against paraquat. A previous study reported a protective effect of GR against paraquat toxicity.42 When we overexpress GR in the cytosol or mitochondria (manuscript in preparation) together with G6PD, we were not able to prevent paraquat-induced cell death. In contrast, we (data not shown) and others have observed that direct supplementation of cell permeable GSH, inhibition of GSH de novo synthesis,43 or glutathione peroxidase44 protects against paraquat toxicity. In addition, a recent manuscript demonstrated that paraquat induces dopamine depletion in the brain of the glutamate-cysteine ligase modifier subunit (Gclm) knockout mice chronically deficient in GSH.45 Thus, GSH exerts a protective effect against paraquat, but its recycling by the GR/NADPH system might be impaired by the depletion of NADPH by paraquat. Interestingly, paraquat induced a slight, but not significant, decrease in NADPH (or NADH) levels, while the NADP+ content remains largely unaffected (Figure 4). While our results demonstrate that paraquat upregulates the PPP and thus the formation of NADPH, the constant use of NADPH by paraquat’s redox cycling mechanism would be expected to prevent any increased accumulation of NADPH in the cell. In any case, the protective effect of 6-AN and the stimulatory role of G6PD overexpression support a role for NADPH metabolism in paraquat toxicity (Figure 7).
Figure 6.
Paraquat-induced cell death and oxidative stress is selectively regulated by G6PD and the PPP pathway. (A) Cells were transduced with Ad-G6PD at distinct MOI for 24 h and G6PD levels were determined by Western blot. (B and C) Cell death and GSH depletion induced by paraquat (0.5 mM), rotenone (4 μM), MPP+ (2.5 mM) or 6-OHDA (50 μM) after 48 h of treatment, was simultaneously evaluated by flow cytometry using PI and mBCl, respectively. Data in (B) is represented as fold increase in the mean PI fluorescence and are means ± SE of 3 independent experiments. *p < 0.05, Empty vs G6PD values. Data in C are represented in a two-dimensional contour plot display of cell death (PI uptake) vs GSH levels (mBCl). Cell death (see Q1–2 quadrants in broken line squares) is observed as an increase in PI fluorescence (y axis), while oxidative stress is reflected by GSH depletion (see Q4 quadrants in dotted lines) and a decrease in mBCl signal (x axis). Percentages in quadrants highlight the changes in the number of cells. (D and E) Cell death and oxidative stress induced by paraquat was evaluated in the presence or absence of 6-aminonicotinamide (6-AN, 1 mM). (D) Cell death is represented in the histograms as an increase in the population of cells (%) with increased PI fluorescence (see broken line squares). (E) GSH depletion and mitochondrial ROS formation were simultaneously evaluated by flow cytometry using mBCl and MitoSOX, respectively. Data are represented in a two-dimensional contour plot display of changes in intracellular GSH (mBCl) vs mitochondrial ROS state levels (MitoSOX). Oxidative stress is observed as a decrease in both GSH content (y axis) and an increase in MitoSOX signal (x axis). Q4 quadrants (broken lines) highlight the population of cells (in %) with both GSH depletion and mitochondrial ROS accumulation. 5% probability contour plots (C and E) and histograms (D) are representative of three independent experiments.
In the cell, G6PD is an important, but not an exclusive source for NADPH, which can also be produced by 6-phosphogluconate dehydrogenase (PGD) (Figure 7), MDH, and isocitrate dehydrogenase (IDH).37 Interestingly, we also found an increase in the expression levels of mitochondrial MDH in cells treated with paraquat (Figure 5C and 7), which might contribute to paraquat’s redox cycle in the mitochondria. 6-AN is taken up by cells and transformed into 6-amino-NADP+ by NAD-glycohydrolase, which acts as an analogue of NADP+. 6-AN acts as a competitive inhibitor of G6PD and PGD, which also requires NADP+. Importantly, 6-AN inhibits PGD with an inhibitor constant (Ki) of 0.13 × 10–6 M, approximately 400-fold lower than the Ki for G6PD.46 Thus, we can consider that 6-AN inhibits the PPP pathway by acting in both PGD and G6PD (Figure 7, highlighted in green). As a result, the reversal of paraquat toxicity by 6-AN cannot be solely attributed to G6PD inhibition. However, because both G6PD and PGD generate NADPH, the protective effects of 6-AN against paraquat toxicity are likely mediated by inhibition of the PPP and NADPH synthesis. It is important to mention that G6PD overexpression/activation would not only provide NADPH for paraquat redox cycling, but it could also increase nitric oxide synthase (NOS) and NADPH-oxidase activities, which have also been suggested to contribute to paraquat-induced oxidative stress.47,48
MPP+-, rotenone-, or 6-OHDA-induced toxicity was not shown to be modulated by G6PD expression/activity. Previous reports have demonstrated that mice overexpressing G6PD are protected against MPTP-induced loss of dopaminergic cells.49 However, an association between G6PD activity levels and PD is still controversial since contradictory results have been published in the literature.50,51 Because of the multifactorial nature of PD pathogenesis, for example, the generation of oxidative stress can be associated with a range of factors such as mitochondrial dysfunction, dopamine toxicity, or exposure to redox cycling agents. It is plausible that a more thorough analysis could reveal an association between alterations in G6PD activity levels or polymorphisms, and an increased risk for developing PD in individuals exposed to redox cycling herbicides. Such studies have already revealed that genetic modifications in glutathione s-transferase M1 (GSTM1) and the dopamine transporter (DAT) are associated with an increased risk of developing PD from paraquat exposure.52,53
Alterations in Dopamine Content
The toxicity of environmental/mitochondrial toxins has also been largely linked to alterations in dopamine metabolism and distribution. Intracellular dopamine content and/or its redistribution from vesicles to the cytoplasm increase the toxicity of paraquat, rotenone, and MPP+. Dopamine levels decrease significantly in paraquat treated mice, which was associated with increased dopamine oxidation.54 Similarly dopamine oxidation has also been shown to mediate rotenone and MPP+ toxicity.55,56 MPP+-, rotenone-, and paraquat- induced DA depletion has also been attributed to an increase in DA release.57−59 In addition, MPTP/MPP+ and 6-OHDA have also been reported to oxidize and inhibit tyrosine hydroxylase, the enzyme that catalyzes the rate limiting step in this synthesis of catecholamines.60,61 Rotenone and MPTP/MPP+ also induce a redistribution of dopamine from vesicles to the cytosol by inhibition or downregulation of the vesicular monoamine transporter 2 (WMAT2).62,63 Accordingly, we found that paraquat induced a decrease in dopamine content (Figure 3A–C). In contrast, MPP+ induced an increase in dopamine content (Supporting Information Figure 1A and B). Interestingly, an increase in tyrosine hydroxylase activity and dopamine levels was found in individual cell bodies in the substantia nigra in a presymptomatic and early symptomatic MPTP mouse model that induce subthreshold and threshold loss of dopaminergic cells, respectively,64 which correlates with our present findings.
Relationship between Oxidative Stress and Alterations in Energy/Redox Metabolism
In the brain, both energy metabolism and redox homeostasis are tightly coupled. Paraquat, MPP+, rotenone, and 6-OHDA induce ROS formation through different mechanisms. MPP+ and rotenone are reported to act as complex I inhibitors, and it is expected that mitochondrial dysfunction results in superoxide anion formation. 6-OHDA has been indicated to produce ROS through enzymatic or nonenzymatic auto-oxidation. In the case of paraquat, ROS formation is mainly generated via its redox cycling. We speculate that NADPH consumption by paraquat39 would be independent from superoxide anion formation, while the increase in citrate via the well reported effect of paraquat inactivating aconitase65 will be ROS dependent. 6-OHDA would be expected to exert its effects by their pro-oxidant nature and/or direct adduction/modification of proteins. In contrast, since rotenone and MPP+ act directly on complex I, ROS formation and metabolic changes can be induced independently. In fact, energy failure has been proposed to mediate MPP+ and rotenone toxicity independent from oxidative stress.8,66−68 In any case, ROS formation induced by these toxins would eventually be expected to impair energy metabolism. Importantly, the different metabolic changes induced by the neurotoxins were not associated with differences in the levels of ROS, because a similar increase in mitochondrial ROS was induced by paraquat, MPP+, and rotenone at the concentrations tested (Supporting Information Figure 4A).
Previous findings have demonstrated that GSH depletion is an important contributor to oxidative stress and dopaminergic cell death induced by environmental/mitochondrial toxins.69 Clinical data also shows that a decrease in the GSH levels is one of the earliest biochemical alterations detected in incidental Lewy body disease, considered an asymptomatic precursor to PD.70 Accordingly, 2D 1H–13C HSQC NMR experiments revealed that a 24 h treatment with paraquat, MPP+, and rotenone induced a decrease in total GSH/GSSG (reduced [GSH] or oxidized [GSSG]) (Supporting Information Figure 2). In contrast, 6-OHDA toxicity was shown to increase total GSH/GSSG content (Supporting Information Figure 1E and F and Figure 2). It has been previously reported that 6-OHDA is oxidized by molecular oxygen to generate reactive oxygen species and 2-hydroxy-5-(2-aminoethyl)-1,4-benzoquinones(p-quinone). Partially substituted quinones (p-quinone) can react with cellular nucleophiles such as thiols (including GSH) forming covalently linked quinone-thiol adducts.6,71 Thus, 6-OHDA-induced GSH depletion could be expected to trigger a compensatory response to increase GSH content. In fact, a previous study demonstrated an early increase in GSH synthesis and γ-glutamylcysteine ligase (the rate-limiting enzyme in GSH synthesis) levels in response to 6-OHDA.72,73 We further aimed to corroborate these results using the enzymatic recycling method. Accordingly 6-OHDA induced a significant increase in total GSH (GSH and GSSG) levels (Supporting Information Figure 4B). In contrast to our 2D 1H–13C HSQC NMR results (Supporting Information Figure 2) paraquat MPP+ or rotenone had no significant effect in total GSH content (Supporting Information Figure 4B). It is important to state that our 2D 1H–13C HSQC NMR experiments only detects metabolites derived from glucose metabolism. In neurons, however, glutamine is required for glutamate synthesis via glutaminase (GLS)74 (Figure 7). Our results then suggest that paraquat, MPP+, and rotenone decrease GSH synthesis directly dependent from glucose metabolism, which correlates with a decrease in the content of the glutathione precursor glutamate (Supporting Information Figure 2 and Figure 7). Then, total GSH content (Supporting Information Figure 4B) might be maintained by glutaminolysis, but additional experiments using isotopically labeled glutamine would be required to clarify this issue. Interestingly, only paraquat induced a significant accumulation of GSSG (Supporting Information Figure 4B), which corroborates our hypothesis that paraquat impairs the GR/NADPH recycling system (Figure 7).
Using flow cytometry we then analyzed the changes in the intracellular content of reduced glutathione (GSH) using monochlorobimane, a GSH-binding dye that forms blue-fluorescent adducts with intracellular GSH after 48 h treatment with neurotoxins. Contour plots in Figure 6C (lower quadrant 4 [Q4] in dotted line) depicts cells with high (basal) levels of intracellular GSH. A 48 h treatment with paraquat (76.4%), MPP+ (68.9%), rotenone (63.5%), and 6-OHDA (78.7%) induces a decrease in this population of cells with high levels of GSH with respect to control (96.9%). The decrease in GSH content was mainly associated with cell death progression as evidenced by the loss of plasma membrane integrity (PI uptake). Thus, while paraquat, MPP+, and rotenone induce a reduction in glucose-dependent glutamate-derived GSH synthesis, 6-OHDA treatment increases it. However, upon cell death progression, GSH concentration depletion parallels cell demise.
In principle, it would seem straightforward to evaluate if the alterations in energy metabolism induced by these neurotoxins are dependent on ROS formation by overexpression of antioxidant enzymes or exposure to antioxidants. However, the exact nature of the ROS and oxidative damage induced by paraquat, MPP+, rotenone, and 6-OHDA is quite complex and the mechanisms are still unclear. For example, superoxide anion has been largely thought to be the primary ROS mediating oxidative damage associated with mitochondrial dysfunction induced MPP+, rotenone, or paraquat. However, we have recently published that overexpression of MnSOD only protects against paraquat-, but not MPP+- or rotenone-induced toxicity.5 Interestingly, we observed that overexpression of MnSOD does not prevent free radical formation induced by MPP+ and rotenone, suggesting that additional mechanisms involved. Indeed, nitric oxide and hydroxyl radical formation has been reported to mediate the toxicity induced by MPP+ and rotenone.75−78
Similarly, while overexpression of MnSOD would scavenge superoxide anion formation induced by paraquat,5 it would lead to an increased accumulation of hydrogen peroxide. When we overexpress catalase or mitochondria-targeted catalase, we have not seen any protection against paraquat toxicity (data not shown). This can also be explained by the fact that catalase also requires NADPH for its proper function, and thus, paraquat redox cycling could be expected to impair catalase activity. Finally, paraquat also impairs NADPH dependent antioxidant systems such as the peroxiredoxins/thioredoxin/thioredoxin reductase (data not shown). 6-OHDA-induced oxidative damage has been linked to depletion/adduction of intracellular thiols (GSH and cysteine) and generation of both extracellular and intracellular ROS.71,79,80 Thus, there is no single antioxidant or antioxidant system that would be expected to efficiently prevent ROS formation and oxidative stress induced by any of these toxins and addressing this issue requires extensive and additional investigation.
In summary, our data demonstrates that paraquat “hijacks” the PPP to produce NADPH, which in turn is used as an electron donor for paraquat’s redox cycle to generate ROS (Figure 7, green square). Paraquat also induced a blockage of glycolysis likely linked to increased citrate accumulation via impaired TCA cycle at the level of aconitase (Figure 7). Another important outcome of this study is that we demonstrate that alterations in energy/redox metabolism, which are specific for distinct environmental toxins, are not bystanders to energy failure but also contribute significantly to cell death progression. Our data supports the notion that by studying metabolic alterations in an integrated and comparative manner, we can reveal novel mechanisms of toxicity specifically associated with different environmental exposures. The differences in the metabolic alterations found between the distinct toxicological models used, exemplifies the concept that the multifactorial nature of PD might require further stratification of cases to identify the specific triggers of dopaminergic cell loss. This information has the potential to contribute to the design of customized therapeutic approaches according to the multifactorial nature of PD.
Methods
Cell Culture, Treatments, Microscopy, Western Immunoblot, Flow Cytometry, and Reagents
The human dopaminergic neuroblastoma cell line SK-N-SH was originally derived from the bone marrow of metastatic neuroblastoma from a female patient. SK-N-SH cells have been reported to express significant levels of dopamine β-hydroxylase, acetylcholinesterase,81 and to have detectable levels of tyrosine hydroxylase activity.82,83 These cells can be differentiated into neuronal cells with retinoic acid (RA). Differentiated cells have been reported to contain higher levels of neuronal markers, such as NMDA receptors.84 We have observed similar levels of dopamine transporter expression (DAT, SLC6A3) between RA differentiated and nondifferentiated cells (unpublished data). In addition, similar to the SH-SY5Y neuroblastoma cells, we have observed that differentiated SK-N-SH become more resistant to neurotoxin treatments, which has been attributed to increased levels of survival signals.85−88 Detailed information on the cell culture procedures for this cell line, neurotoxin treatments, and Western blot procedures can be found in previous work from our group.89 Anti-glucose-6-phosphate dehydrogenase (G6PD) was from Abcam Ab993. 6-OHDA was prepared in ddH2O containing 0.01% ascorbic acid to avoid confounding effects mediated by its oxidized breakdown products.90,91 Phase contrast images of cells were taken using a Zeiss 20×/0.3 LD-A-Plan Ph1 objective and a Moticam 580 (5.0 MP) camera. The construction of adenoviral-human G6PD expression vector has been described previously.92 Recombinant adenovirus amplification, titration, and infection procedure of SK-N-SH cells were also described elsewhere.89 Loss of cell viability was determined by propidium iodide uptake (PI) as a marker for compromised plasma membrane integrity. Oxidative stress was assessed by simultaneous determination of mitochondrial reactive oxygen species (ROS) levels using MitoSOX Red and intracellular GSH levels using monochlorobimane (mBCl) (Invitrogen). Flow cytometric approaches have been previously explained in detail.5,93
NMR and MS Metabolomics Data Collection
A detailed description of the protocol and optimization of the combined use of NMR and direct-infusion electrospray ionization mass spectrometry (DI-ESI-MS) for the analysis of the metabolome is described in another manuscript (Marshall et al., Combining MS and NMR Data Sets for Metabolomics Profiling, in revision). Briefly, a single cell lysate sample was prepared for both NMR and DI-ESI-MS analysis. Cells were treated as indicated. Six replicates were prepared for each treatment class for the analysis of global changes in the metabolome (1D 1H NMR), while three replicates were prepared for metabolite identification using 2D 1H–13C HSQC experiment, where 12C-glucose in the medium was replaced with 13C-glucose (3.5 g/L). Cells were washed twice with ice-cold PBS to discard dead cells. Metabolites were extracted with cold methanol (−80 °C), followed by 100% ddH2O. The supernatants from the three extractions were used for NMR and DI-ESI-MS analysis. Reserpine (20 μM) was used as internal mass reference for DI-ESI-MS. 3-(trimethylsilyl)propionic acid-2,2,3,3-d4 (TMSP) was used in the1D 1H NMR (50 μM) and 2D1H–13C HSQC (500 μM) experiments for chemical shift referencing. The 1D 1H NMR and 2D 1H–13C HSQC spectra were collected on a Bruker DRX Avance 500-MHz spectrometer equipped with a 5 mm triple-resonance cryoprobe (1H, 13C, and 15N) with a z-axis gradient, a BACS-120 sample changer, Bruker ICON-NMR, and an automatic tuning and matching (ATM) unit, and analyzed, as previously described.94
DI-ESI-MS was performed on a Synapt G2 HDMS quadrupole time-of-flight instrument (Waters Corp., Milford, MA). The spectra for multivariate analysis were acquired for 0.5 min with a mass range of m/z 50 to 1200 using optimized ESI and nESI source conditions (Marshall et al., Combining MS and NMR Data Sets for Metabolomics Profiling, in revision). DI-ESI-MS spectra were processed using MassLynx V4.1 (Waters).
Multivariate Analysis of NMR and DI-ESI-MS Metabolomics Data Set
To prepare an input data set for the multivariate analysis, the 1D 1H NMR spectra were preprocessed by our MVAPACK software suite (http://bionmr.unl.edu/mvapack.php).95 The NMR spectra were preprocessed by exponential apodization and zero-filling prior to Fourier transformation and then were automatically phased, PSC-normalized,95 and binned.96 The DI-ESI-MS spectra were binned using a uniform bin size of 0.5 m/z and probabilistic quotient (PQ)-normalized97 by the MVAPCK software suite. A manual noise removal step was performed on both 1D 1H NMR spectra and DI-ESI-MS spectra. The LDA plots, MB-PLS-DA S-plots, and MB-PLS-DA SUS plots were generated using the MVAPACK software suite. Specifically, the results of multivariate analysis on the combination of the 1D 1H NMR and DI-ESI-MS data sets were obtained by using a multiblock structure and PCA and PLS modeling functions in MVAPACK. Importantly, the two blocks of NMR and MS data sets were scaled by the square root of its respective variable count in order to avoid the unequal contribution of each block to the combined model, caused by large differences in the variable count between blocks. The detailed process procedures can be found in the manuscript (Marshall et al., Combining MS and NMR Data Sets for Metabolomics Profiling, in revision). Hotelling 95% confidence ellipses, MB-PCA scores dendrograms and corresponding Mahalanobis p-values were generated using our PCA/PLS-DA utilities (http://bionmr.unl.edu/pca-utils.php).98,99 An observed p-value ≤0.05 between two clusters indicates a statistically significant difference between clusters. The MB-PLS-DA models were validated using CV-ANOVA100 7-fold Monte Carlo single cross-validation.101
Metabolite Identification
The Platform for RIKEN Metabolomics (PRIMe, http://prime.psc.riken.jp/),102 Human Metabolome Database (HMDB, http://www.hmdb.ca/),103 Madison Metabolomics Consortium Database (http://mmcd.nmrfam.wisc.edu/),104 Metabominer (http://wishart.biology.ualberta.ca/metabominer/),105 and BiomagResBank (BMRB, http://www.bmrb.wisc.edu/)106 were used for NMR peak annotation using an error tolerance of 0.08 and 0.25 ppm for 1H and 13C chemical shifts, respectively. The intensities of all peaks assigned to a metabolite were then used to report the average peak intensity, and intensity (concentration) changes between treatment classes. Accurate mass experiments were also used to assist in the identification of metabolites associated with class differentiation. All metabolite mass spectra from the accurate mass experiments were smoothed, centroided, and internally mass corrected relative to the [M + H]+ ion for reserpine (m/z 609.2812) using MassLynx V4.1. The accurate masses were searched against the online metabolite DI-ESI-MS databases Human Metabolome (HMDB, http://www.hmdb.ca/)107 and Metabolite and Tandem DI-ESI-MS Database (METLIN, http://metlin.scripps.edu)108 with a threshold window of 20 ppm.
Proteomics
Proteomic analysis was done as explained in ref (7). Heat maps of proteins with a significant up- or downregulation of at least 25% were created using GENE-E software (http://www.broadinstitute.org/cancer/software/GENE-E/). Venn diagrams were created using Venn Diagram Plotter software (PNNL, Richland, WA).
Reduced Glutathione (GSH) and Glutathione Disulfide (GSSG, or Oxidized Glutathione) Content
Cells (>1 × 107) were harvested and washed with PBS. Acid deproteinization was performed in 5% metaphosphoric acid. GSSG samples were prepared by adding 10 μL M2VP (1-methyl-2-vinylpyridium trifluoromethanesulfonate, a thiol scavenger). GSH and GSSG quantification was using a Bioxytech GSH/GSSG-412 assay kit (Oxis Research Assay Service, Portland, OR). The method uses Ellman’s reagent (5,5′-dithiobis-2-nitrobenzoic acid or DTNB), which reacts with GSH to form a spectrophotometrically detectable product at 412 nm. GSSG was determined by reduction of GSSG to GSH via GR. Data was normalized by protein concentration. The GSH/GSSH ratio was calculated by dividing the difference between the total GSH (GSH and GSSG) and GSSG concentrations (reduced GSH) by the concentration of GSSG.
Acknowledgments
This work was supported by the National Institutes of Health Grants P20RR17675 and P30 GM103335, Centers of Biomedical Research Excellence (COBRE), the Scientist Development Grant of the American Heart Association (12SDG12090015), the Research Council Interdisciplinary Grant, and the Life Sciences Grant Program of the University of Nebraska—Lincoln. Research was performed in facilities renovated with support from the NIH under Grant RR015468-01. We thank the Flow Cytometry Core Facility at the Nebraska Center for Virology for the access to flow cytometry instrumentation. We also acknowledge B. Worley and D. D. Marshall for assisting in the preparation of the manuscript.
Supporting Information Available
Supplementary figures. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gao H. M.; Hong J. S. (2011) Gene-environment interactions: Key to unraveling the mystery of Parkinson’s disease. Prog. Neurobiol. 94, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner C. M.; Kamel F.; Ross G. W.; Hoppin J. A.; Goldman S. M.; Korell M.; Marras C.; Bhudhikanok G. S.; Kasten M.; Chade A. R.; Comyns K.; Richards M. B.; Meng C.; Priestley B.; Fernandez H. H.; Cambi F.; Umbach D. M.; Blair A.; Sandler D. P.; Langston J. W. (2011) Rotenone, paraquat, and Parkinson’s disease. Environ. Health Perspect 119, 866–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez T. N.; Greenamyre J. T. (2012) Toxin models of mitochondrial dysfunction in Parkinson’s disease. Antioxid Redox Signal 16, 920–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola N.; Morelli M.; Carta A. R. (2007) The 6-hydroxydopamine model of Parkinson’s disease. Neurotox Res. 11, 151–167. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rocha H.; Garcia-Garcia A.; Pickett C.; Li S.; Jones J.; Chen H.; Webb B.; Choi J.; Zhou Y.; Zimmerman M. C.; Franco R. (2013) Compartmentalized oxidative stress in dopaminergic cell death induced by pesticides and complex I inhibitors: Distinct roles of superoxide anion and superoxide dismutases. Free Radic Biol. Med. 61C, 370–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi Y.; Sawada H.; Sakka N.; Yamamoto N.; Kume T.; Katsuki H.; Shimohama S.; Akaike A. (2005) p-Quinone mediates 6-hydroxydopamine-induced dopaminergic neuronal death and ferrous iron accelerates the conversion of p-quinone into melanin extracellularly. J. Neurosci Res. 79, 849–860. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Rocha H.; Garcia Garcia A.; Zavala-Flores L.; Li S.; Madayiputhiya N.; Franco R. (2012) Glutaredoxin 1 protects dopaminergic cells by increased protein glutathionylation in experimental Parkinson’s disease. Antioxid Redox Signal 17, 1676–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranka B. P.; Zielonka J.; Kanthasamy A. G.; Kalyanaraman B. (2012) Alterations in bioenergetic function induced by Parkinson’s disease mimetic compounds: Lack of correlation with superoxide generation. J. Neurochem. 122, 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopert P.; Day B. J.; Patel M. (2012) Thioredoxin reductase deficiency potentiates oxidative stress, mitochondrial dysfunction, and cell death in dopaminergic cells. PLoS One 7, e50683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. S.; Kruse S. E.; Palmiter R. D.; Xia Z. (2008) Mitochondrial complex I inhibition is not required for dopaminergic neuron death induced by rotenone, MPP+, or paraquat. Proc. Natl. Acad. Sci. U.S.A. 105, 15136–15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartelli D.; Ronchi C.; Maggioni M. G.; Rodighiero S.; Giavini E.; Cappelletti G. (2010) Microtubule dysfunction precedes transport impairment and mitochondria damage in MPP+-induced neurodegeneration. J. Neurochem. 115, 247–258. [DOI] [PubMed] [Google Scholar]
- Giordano S.; Lee J.; Darley-Usmar V. M.; Zhang J. (2012) Distinct effects of rotenone, 1-methyl-4-phenylpyridinium and 6-hydroxydopamine on cellular bioenergetics and cell death. PLoS One 7, e44610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekke E. M.; Walls A. B.; Schousboe A.; Waagepetersen H. S.; Sonnewald U. (2012) Quantitative importance of the pentose phosphate pathway determined by incorporation of 13C from [2–13C]- and [3–13C]glucose into TCA cycle intermediates and neurotransmitter amino acids in functionally intact neurons. J. Cereb. Blood Flow Metab. 32, 1788–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman J. N.; Sanchez-Padilla J.; Wokosin D.; Kondapalli J.; Ilijic E.; Schumacker P. T.; Surmeier D. J. (2010) Oxidant stress evoked by pacemaking in dopaminergic neurons is attenuated by DJ-1. Nature 468, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perier C.; Vila M. (2012) Mitochondrial biology and Parkinson’s disease. Cold Spring Harbor Perspect. Med. 2, a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson G. E.; Kingsbury A. E.; Xu H.; Lindsay J. G.; Daniel S.; Foster O. J.; Lees A. J.; Blass J. P. (2003) Deficits in a tricarboxylic acid cycle enzyme in brains from patients with Parkinson’s disease. Neurochem. Int. 43, 129–135. [DOI] [PubMed] [Google Scholar]
- Palombo E.; Porrino L. J.; Bankiewicz K. S.; Crane A. M.; Sokoloff L.; Kopin I. J. (1990) Local cerebral glucose utilization in monkeys with hemiparkinsonism induced by intracarotid infusion of the neurotoxin MPTP. J. Neurosci. 10, 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberling J. L.; Richardson B. C.; Reed B. R.; Wolfe N.; Jagust W. J. (1994) Cortical glucose metabolism in Parkinson’s disease without dementia. Neurobiol Aging 15, 329–335. [DOI] [PubMed] [Google Scholar]
- Henchcliffe C.; Shungu D. C.; Mao X.; Huang C.; Nirenberg M. J.; Jenkins B. G.; Beal M. F. (2008) Multinuclear magnetic resonance spectroscopy for in vivo assessment of mitochondrial dysfunction in Parkinson’s disease. Ann. N.Y. Acad. Sci. 1147, 206–220. [DOI] [PubMed] [Google Scholar]
- Dunn L.; Allen G. F.; Mamais A.; Ling H.; Li A.; Duberley K. E.; Hargreaves I. P.; Pope S.; Holton J. L.; Lees A.; Heales S. J.; Bandopadhyay R. (2014) Dysregulation of glucose metabolism is an early event in sporadic Parkinson’s disease. Neurobiol. Aging 35, 1111–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke C. J.; Haselden J. N. (2008) Metabolic profiling as a tool for understanding mechanisms of toxicity. Toxicol. Pathol. 36, 140–147. [DOI] [PubMed] [Google Scholar]
- Hong Z.; Shi M.; Chung K. A.; Quinn J. F.; Peskind E. R.; Galasko D.; Jankovic J.; Zabetian C. P.; Leverenz J. B.; Baird G.; Montine T. J.; Hancock A. M.; Hwang H.; Pan C.; Bradner J.; Kang U. J.; Jensen P. H.; Zhang J. (2010) DJ-1 and α-synuclein in human cerebrospinal fluid as biomarkers of Parkinson’s disease. Brain 133, 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Pan Z.; Talaty N.; Raftery D.; Cooks R. (2006) Combining desorption electrospray ionization mass spectrometry and nuclear magnetic resonance for differential metabolomics without sample preparation. Rapid Commun. Mass Spectrom. 20, 1577–1584. [DOI] [PubMed] [Google Scholar]
- Cantu D.; Fulton R. E.; Drechsel D. A.; Patel M. (2011) Mitochondrial aconitase knockdown attenuates paraquat-induced dopaminergic cell death via decreased cellular metabolism and release of iron and H(2)O(2). J. Neurochem. 118, 79–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling P. L.; Smith L. L.; Aldridge W. N. (1982) The formation of mixed disulphides in rat lung following paraquat administration. Correlation with changes in intermediary metabolism. Biochim. Biophys. Acta 716, 249–257. [DOI] [PubMed] [Google Scholar]
- Newsholme E. A.; Sugden P. H.; Williams T. (1977) Effect of citrate on the activities of 6-phosphofructokinase from nervous and muscle tissues from different animals and its relationships to the regulation of glycolysis. Biochem. J. 166, 123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W.; Clark P. M.; Mason D. E.; Keenan M. C.; Hill C.; Goddard W. A. 3rd; Peters E. C.; Driggers E. M.; Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baykal A. T.; Jain M. R.; Li H. (2008) Aberrant regulation of choline metabolism by mitochondrial electron transport system inhibition in neuroblastoma cells. Metabolomics 4, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie K.; Zhang Y.; Huang B.; Wang L.; Zhao J.; Huang Z.; Gan R. (2013) Marked N-acetylaspartate and choline metabolite changes in Parkinson’s disease patients with mild cognitive impairment. Parkinsonism Relat. Disord. 19, 329–334. [DOI] [PubMed] [Google Scholar]
- Ellis C. M.; Lemmens G.; Williams S. C.; Simmons A.; Dawson J.; Leigh P. N.; Chaudhuri K. R. (1997) Changes in putamen N-acetylaspartate and choline ratios in untreated and levodopa-treated Parkinson’s disease: A proton magnetic resonance spectroscopy study. Neurology 49, 438–444. [DOI] [PubMed] [Google Scholar]
- Xu Q. W.; Vu H.; Liu L. P.; Wang T. C.; Schaefer W. H. (2011) Metabolic profiles show specific mitochondrial toxicities in vitro in myotube cells. J. Biomol. NMR 49, 207–219. [DOI] [PubMed] [Google Scholar]
- Ahmed S. S.; Santosh W.; Kumar S.; Christlet H. T. (2009) Metabolic profiling of Parkinson’s disease: Evidence of biomarker from gene expression analysis and rapid neural network detection. J. Biomed Sci. 16, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H. C.; Zhu H.; Song C. Y.; Lin L.; Xiang Y.; Yan Z. H.; Bai G. H.; Ye F. Q.; Li X. K. (2013) Metabolic changes detected by ex vivo high resolution 1H NMR spectroscopy in the striatum of 6-OHDA-induced Parkinson’s rat. Mol. Neurobiol. 47, 123–130. [DOI] [PubMed] [Google Scholar]
- Chassain C.; Bielicki G.; Durand E.; Lolignier S.; Essafi F.; Traore A.; Durif F. (2008) Metabolic changes detected by proton magnetic resonance spectroscopy in vivo and in vitro in a murin model of Parkinson’s disease, the MPTP-intoxicated mouse. J. Neurochem 105, 874–882. [DOI] [PubMed] [Google Scholar]
- Mally J.; Szalai G.; Stone T. W. (1997) Changes in the concentration of amino acids in serum and cerebrospinal fluid of patients with Parkinson’s disease. J. Neurol Sci. 151, 159–162. [DOI] [PubMed] [Google Scholar]
- Jimenez-Jimenez F. J.; Molina J. A.; Vargas C.; Gomez P.; Navarro J. A.; Benito-Leon J.; Orti-Pareja M.; Gasalla T.; Cisneros E.; Arenas J. (1996) Neurotransmitter amino acids in cerebrospinal fluid of patients with Parkinson’s disease. J. Neurol Sci. 141, 39–44. [DOI] [PubMed] [Google Scholar]
- Stanton R. C. (2012) Glucose-6-phosphate dehydrogenase, NADPH, and cell survival. IUBMB Life 64, 362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fico A.; Paglialunga F.; Cigliano L.; Abrescia P.; Verde P.; Martini G.; Iaccarino I.; Filosa S. (2004) Glucose-6-phosphate dehydrogenase plays a crucial role in protection from redox-stress-induced apoptosis. Cell Death Differ. 11, 823–831. [DOI] [PubMed] [Google Scholar]
- Fabregat I.; Vitorica J.; Satrustegui J.; Machado A. (1985) The pentose phosphate cycle is regulated by NADPH/NADP ratio in rat liver. Arch. Biochem. Biophys. 236, 110–118. [DOI] [PubMed] [Google Scholar]
- Lee T. C.; Lai G. J.; Kao S. L.; Ho I. C.; Wu C. W. (1993) Protection of a rat tracheal epithelial cell line from paraquat toxicity by inhibition of glucose-6-phosphate dehydrogenase. Biochem. Pharmacol. 45, 1143–1147. [DOI] [PubMed] [Google Scholar]
- Kuo W. Y.; Tang T. K. (1998) Effects of G6PD overexpression in NIH3T3 cells treated with tert-butyl hydroperoxide or paraquat. Free Radic Biol. Med. 24, 1130–1138. [DOI] [PubMed] [Google Scholar]
- Djukic M. M.; Jovanovic M. D.; Ninkovic M.; Stevanovic I.; Ilic K.; Curcic M.; Vekic J. (2012) Protective role of glutathione reductase in paraquat induced neurotoxicity. Chem. Biol. Interact 199, 74–86. [DOI] [PubMed] [Google Scholar]
- Nakagawa I.; Suzuki M.; Imura N.; Naganuma A. (1998) Involvement of oxidative stress in paraquat-induced metallothionein synthesis under glutathione depletion. Free Radic Biol. Med. 24, 1390–1395. [DOI] [PubMed] [Google Scholar]
- Cheng W.; Fu Y. X.; Porres J. M.; Ross D. A.; Lei X. G. (1999) Selenium-dependent cellular glutathione peroxidase protects mice against a pro-oxidant-induced oxidation of NADPH, NADH, lipids, and protein. FASEB J. 13, 1467–1475. [DOI] [PubMed] [Google Scholar]
- Liang L. P.; Kavanagh T. J.; Patel M. (2013) Glutathione deficiency in Gclm null mice results in complex I inhibition and dopamine depletion following paraquat administration. Toxicol. Sci. 134, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hothersall J. S.; Gordge M.; Noronha-Dutra A. A. (1998) Inhibition of NADPH supply by 6-aminonicotinamide: Effect on glutathione, nitric oxide, and superoxide in J774 cells. FEBS Lett. 434, 97–100. [DOI] [PubMed] [Google Scholar]
- Day B. J.; Patel M.; Calavetta L.; Chang L. Y.; Stamler J. S. (1999) A mechanism of paraquat toxicity involving nitric oxide synthase. Proc. Natl. Acad. Sci. U.S.A. 96, 12760–12765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristovao A. C.; Choi D. H.; Baltazar G.; Beal M. F.; Kim Y. S. (2009) The role of NADPH oxidase 1-derived reactive oxygen species in paraquat-mediated dopaminergic cell death. Antioxid. Redox Signaling 11, 2105–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejias R.; Villadiego J.; Pintado C. O.; Vime P. J.; Gao L.; Toledo-Aral J. J.; Echevarria M.; Lopez-Barneo J. (2006) Neuroprotection by transgenic expression of glucose-6-phosphate dehydrogenase in dopaminergic nigrostriatal neurons of mice. J. Neurosci. 26, 4500–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham S.; Soundararajan C. C.; Vivekanandhan S.; Behari M. (2005) Erythrocyte antioxidant enzymes in Parkinson’s disease. Indian J. Med. Res. 121, 111–115. [PubMed] [Google Scholar]
- Gao L.; Mir P.; Diaz-Corrales F. J.; Mejias R.; Carrillo F.; Vime P. J.; Diaz-Martin J.; Palomino A.; Carballo M.; Pintado E.; Lucas M.; Lopez-Barneo J. (2008) Glucose-6-phosphate dehydrogenase activity in Parkinson’s disease. J. Neurol 255, 1850–1851. [DOI] [PubMed] [Google Scholar]
- Goldman S. M.; Kamel F.; Ross G. W.; Bhudhikanok G. S.; Hoppin J. A.; Korell M.; Marras C.; Meng C.; Umbach D. M.; Kasten M.; Chade A. R.; Comyns K.; Richards M. B.; Sandler D. P.; Blair A.; Langston J. W.; Tanner C. M. (2012) Genetic modification of the association of paraquat and Parkinson’s disease. Mov Disord 27, 1652–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritz B. R.; Manthripragada A. D.; Costello S.; Lincoln S. J.; Farrer M. J.; Cockburn M.; Bronstein J. (2009) Dopamine transporter genetic variants and pesticides in Parkinson’s disease. Environ. Health Perspect. 117, 964–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. J.; Gil S. J.; Koh H. C. (2009) Paraquat induces alternation of the dopamine catabolic pathways and glutathione levels in the substantia nigra of mice. Toxicol. Lett. 188, 148–152. [DOI] [PubMed] [Google Scholar]
- Liu H. Q.; Zhu X. Z.; Weng E. Q. (2005) Intracellular dopamine oxidation mediates rotenone-induced apoptosis in PC12 cells. Acta Pharmacol Sin 26, 17–26. [DOI] [PubMed] [Google Scholar]
- Lotharius J.; O’Malley K. L. (2000) The parkinsonism-inducing drug 1-methyl-4-phenylpyridinium triggers intracellular dopamine oxidation. A novel mechanism of toxicity. J. Biol. Chem. 275, 38581–38588. [DOI] [PubMed] [Google Scholar]
- Obata T. (2002) Dopamine efflux by MPTP and hydroxyl radical generation. J. Neural Transm 109, 1159–1180. [DOI] [PubMed] [Google Scholar]
- Shimizu K.; Matsubara K.; Ohtaki K.; Fujimaru S.; Saito O.; Shiono H. (2003) Paraquat induces long-lasting dopamine overflow through the excitotoxic pathway in the striatum of freely moving rats. Brain Res. 976, 243–252. [DOI] [PubMed] [Google Scholar]
- Milusheva E.; Baranyi M.; Kormos E.; Hracsko Z.; Sylvester Vizi E.; Sperlagh B. (2010) The effect of anti-Parkinsonian drugs on oxidative stress induced pathological [3H]dopamine efflux after in vitro rotenone exposure in rat striatal slices. Neuropharmacology 58, 816–825. [DOI] [PubMed] [Google Scholar]
- Pong K.; Doctrow S. R.; Baudry M. (2000) Prevention of 1-methyl-4-phenylpyridinium- and 6-hydroxydopamine-induced nitration of tyrosine hydroxylase and neurotoxicity by EUK-134, a superoxide dismutase and catalase mimetic, in cultured dopaminergic neurons. Brain Res. 881, 182–189. [DOI] [PubMed] [Google Scholar]
- Ara J.; Przedborski S.; Naini A. B.; Jackson-Lewis V.; Trifiletti R. R.; Horwitz J.; Ischiropoulos H. (1998) Inactivation of tyrosine hydroxylase by nitration following exposure to peroxynitrite and 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP). Proc. Natl. Acad. Sci. U.S.A. 95, 7659–7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe M.; Nakaki T. (2008) Mitochondrial complex I inhibitor rotenone inhibits and redistributes vesicular monoamine transporter 2 via nitration in human dopaminergic SH-SY5Y cells. Mol. Pharmacol. 74, 933–940. [DOI] [PubMed] [Google Scholar]
- Chen M. K.; Kuwabara H.; Zhou Y.; Adams R. J.; Brasic J. R.; McGlothan J. L.; Verina T.; Burton N. C.; Alexander M.; Kumar A.; Wong D. F.; Guilarte T. R. (2008) VMAT2 and dopamine neuron loss in a primate model of Parkinson’s disease. J. Neurochem. 105, 78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozina E. A.; Khakimova G. R.; Khaindrava V. G.; Kucheryanu V. G.; Vorobyeva N. E.; Krasnov A. N.; Georgieva S. G.; Kerkerian-Le Goff L.; Ugrumov M. V. (2014) Tyrosine hydroxylase expression and activity in nigrostriatal dopaminergic neurons of MPTP-treated mice at the presymptomatic and symptomatic stages of Parkinsonism. J. Neurol Sci. 340, 198–207. [DOI] [PubMed] [Google Scholar]
- Patel M.; Day B. J.; Crapo J. D.; Fridovich I.; McNamara J. O. (1996) Requirement for superoxide in excitotoxic cell death. Neuron 16, 345–355. [DOI] [PubMed] [Google Scholar]
- Fonck C.; Baudry M. (2001) Toxic effects of MPP(+) and MPTP in PC12 cells independent of reactive oxygen species formation. Brain Res. 905, 199–206. [DOI] [PubMed] [Google Scholar]
- Lee H. S.; Park C. W.; Kim Y. S. (2000) MPP(+) increases the vulnerability to oxidative stress rather than directly mediating oxidative damage in human neuroblastoma cells. Exp. Neurol. 165, 164–171. [DOI] [PubMed] [Google Scholar]
- Nakamura K.; Bindokas V. P.; Marks J. D.; Wright D. A.; Frim D. M.; Miller R. J.; Kang U. J. (2000) The selective toxicity of 1-methyl-4-phenylpyridinium to dopaminergic neurons: The role of mitochondrial complex I and reactive oxygen species revisited. Mol. Pharmacol. 58, 271–278. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia A.; Zavala-Flores L.; Rodriguez-Rocha H.; Franco R. (2012) Thiol-redox signaling, dopaminergic cell death, and Parkinson’s disease. Antioxid Redox Signal 17, 1764–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry T. L.; Yong V. W. (1986) Idiopathic Parkinson’s disease, progressive supranuclear palsy, and glutathione metabolism in the Substantia nigra of patients. Neurosci. Lett. 67, 269–274. [DOI] [PubMed] [Google Scholar]
- Saito Y.; Nishio K.; Ogawa Y.; Kinumi T.; Yoshida Y.; Masuo Y.; Niki E. (2007) Molecular mechanisms of 6-hydroxydopamine-induced cytotoxicity in PC12 cells: Involvement of hydrogen peroxide-dependent and -independent action. Free Radic Biol. Med. 42, 675–685. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Hu J.; Ding J. H.; Yao H. H.; Hu G. (2005) 6-Hydroxydopamine-induced glutathione alteration occurs via glutathione enzyme system in primary cultured astrocytes. Acta Pharmacol Sin 26, 799–805. [DOI] [PubMed] [Google Scholar]
- Shimizu E.; Hashimoto K.; Komatsu N.; Iyo M. (2002) Roles of endogenous glutathione levels on 6-hydroxydopamine-induced apoptotic neuronal cell death in human neuroblastoma SK-N-SH cells. Neuropharmacology 43, 434–443. [DOI] [PubMed] [Google Scholar]
- El Hage M.; Masson J.; Conjard-Duplany A.; Ferrier B.; Baverel G.; Martin G. (2012) Brain slices from glutaminase-deficient mice metabolize less glutamine: A cellular metabolomic study with carbon 13 NMR. J. Cereb Blood Flow Metab 32, 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T. (2002) Role of hydroxyl radical formation in neurotoxicity as revealed by in vivo free radical trapping. Toxicol. Lett. 132, 83–93. [DOI] [PubMed] [Google Scholar]
- Madathil S. K.; Karuppagounder S. S.; Mohanakumar K. P. (2013) Sodium salicylate protects against rotenone-induced Parkinsonism in rats. Synapse 67, 502–514. [DOI] [PubMed] [Google Scholar]
- He Y.; Imam S. Z.; Dong Z.; Jankovic J.; Ali S. F.; Appel S. H.; Le W. (2003) Role of nitric oxide in rotenone-induced nigro-striatal injury. J. Neurochem 86, 1338–1345. [DOI] [PubMed] [Google Scholar]
- Hantraye P.; Brouillet E.; Ferrante R.; Palfi S.; Dolan R.; Matthews R. T.; Beal M. F. (1996) Inhibition of neuronal nitric oxide synthase prevents MPTP-induced parkinsonism in baboons. Nat. Med. 2, 1017–1021. [DOI] [PubMed] [Google Scholar]
- Hanrott K.; Gudmunsen L.; O’Neill M. J.; Wonnacott S. (2006) 6-hydroxydopamine-induced apoptosis is mediated via extracellular auto-oxidation and caspase 3-dependent activation of protein kinase Cδ. J. Biol. Chem. 281, 5373–5382. [DOI] [PubMed] [Google Scholar]
- Granot J.; Rotman A. (1978) Nuclear magnetic resonance studies of 6-hydroxydopamine and its interactions with SH-containing model compounds. Evaluation of possible mechanism for neurocytotoxicity. Biochemistry 17, 2370–2374. [DOI] [PubMed] [Google Scholar]
- Biedler J. L.; Helson L.; Spengler B. A. (1973) Morphology and growth, tumorigenicity, and cytogenetics of human neuroblastoma cells in continuous culture. Cancer Res. 33, 2643–2652. [PubMed] [Google Scholar]
- Klongpanichapak S.; Phansuwan-Pujito P.; Ebadi M.; Govitrapong P. (2008) Melatonin inhibits amphetamine-induced increase in α-synuclein and decrease in phosphorylated tyrosine hydroxylase in SK-N-SH cells. Neurosci. Lett. 436, 309–313. [DOI] [PubMed] [Google Scholar]
- West G. J.; Uki J.; Herschman H. R.; Seeger R. C. (1977) Adrenergic, cholinergic, and inactive human neuroblastoma cell lines with the action-potential Na+ ionophore. Cancer Res. 37, 1372–1376. [PubMed] [Google Scholar]
- Pizzi M.; Boroni F.; Bianchetti A.; Moraitis C.; Sarnico I.; Benarese M.; Goffi F.; Valerio A.; Spano P. (2002) Expression of functional NR1/NR2B-type NMDA receptors in neuronally differentiated SK-N-SH human cell line. Eur. J. Neurosci 16, 2342–2350. [DOI] [PubMed] [Google Scholar]
- Jantas D.; Roman A.; Kusmierczyk J.; Lorenc-Koci E.; Konieczny J.; Lenda T.; Lason W. (2013) The extent of neurodegeneration and neuroprotection in two chemical in vitro models related to Parkinson’s disease is critically dependent on cell culture conditions. Neurotox Res. 24, 41–54. [DOI] [PubMed] [Google Scholar]
- Wenker S. D.; Chamorro M. E.; Vota D. M.; Callero M. A.; Vittori D. C.; Nesse A. B. (2010) Differential antiapoptotic effect of erythropoietin on undifferentiated and retinoic acid-differentiated SH-SY5Y cells. J. Cell Biochem 110, 151–161. [DOI] [PubMed] [Google Scholar]
- Xie H. R.; Hu L. S.; Li G. Y. (2010) SH-SY5Y human neuroblastoma cell line: In vitro cell model of dopaminergic neurons in Parkinson’s disease. Chin Med. J. (Engl) 123, 1086–1092. [PubMed] [Google Scholar]
- Cheung Y. T.; Lau W. K.; Yu M. S.; Lai C. S.; Yeung S. C.; So K. F.; Chang R. C. (2009) Effects of all-trans-retinoic acid on human SH-SY5Y neuroblastoma as in vitro model in neurotoxicity research. Neurotoxicology 30, 127–135. [DOI] [PubMed] [Google Scholar]
- Garcia-Garcia A.; Anandhan A.; Burns M.; Chen H.; Zhou Y.; Franco R. (2013) Impairment of Atg5-dependent autophagic flux promotes paraquat- and MPP+-induced apoptosis but not rotenone or 6-hydroxydopamine toxicity. Toxicol. Sci. 136, 166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement M. V.; Long L. H.; Ramalingam J.; Halliwell B. (2002) The cytotoxicity of dopamine may be an artefact of cell culture. J. Neurochem 81, 414–421. [DOI] [PubMed] [Google Scholar]
- Ding Y. M.; Jaumotte J. D.; Signore A. P.; Zigmond M. J. (2004) Effects of 6-hydroxydopamine on primary cultures of Substantia nigra: Specific damage to dopamine neurons and the impact of glial cell line-derived neurotrophic factor. J. Neurochem. 89, 776–787. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Liew C. W.; Handy D. E.; Zhang Y.; Leopold J. A.; Hu J.; Guo L.; Kulkarni R. N.; Loscalzo J.; Stanton R. C. (2010) High glucose inhibits glucose-6-phosphate dehydrogenase, leading to increased oxidative stress and β-cell apoptosis. FASEB J. 24, 1497–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco R.; Panayiotidis M. I.; Cidlowski J. A. (2007) Glutathione depletion is necessary for apoptosis in lymphoid cells independent of reactive oxygen species formation. J. Biol. Chem. 282, 30452–30465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaika N. V.; Gebregiworgis T.; Lewallen M. E.; Purohit V.; Radhakrishnan P.; Liu X.; Zhang B.; Mehla K.; Brown R. B.; Caffrey T.; Yu F.; Johnson K. R.; Powers R.; Hollingsworth M. A.; Singh P. K. (2012) MUC1 mucin stabilizes and activates hypoxia-inducible factor 1 alpha to regulate metabolism in pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 109, 13787–13792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B.; Powers R. (2014) MVAPACK: A Complete Data Handling Package for NMR Metabolomics. ACS Chem. Biol. 9, 1138–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Meyer T.; Sinnaeve D.; Van Gasse B.; Tsiporkova E.; Rietzschel E. R.; De Buyzere M. L.; Gillebert T. C.; Bekaert S.; Martins J. C.; Van Criekinge W. (2008) NMR-based characterization of metabolic alterations in hypertension using an adaptive, intelligent binning algorithm. Anal. Chem. 80, 3783–3790. [DOI] [PubMed] [Google Scholar]
- Dieterle F.; Ross A.; Schlotterbeck G.; Senn H. (2006) Probabilistic quotient normalization as robust method to account for dilution of complex biological mixtures. Application in 1H NMR metabonomics. Anal. Chem. 78, 4281–4290. [DOI] [PubMed] [Google Scholar]
- Worley B.; Halouska S.; Powers R. (2013) Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 433, 102–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werth M. T.; Halouska S.; Shortridge M. D.; Zhang B.; Powers R. (2010) Analysis of metabolomic PCA data using tree diagrams. Anal. Biochem. 399, 58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson L.; Trygg J.; Wold S. (2008) CV-ANOVA for significance testing of PLS and OPLS (R) models. J. Chemometr 22, 594–600. [Google Scholar]
- Shao J. (1993) Linear-model selection by cross-validation. J. Am. Stat Assoc 88, 486–494. [Google Scholar]
- Akiyama K.; Chikayama E.; Yuasa H.; Shimada Y.; Tohge T.; Shinozaki K.; Hirai M. Y.; Sakurai T.; Kikuchi J.; Saito K. (2008) PRIMe: A Web site that assembles tools for metabolomics and transcriptomics. In Silico Biol. 8, 339–345. [PubMed] [Google Scholar]
- Wishart D. S.; Knox C.; Guo A. C.; Eisner R.; Young N.; Gautam B.; Hau D. D.; Psychogios N.; Dong E.; Bouatra S.; Mandal R.; Sinelnikov I.; Xia J.; Jia L.; Cruz J. A.; Lim E.; Sobsey C. A.; Shrivastava S.; Huang P.; Liu P.; Fang L.; Peng J.; Fradette R.; Cheng D.; Tzur D.; Clements M.; Lewis A.; De Souza A.; Zuniga A.; Dawe M.; Xiong Y.; Clive D.; Greiner R.; Nazyrova A.; Shaykhutdinov R.; Li L.; Vogel H. J.; Forsythe I. (2009) HMDB: A knowledgebase for the human metabolome. Nucleic Acids Res. 37, D603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Q.; Lewis I. A.; Hegeman A. D.; Anderson M. E.; Li J.; Schulte C. F.; Westler W. M.; Eghbalnia H. R.; Sussman M. R.; Markley J. L. (2008) Metabolite identification via the Madison Metabolomics Consortium Database. Nat. Biotechnol. 26, 162–164. [DOI] [PubMed] [Google Scholar]
- Xia J.; Bjorndahl T. C.; Tang P.; Wishart D. S. (2008) MetaboMiner—Semi-automated identification of metabolites from 2D NMR spectra of complex biofluids. BMC Bioinformatics 9, 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich E. L.; Akutsu H.; Doreleijers J. F.; Harano Y.; Ioannidis Y. E.; Lin J.; Livny M.; Mading S.; Maziuk D.; Miller Z.; Nakatani E.; Schulte C. F.; Tolmie D. E.; Kent Wenger R.; Yao H.; Markley J. L. (2008) BioMagResBank. Nucleic Acids Res. 36, D402–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D. S.; Jewison T.; Guo A. C.; Wilson M.; Knox C.; Liu Y.; Djoumbou Y.; Mandal R.; Aziat F.; Dong E.; Bouatra S.; Sinelnikov I.; Arndt D.; Xia J.; Liu P.; Yallou F.; Bjorndahl T.; Perez-Pineiro R.; Eisner R.; Allen F.; Neveu V.; Greiner R.; Scalbert A. (2013) HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 41, D801–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. A.; O’Maille G.; Want E. J.; Qin C.; Trauger S. A.; Brandon T. R.; Custodio D. E.; Abagyan R.; Siuzdak G. (2005) METLIN: A metabolite mass spectral database. Ther. Drug Monit. 27, 747–751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.