Abstract
In this paper, a reverse-transcriptase PCR-based protocol suitable for efficient expression analysis of multigene families is presented. The method combines restriction fragment length polymorphism (RFLP) technology with a gene family-specific version of mRNA differential display and hence is called "RFLP-coupled domain-directed differential display. "With this method, expression of all members of a multigene family at many different developmental stages, in diverse tissues and even in different organisms, can be displayed on one gel. Moreover, bands of interest, representing gene family members, are directly accessible to sequence analysis, without the need for subcloning. The method thus enables a detailed, high-resolution expression analysis of known gene family members as well as the identification and characterization of new ones. Here the technique was used to analyze differential expression of MADS-box genes in male and female inflorescences of maize (Zea mays ssp. mays). Six different MADS-box genes could be identified, being either specifically expressed in the female sex or preferentially expressed in male or female inflorescences, respectively. Other possible applications of the method are discussed.
Full text
PDF


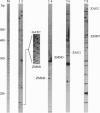
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angenent G. C., Busscher M., Franken J., Mol J. N., van Tunen A. J. Differential expression of two MADS box genes in wild-type and mutant petunia flowers. Plant Cell. 1992 Aug;4(8):983–993. doi: 10.1105/tpc.4.8.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angenent G. C., Franken J., Busscher M., Colombo L., van Tunen A. J. Petal and stamen formation in petunia is regulated by the homeotic gene fbp1. Plant J. 1993 Jul;4(1):101–112. doi: 10.1046/j.1365-313x.1993.04010101.x. [DOI] [PubMed] [Google Scholar]
- Bauer D., Müller H., Reich J., Riedel H., Ahrenkiel V., Warthoe P., Strauss M. Identification of differentially expressed mRNA species by an improved display technique (DDRT-PCR). Nucleic Acids Res. 1993 Sep 11;21(18):4272–4280. doi: 10.1093/nar/21.18.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell I. D., Downing A. K. Building protein structure and function from modular units. Trends Biotechnol. 1994 May;12(5):168–172. doi: 10.1016/0167-7799(94)90078-7. [DOI] [PubMed] [Google Scholar]
- Davies B., Schwarz-Sommer Z. Control of floral organ identity by homeotic MADS-box transcription factors. Results Probl Cell Differ. 1994;20:235–258. doi: 10.1007/978-3-540-48037-2_11. [DOI] [PubMed] [Google Scholar]
- Dellaporta S. L., Calderon-Urrea A. The sex determination process in maize. Science. 1994 Dec 2;266(5190):1501–1505. doi: 10.1126/science.7985019. [DOI] [PubMed] [Google Scholar]
- Flanagan C. A., Ma H. Spatially and temporally regulated expression of the MADS-box gene AGL2 in wild-type and mutant arabidopsis flowers. Plant Mol Biol. 1994 Oct;26(2):581–595. doi: 10.1007/BF00013745. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser P., Klein J., Lönnig W. E., Meijer H., Saedler H., Sommer H. Bracteomania, an inflorescence anomaly, is caused by the loss of function of the MADS-box gene squamosa in Antirrhinum majus. EMBO J. 1992 Apr;11(4):1239–1249. doi: 10.1002/j.1460-2075.1992.tb05168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees E. M., Harlow E. Sequences within the conserved cyclin box of human cyclin A are sufficient for binding to and activation of cdc2 kinase. Mol Cell Biol. 1993 Feb;13(2):1194–1201. doi: 10.1128/mcb.13.2.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Barnathan E. S., Karikó K. Rapid method for screening and cloning cDNAs generated in differential mRNA display: application of northern blot for affinity capturing of cDNAs. Nucleic Acids Res. 1994 May 11;22(9):1764–1765. doi: 10.1093/nar/22.9.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Averboukh L., Pardee A. B. Distribution and cloning of eukaryotic mRNAs by means of differential display: refinements and optimization. Nucleic Acids Res. 1993 Jul 11;21(14):3269–3275. doi: 10.1093/nar/21.14.3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang P., Pardee A. B. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992 Aug 14;257(5072):967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lozovskaya E. R., Petrov D. A., Hartl D. L. A combined molecular and cytogenetic approach to genome evolution in Drosophila using large-fragment DNA cloning. Chromosoma. 1993 Mar;102(4):253–266. doi: 10.1007/BF00352399. [DOI] [PubMed] [Google Scholar]
- Ma H., Yanofsky M. F., Meyerowitz E. M. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991 Mar;5(3):484–495. doi: 10.1101/gad.5.3.484. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Hunt T. Cell cycle. Dams and sluices. Nature. 1993 Dec 16;366(6456):634–635. doi: 10.1038/366634a0. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Pnueli L., Abu-Abeid M., Zamir D., Nacken W., Schwarz-Sommer Z., Lifschitz E. The MADS box gene family in tomato: temporal expression during floral development, conserved secondary structures and homology with homeotic genes from Antirrhinum and Arabidopsis. Plant J. 1991 Sep;1(2):255–266. [PubMed] [Google Scholar]
- Pnueli L., Hareven D., Rounsley S. D., Yanofsky M. F., Lifschitz E. Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell. 1994 Feb;6(2):163–173. doi: 10.1105/tpc.6.2.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Anisowicz A., Neveu M., Liang P., Sotiropoulou G. Identification by differential display of alpha 6 integrin as a candidate tumor suppressor gene. FASEB J. 1993 Jul;7(10):964–970. doi: 10.1096/fasebj.7.10.8344495. [DOI] [PubMed] [Google Scholar]
- Schmidt R. J., Veit B., Mandel M. A., Mena M., Hake S., Yanofsky M. F. Identification and molecular characterization of ZAG1, the maize homolog of the Arabidopsis floral homeotic gene AGAMOUS. Plant Cell. 1993 Jul;5(7):729–737. doi: 10.1105/tpc.5.7.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz-Sommer Z., Huijser P., Nacken W., Saedler H., Sommer H. Genetic Control of Flower Development by Homeotic Genes in Antirrhinum majus. Science. 1990 Nov 16;250(4983):931–936. doi: 10.1126/science.250.4983.931. [DOI] [PubMed] [Google Scholar]
- Sommer H., Beltrán J. P., Huijser P., Pape H., Lönnig W. E., Saedler H., Schwarz-Sommer Z. Deficiens, a homeotic gene involved in the control of flower morphogenesis in Antirrhinum majus: the protein shows homology to transcription factors. EMBO J. 1990 Mar;9(3):605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Strater T., Fischer A., Saedler H. Structural characterization, chromosomal localization and phylogenetic evaluation of two pairs of AGAMOUS-like MADS-box genes from maize. Gene. 1995 Apr 24;156(2):155–166. doi: 10.1016/0378-1119(95)00020-7. [DOI] [PubMed] [Google Scholar]
- Tröbner W., Ramirez L., Motte P., Hue I., Huijser P., Lönnig W. E., Saedler H., Sommer H., Schwarz-Sommer Z. GLOBOSA: a homeotic gene which interacts with DEFICIENS in the control of Antirrhinum floral organogenesis. EMBO J. 1992 Dec;11(13):4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990 Jul 5;346(6279):35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- Zimmermann J. W., Schultz R. M. Analysis of gene expression in the preimplantation mouse embryo: use of mRNA differential display. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5456–5460. doi: 10.1073/pnas.91.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]