Abstract
Natural products remain an important source of drug candidates, but the difficulties inherent to traditional isolation, coupled with unacceptably high rates of compound rediscovery, limit the pace of natural product detection. Here we describe a reactivity-based screening method to rapidly identify exported bacterial metabolites that contain dehydrated amino acids (i.e., carbonyl- or imine-activated alkenes), a common motif in several classes of natural products. Our strategy entails the use of a commercially available thiol, dithiothreitol, for the covalent labeling of activated alkenes by nucleophilic 1,4-addition. Modification is easily discerned by comparing mass spectra of reacted and unreacted cell surface extracts. When combined with bioinformatic analysis of putative natural product gene clusters, targeted screening and isolation can be performed on a prioritized list of strains. Moreover, known compounds are easily dereplicated, effectively eliminating superfluous isolation and characterization. As a proof of principle, this labeling method was used to identify known natural products belonging to the thiopeptide, lanthipeptide, and linaridin classes. Further, upon screening a panel of only 23 actinomycetes, we discovered and characterized a novel thiopeptide antibiotic, cyclothiazomycin C.
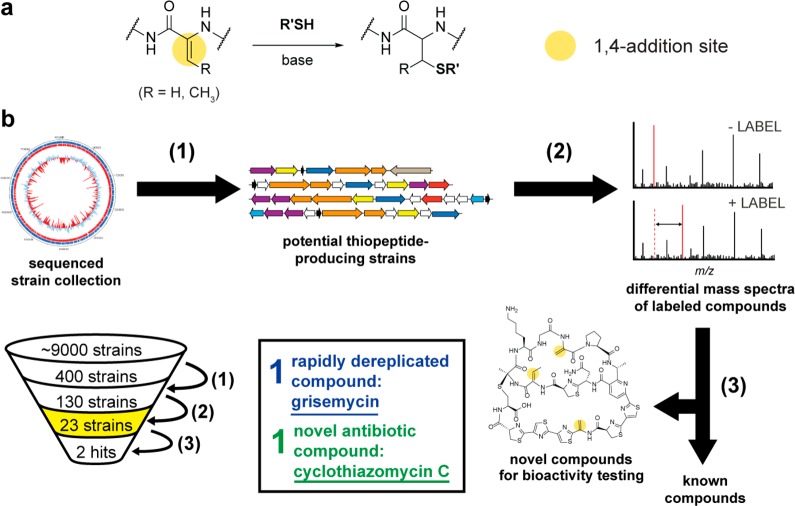
Bacteria have historically been a rich reservoir of architecturally complex natural products exhibiting antibiotic activity.1 However, the traditional approach to natural product discovery, bioassay-guided isolation of compounds from extracts, is limited by high rates of compound rediscovery.2 As such, the potential value of novel natural products to advance the treatment of disease and in particular to address the issue of antibiotic resistance3 warrants the development of alternative strategies to discover novel compounds. The advent of widely available genome sequences makes bioinformatics-driven methods increasingly appealing, since the enzymatic machinery responsible for natural product biosynthesis can be readily identified.4,5 Consequently, a number of strategies have emerged that aid in connecting biosynthetic gene clusters to their products, including selective enzymatic derivatization,6 chemoselective enrichment,7 mass spectrometry-based network analysis,8 and PCR prioritization9 among others. Another approach to address the innovation gap in natural product discovery is to utilize the intrinsic chemical reactivity of functional groups that are enriched in a target class of metabolites. Here, we report the development of a reactivity-based screening method to identify, isolate, dereplicate, and characterize novel natural products using a combination of bioinformatics and a simple chemical probe for modifying a reactive functional group (Figure 1).
Figure 1.
Strategy for natural product discovery by bioinformatics-guided prioritization and nucleophilic 1,4-addition chemistry. (a) Reaction scheme for the thiol (DTT/DIPEA) labeling method with 1,4-addition sites indicated with yellow circles. (b) Work flow for the bioinformatics-based strain prioritization, subsequent DTT-labeling, and MS screening (reactivity-based screening). (1) Prediction of DHAA-containing thiopeptide biosynthetic gene clusters from 400 in-house sequenced genomes (all from the USDA ARS Actinobacteria collection, which totals ∼9000 unique strains). More information on strain prioritization is given in Supplemental Figure 3. (2) DHAAs on exported bacterial metabolites that are reactive toward nucleophilic 1,4-additions (by DTT/DIPEA) are identified by differential mass spectrometry. (3) Compound isolation and characterization after dereplication. Compounds are dereplicated, taking only potentially novel compounds through the time-consuming characterization steps. Of the 400 sequenced genomes, 130 strains were prioritized, 23 strains were screened, 1 compound was rapidly dereplicated, and 1 compound was predicted to be novel and thus further characterized.
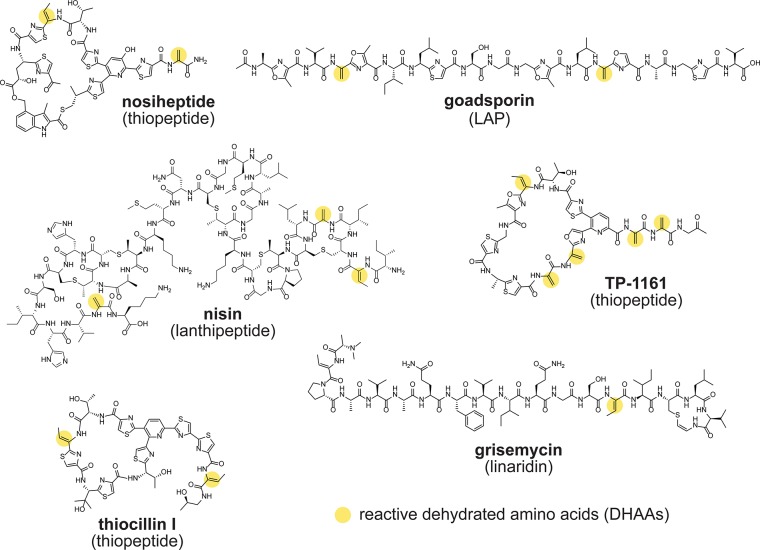
The dehydrated amino acids (DHAAs) dehydroalanine and dehydrobutyrine are frequently found in natural products,10 including thiopeptides,11 lanthipeptides,12,13 and linaridins14−16 among others (Figure 2). We thus envisioned DHAAs serving as a useful chemical handle for the discovery of natural products. It has been demonstrated that thiol nucleophiles participate in 1,4-addition into α,β-unsaturated carbonyl/imine DHAAs under mild conditions to yield covalent thioether adducts (Figure 1a).17 This reactivity has been exploited previously in the chemical modification of thiostrepton,18−20 the mapping of Ser/Thr modifications in proteins,21 the design of solid-phase capture resins,22 and the identification of lanthipeptides.23 Thus, we sought to employ this well-established, reliable chemistry as part of a novel tandem bioinformatics/reactivity-based screening effort.
Figure 2.
Representative natural products bearing dehydrated amino acids (DHAAs). Structures of example molecules that contain DHAAs suitable for nucleophilic addition are shown. The sites of potential nucleophilic reactivity (i.e., the DHAA alkenes, often in the form of an α,β-unsaturated carbonyl) are indicated with yellow circles. LAP, linear azol(in)e-containing peptide.
Many classes of DHAA-bearing natural products are ribosomally produced, rendering them ideal for genome-guided discovery. The availability of genome sequences has revealed a tremendous biosynthetic capability among diverse microbial species.24 It has become apparent that even well-characterized bacteria harbor the potential to produce an abundance of yet-uncharacterized natural products.25 To overcome the burden of rediscovery,26 knowledge of biosynthetic gene sequences can be used to preselect bacterial strains for screening to include only the organisms with the theoretical capacity to produce a particular type of natural product.9 However, even with the bioinformatic identification of promising biosynthetic gene clusters, the detection and isolation of the resultant natural products often proves to be difficult given that the products of most biosynthetic pathways are present in extremely low quantities (if present at all) during laboratory cultivation.27 Accordingly, a broadly applicable companion strategy to genome mining that would allow the determination of whether a natural product of interest is produced at a detectable level would be valuable. We thus reasoned that a combination of bioinformatics- and reactivity-based screening (i.e., nucleophilic 1,4-addition to DHAAs) would streamline natural product discovery efforts.
Results and Discussion
Rationale and Overview of a New Natural Product Discovery Method
Herein we have utilized the combination of bioinformatics and nucleophilic 1,4-addition chemistry for the rapid labeling, discovery, and dereplication of DHAA-containing natural products (Figure 1b) by reactivity-based screening. Our discovery pipeline begins with a bioinformatic survey for strains of Actinobacteria predicted to be capable of producing a DHAA-containing natural product (Figure 1b, Step 1, vide infra for specifics on the bioinformatics-based strain prioritization). After cultivation, the exported metabolites from the prioritized Actinobacteria are extracted with organic solvent using a nonlytic procedure (see Methods). A portion of this cell-surface extract then undergoes treatment with dithiothreitol (DTT) in the presence of base. DTT was chosen as the thiol probe owing to its low cost and ubiquity in natural product discovery laboratories. If reactive DHAA moieties are present in the cell-surface extract, the resulting DTT adducts increase the mass of the exported metabolite by multiples of 154.0 Da (Figure 1b, Step 2). Differential mass spectrometry between the unreacted control and the DTT-reacted extracts readily identifies the compounds containing DHAAs within a predetermined mass range. The molecular mass, number of DTT additions, and analysis of tandem mass spectra, combined with the initial bioinformatic prediction of DHAA-containing natural products, permits a rapid determination of compound novelty. At this step, every DTT-labeled compound can be analyzed, irrespective of whether the mass corresponds to a predicted biosynthetic gene cluster. Known compounds are removed from further analysis at this step, leaving only compounds with a high probability of novelty for further structural and functional characterization, which is considerably more time-consuming (Figure 1b, Step 3). To determine if the above proposed discovery pipeline was viable, we sought to discover a novel DHAA-containing thiopeptide via bioinformatic prioritization and reactivity-based screening utilizing nucleophilic 1,4-addition chemistry.
Validation of the DTT-Labeling Strategy
With the ultimate goal of using the above-described DTT-labeling method to discover a new natural product, we first sought to establish an operationally simple route to rapidly screen organic extracts for compounds of interest. We utilized two DHAA-containing natural products, thiostrepton and geobacillin I, for method development and validation.
Thiostrepton, whose biosynthetic gene cluster was identified in 2009,11 is a thiopeptide produced by Streptomyces azureus ATCC 14921 (among others).28 Notably, the highly modified scaffold of thiostrepton contains four DHAAs where labeling can occur: three dehydroalanine residues and one dehydrobutyrine (Figure 3a).29 To test the method, reactions were conducted using commercially obtained thiostrepton, DTT, and either diisopropylethylamine (DIPEA) or no base at 23 °C for 16 h in a 1:1 mixture of chloroform and methanol. The authentic thiostrepton standard and the DTT-reacted samples were then subjected to matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) analysis. The peaks corresponding to unmodified thiostrepton (m/z 1664.4 Da) were supplanted in the DTT-reacted sample by peaks corresponding to the addition of multiple DTT labels. The tertiary adduct was the most prominent, suggesting the successful addition of DTT into the reactive alkenes (Supplemental Figure 1). The addition of DIPEA enhanced the DTT-labeling reaction. Other bases, including triethylamine and 1,8-diazabicycloundec-7-ene (DBU), were tested, and labeling occurred similarly to the reactions using DIPEA. A range of DIPEA concentrations were tested (10–50 mM), and the extent of labeling did not greatly vary. Therefore, all further experiments employed 10 mM DIPEA.
Figure 3.
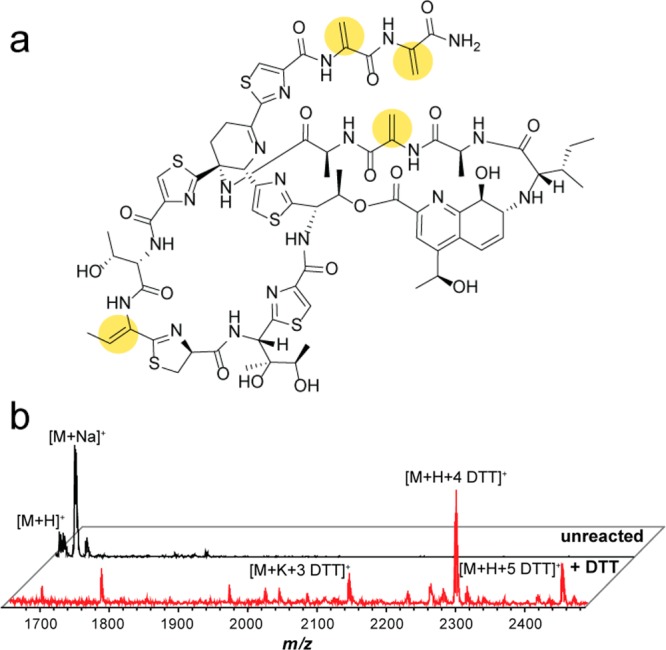
DTT-labeling of thiostrepton as a proof of principle. (a) Structure of thiostrepton with DHAAs suitable for nucleophilic addition highlighted with yellow circles. (b) MALDI-TOF MS of thiostrepton labeling performed in the context of an organic, cell-surface extract of Streptomyces azureus ATCC 14921. The black spectrum (top) is an unreacted control, while the red spectrum (bottom) resulted from DTT-labeling. Thiostrepton was visibly labeled by 1–5 DTT moieties, with the 4 DTT adduct being the majority product.
To confirm DTT-labeling of thiostrepton could be observed by MALDI-TOF MS in the context of a more complex biological mixture, we subjected an organic cell-surface extract of S. azureus ATCC 14921 (thiostrepton producer) to the above labeling reaction. Analogous to the pure thiostrepton sample, comparison of the crude extract with the DTT-labeled extraction again showed the appearance of multiple DTT adducts, this time with the tetra-adduct being the primary species; a higher extent of labeling was seen here due to the larger relative excess of the labeling reagents in the context of a biological extract (Figure 3b). Although thiostrepton contains only 4 reactive DHAA sites, a minor fifth adduct was observed in both the commercially available and extracted samples, presumably from reaction with another electrophilic site. Thiostrepton possesses an additional alkene that is conjugated to pyridine within the quinaldic acid moiety; we hypothesize that addition of DTT may have occurred at this site, given the literature precedent for addition of thiols to aromatic-conjugated alkenes.30 Importantly, the appearance of this low-intensity ion does not complicate detection or interpretation of the labeled analyte.
Lanthipeptides are ribosomally synthesized and post-translationally modified peptide natural products (RiPPs) that are easily identified using bioinformatics and frequently contain DHAAs.5,12,13,31 To test if the reactivity-based screening method could also be used to identify other classes of natural products in varied bacterial extracts, we attempted to label the lanthipeptide geobacillin I. Geobacillin I, a nisin analogue, is produced by Geobacillus sp. M10EXG (Supplemental Figure 2a).33,34 Upon subjecting an organic cell-surface extract from Geobacillus sp. M10EXG to our labeling conditions, a mass corresponding to two DTT adducts was prominently observed; a third adduct was visible but of very low intensity (Supplemental Figure 2b). Only two reactive DHAA sites are present in geobacillin I: a dehydroalanine and a dehydrobutyrine. However, transient DHAA sites occur in the biosynthesis of the lanthionine rings, which are formed by intramolecular 1,4-addition of cysteines to DHAAs.13 We hypothesize, accordingly, that a small percentage of the geobacillin present in the extract may have an unformed lanthionine ring, leaving a free reactive site available for DTT-labeling. Again, even under stoichiometrically forcing conditions, this extract adduct was of only minor abundance and thus did not interfere with compound detection or analysis.
Bioinformatics-Guided Strain Prioritization
Like lanthipeptides, thiopeptides are RiPPs, and the biosynthetic genes responsible for their production are often clustered, rendering them identifiable by sequence similarity searching. From the perspective of the present study, we sought to prioritize bacterial strains for subsequent screening based on the presence of biosynthetic genes capable of installing DHAAs (often misleadingly annotated as “lantibiotic dehydratases”).12 These genes, however, can be found in a variety of other natural product gene clusters and not exclusively in thiopeptide clusters. Therefore, we first identified clusters that encode for the YcaO cyclodehydratase protein that is necessary for the biosynthesis of all thiazole/oxazole-modified microcin natural products, of which thiopeptides can be broadly categorized. Strains containing a YcaO cyclodehydratase were analyzed further for the local co-occurrence of genes encoding a “lantibiotic dehydratase” (for the production of DHAAs) and a thiopeptide-like precursor peptide (Supplemental Figure 3a).35 A total of 130 unique strains of recently sequenced (in-house) Actinobacteria from the Northern Regional Research Laboratory collection (NRRL), which is curated by the Agricultural Research Service under the supervision of the U.S. Department of Agriculture (USDA/ARS), were predicted to have the genetic capacity to produce a DHAA-containing thiopeptide (Figure 1b). The precursor peptide sequences from these clusters were then used to estimate the masses of the final natural products for dereplication and characterization purposes (Supplemental Figure 3b). These strains were then subjected to reactivity-based screening with DTT and DIPEA to discover a novel thiopeptide.
MS-Based Screening of Prioritized Strains
Twenty-three of the prioritized strains with novel precursor peptide sequences were selected for screening by DTT-labeling (Supplemental Figure 4). We first noticed a sample containing 1–2 DTT adducts on an exported metabolite with a mass of [M + H]+, m/z 1855.0 Da. While we were intentionally blind to which of the Actinobacteria strains were undergoing analysis, after labeling we established that this particular extract originated from Streptomyces griseus subsp. griseus, and the labeled mass did not correlate with the expected mass of the predicted thiopeptide from this strain. However, Streptomyces griseus subsp. griseus is a known producer of grisemycin, with which the mass of the labeled natural product did correlate (Figure 4a,b). MS/MS fragmentation analysis yielded a seven amino acid sequence tag, confirming the identity of the compound as grisemycin (Figure 4c).15 The labeling and identification of grisemycin, a member of the linaridin class of natural products, further validated our reactivity-based screen while also highlighting the usefulness of bioinformatic integration to rapidly dereplicate known compounds.
Figure 4.
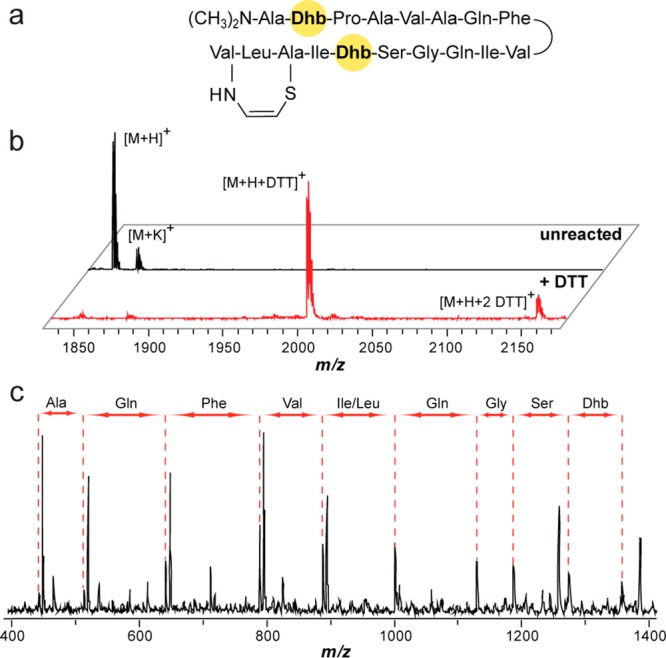
Grisemycin DTT-labeling and dereplication. (a) Structure of grisemycin. Dhb, dehydrobutyrine. (b) MALDI-TOF MS analysis of unreacted grisemycin (black spectrum, top) and DTT-labeled grisemycin (red spectrum, bottom) from an organic, cell-surface extract showing 1–2 DTT adducts. (c) MS/MS analysis of grisemycin with the discerned sequence tag listed above the spectrum.
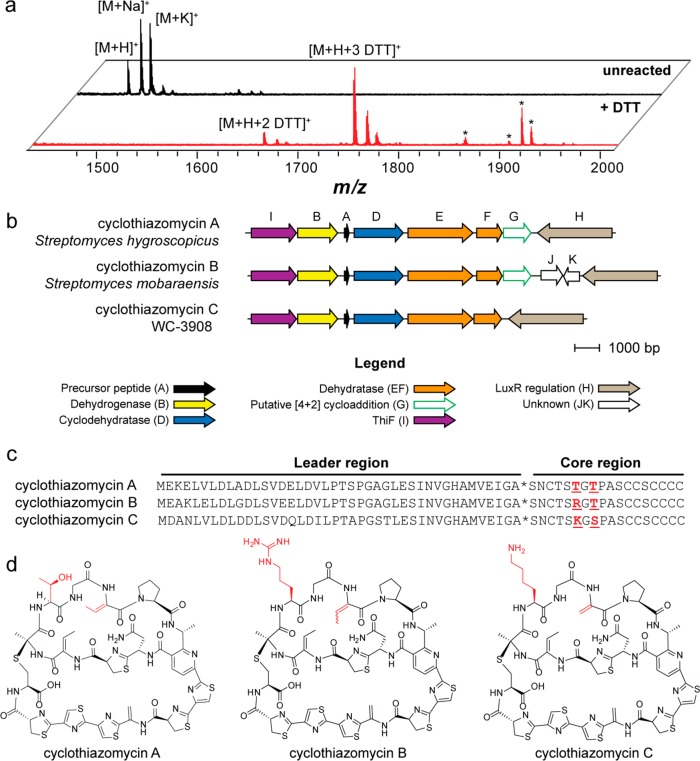
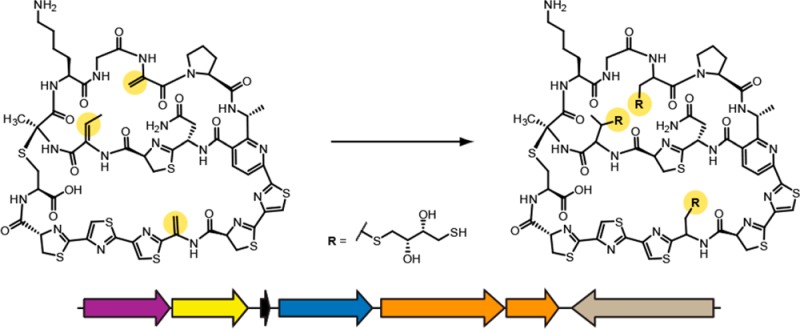
The organic cell-surface extract from a separate sample contained a compound ([M + H]+, m/z 1486.3 Da) that underwent labeling to contain primarily three DTT adducts (Figure 5a). This mass correlated well with the predicted mass of a hypothetical thiopeptide from NRRL strain WC-3908. The thiopeptide gene cluster from WC-3908 was similar to the gene clusters responsible for the production of the thiopeptides cyclothiazomycin A, originally termed 5102-I,36,37 and cyclothiazomycin B (Figure 5b). The core region of the precursor peptide (i.e., the portion that undergoes enzymatic tailoring to yield the mature natural product)31,38 from WC-3908 differed by two amino acids from the precursor peptides of cyclothiazomycin A and B (Figure 5c). Accordingly, we designated the WC-3908 thiopeptide cyclothiazomycin C. Given that the structures of cyclothiazomycin A and B have been reported,39−42 we could accurately predict the structure of cyclothiazomycin C, which was in agreement with the labeling results (Figure 5d).
Figure 5.
Identification, genetics, and structure of cyclothiazomycin C. (a) MALDI-TOF MS analysis showing spectra of unreacted (black spectrum, top) and DTT-labeled (red spectrum, bottom) extracts of WC-3908, the producer of cyclothiazomycin C. Peaks labeled with an asterisk do not correspond to DTT-labeled cyclothiazomycin C. (b) Conserved open-reading frames from each of the three cyclothiazomycin gene clusters (precise cluster boundaries are not yet established). Genes are color-coded with proposed functions given in the legend. The strain used for the comparison of cyclothiazomycin A is Streptomyces hygroscopicus subsp. jinggangensis 5008, and cyclothiazomycin B is Streptomyces mobaraensis. (c) Precursor peptide sequences of cyclothiazomycins A, B, and C. Highlighted in red are residues that differ in the core region of the peptide. The asterisk denotes the leader peptide cleavage site. (d) Structures of cyclothiazomycins A, B, and C.
Verification of the Cyclothiazomycin C Structure
Prior to detailed structural characterization, cyclothiazomycin C was purified by MPLC and HPLC (Supplemental Figure 5). The mass spectrum of purified cyclothiazomycin C revealed an [M + H]+ ion of m/z 1486.3309 Da (Supplemental Figure 6a), supporting the molecular formula for the predicted structure of cyclothiazomycin C (C60H67N19O13S7). Analysis of the collision-induced dissociation (CID) mass spectrum corroborated the amino acid sequence of the precursor peptide, strongly connecting the predicted gene cluster to the mature natural product (Supplemental Figure 6b). NMR spectroscopy was then used to confirm the predicted structure of cyclothiazomycin C (Supplemental Figures 7–8). Bond connectivity was established using 1H–1H COSY, 1H–1H TOCSY, 1H–13C HSQC, and 1H–13C HMBC experiments. Chemical shifts were assigned from this information and by comparison to the reported values for cyclothiazomycin B.40 Due to the spectral similarity to cyclothiazomycin B, we have assigned the stereochemistry of cyclothiazomycin C analogously to that of the reported compound.
Conservation Analysis of the Cyclothiazomycin C Biosynthetic Gene Cluster
To provide additional evidence that the thiopeptide gene cluster from WC-3908 was responsible for the production of cyclothiazomycin C, conservation analysis was performed with the cyclothiazomycin A, B, and C (putative) gene clusters. The cyclothiazomycin A biosynthetic genes derived from Streptomyces hygroscopicus subsp. jinggangensis 5008, while the cyclothiazomycin B genes were from Streptomyces mobaraensis. A subset of the genes predicted for the production of cyclothiazomycin B37 was conserved among the three clusters (Figure 5b). All three clusters contain a short open reading frame, here designated ctmA, encoding the precursor peptide. CtmD encodes a “fused” TOMM cyclodehydratase (E1 ubiquitin-activating enzyme/MccB-like and YcaO domains), which implicates CtmD in the formation of thiazolines.43,44CtmB encodes a flavin mononucleotide-dependent protein, putatively responsible for the dehydrogenation of the thiazolines to thiazoles.45CtmE and ctmF encode homologues of a split lanthipeptide dehydratase, which performs the dehydration of serine and threonine to dehydroalanine and dehydrobutyrine.12,13 Like all thiopeptides, cyclothiazomycin C has a substituted six-membered, nitrogen-containing central heterocycle (here a pyridine). In the case of cyclothiazomycins A and B, the pyridine moiety is likely formed by the gene product of ctmG, given the homology to tclM, which has been implicated in the formal [4 + 2] cycloaddition reaction during thiocillin biosynthesis (Supplemental Figure 9).46 For cyclothiazomycin C, a gene with high similarity to ctmG from the cyclothiazomycin A and B clusters is present but distantly located in the genome, indicating that the cyclothiazomycin C gene cluster is fragmented. Interestingly, ctmG from WC-3908 is found next to a gene duplication of ctmF, which is suggestive of paralogous duplication (Supplemental Figure 9). CtmI, which is present in all three clusters, encodes a ThiF-like protein. ThiF-like proteins have been implicated in the biosynthesis of thiamine diphosphate in E. coli.47 However, the function of ThiF-like proteins in the context of TOMM biosynthesis remains to be established. Other local genes include ctmH, which is a LuxR-type regulatory gene, and ctmJK, which are omitted from the cyclothiazomycin A and C clusters and have no known function (Figure 5). We further note that the genes flanking the conserved region are highly disparate between the three clusters (Supplemental Figure 10). This subset of genes, ctmA-G and ctmI from Streptomyces hygroscopicus subsp. jinggangensis 5008, were recently shown to be regulated by the LuxR-type regulatory gene ctmH. Furthermore, the deletion of ctmA, ctmD, ctmF, and ctmG abolished the production of cyclothiazomycin A.48 These data further support the gene cluster prediction for cyclothiazomycin C from WC-3908.
Assessment of Cyclothiazomycin Bioactivity
Previous reports on cyclothiazomycins A and B describe a wide range of bioactivities, including renin inhibition,39 RNA polymerase inhibition,40 and antifungal activity.49 We found that purified cyclothiazomycin C exhibited growth inhibitory action toward several Gram-positive (Firmicutes) bacteria but was inactive against all tested Gram-negative (Proteobacteria) organisms (Table 1). The greatest inhibitory activity was observed toward the genus Bacillus. On the basis of prior reports, we decided to also evaluate if cyclothiazomycin C exhibited growth inhibitory action toward a variety of fungal strains, but none was observed.
Table 1. Antimicrobial Activity of Cyclothiazomycins B and C toward a Panel of Diverse Bacteria and Fungi.
| MICb |
||
|---|---|---|
| speciesa | cyclothiazomycin B | cyclothiazomycin C |
| Bacillus anthracis | 1 | 1 |
| Bacillus subtilis | 2 | 4 |
| Enterococcus faecalis | 32 | 32–64 |
| Listeria monocytogenes | 8 | 16 |
| Staphylococcus aureus | 4 | 16 |
| Escherichia coli | 64 | >64 |
| Neisseria sicca | >64 | >64 |
| Pseudomonas putida | >64 | >64 |
| Aspergillus niger | >64 | >64 |
| Fusarium virguliforme | 64 | >64 |
| Saccharomyces cerevisiae | 64 | >64 |
| Talaromyces stipitatus | 64 | >64 |
The top five species are Gram-positive bacteria from the Firmicutes phylum. The next three species are Gram-negative bacteria from the Proteobacteria phylum. The lowest 4 species are fungi from the Ascomycota phylum.
All minimum inhibitory concentrations (MIC) were determined by the microbroth dilution method and are presented in μg/mL.
To further clarify cyclothiazomycin bioactivity, we obtained a cyclothiazomycin B producer, strain NRRL B-3306, and purified cyclothiazomycin B in a manner analogous to that employed for cyclothiazomycin C (Supplemental Figures 11 and 12). As above, we assessed cyclothiazomycin B for antibiotic and antifungal activity. Cyclothiazomycin B also had the greatest inhibitory activity toward the genus Bacillus, with little to no activity against a panel of Gram-negatives and fungal strains (Table 1). This activity does not align with previous reports;40,49 however, additional fungal strains will need to be tested to more concretely establish cyclothiazomycin’s spectrum of activity. The antibiotic activity of cyclothiazomycin B and C is similar to that of known thiopeptides, which act as translation inhibitors by binding to either the 50S subunit or EF-Tu.50 It is possible that the cyclothiazomycins act in a similar manner, but the determination of the precise mode of action will require further exploration.
Conclusion and Outlook
In summary, we have described a reactivity-based screening method to conveniently identify natural products containing dehydrated amino acids (DHAAs). This method employs ubiquitous reagents and instrumentation, making it a broadly accessible strategy for natural product discovery. Three characteristics make the nucleophilic 1,4-addition labeling procedure operationally straightforward: (a) anhydrous solvents are unnecessary, meaning the reaction is performed under ambient atmosphere; (b) the reagents employed are common in most laboratories and easily handled; and (c) the large excess of labeling reagent relative to the substrate means that precise stoichiometric calculations for each reaction are unnecessary. Although under these excess labeling conditions we often observe minor peaks related to non-DHAA labeling, these species never convoluted spectral interpretation. Including a rapidly dereplicated example, we validated the use of nucleophilic 1,4-additions for natural product discovery with the labeling of three previously characterized natural products: thiostrepton, grisemycin, and geobacillin I. This reactivity-based screen was combined with bioinformatics and mass spectrometry to increase the rate of natural product discovery. Often, natural products are present only at trace quantities. By capitalizing on the remarkable sensitivity of mass spectrometry, the compound(s) to be discovered do not need to be present at bioactive concentrations, they only need to be detectable upon labeling. After screening the organic extracts of only 23 Actinobacteria, we report on a new thiopeptide, cyclothiazomycin C. The structure of cyclothiazomycin C was established through MS and NMR, along with confirmed bioactivity toward Gram-positive bacteria. When compared to traditional bioassay-guided isolation, which can require many thousands of samples to be screened to find new compounds, our discovery rate (1 out of 23 strains) highlights the potential of this tandem strategy. With the substantial rise of available genomic sequences, we anticipate that the combination of bioinformatics and simple chemoselective reactivity-based labeling will provide a powerful tool to identify novel natural products, while dramatically reducing the time invested on the unfruitful rediscovery of known compounds.
Methods
Preparation of Cell Extracts for Screening
Actinomycete strains were grown in 10 mL of MS medium (1 L contains 20 g mannitol, 20 g roasted soy flour) at 30 °C for 7 d. Exported metabolites were extracted from the cultures using 2 mL of n-BuOH at RT. For thiostrepton production, Streptomyces azureus was grown in 10 mL of ISP4 medium (1 L contains 10 g soluble starch, 1 g K2HPO4, 1 g MgSO4, 1 g NaCl, 2 g Na2SO4, 2 g CaCO3, 1 mg FeSO4, 1 mg ZnSO4 heptahydrate, 1 mg MnCl2 heptahydrate) for 7 d at 30 °C. Thiostrepton was extracted with 1 mL of CHCl3 at 23 °C. Both extracts were agitated for 1 min by vortex and submitted to centrifugation (4000 × g, 5 min), and the organic layer was removed from the intact, harvested cells. For geobacillin I production, Geobacillus sp. M10EXG was grown on modified LB agar (1 L contains 10 g casein enzymatic hydrolysate, 5 g yeast extract, 5 g NaCl, and 10 g agar) at 50 °C for 60 h. Cells were removed from the plates with 10 mL of 70% aq i-PrOH and agitated by rocking for 24 h at 23 °C. The intact cells were then removed from the extract by centrifugation (4000 × g, 5 min). An aliquot (1 μL) of the extract was then mixed with 9 μL of satd α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution in 1:1 MeCN/H2O containing 0.1% trifluoroacetic acid (TFA). One microliter was spotted onto a MALDI plate for subsequent MALDI-TOF MS analysis.
DTT-Labeling
For commercially obtained thiostrepton (Calbiochem, 99%), a 20 μL volume of 10.5 mM thiostrepton, 500 mM DTT, and 10 mM DIPEA in 1:1 CHCl3/MeOH was allowed to react at 23 °C for 16 h. For the no-base reaction, thiostrepton and DTT were added similarly to above, and MeOH (without DIPEA) was added to establish a 1:1 CHCl3/MeOH. The sample was then analyzed for DTT incorporation by MALDI-TOF MS (see below). For thiostrepton produced by Streptomyces azureus (and thus labeling occurred in the context of the crude cell-surface extract), 14 μL of the extract was mixed with DTT (in MeOH) and DIPEA (in MeOH) to generate a final volume of 20 μL with a final concentration of 500 mM DTT and 10 mM DIPEA, in 7:3 CHCl3/MeOH, and the mixture was allowed to proceed for 16 h at 23 °C. An aliquot (1 μL) of the extract was then mixed with 9 μL of satd α-cyano-4-hydroxycinnamic acid (CHCA) matrix solution in 1:1 MeCN/H2O containing 0.1% TFA. One microliter was spotted onto a MALDI plate for subsequent MALDI-TOF MS analysis.
Bioinformatics-Based Strain Prioritization
A previously reported profile Hidden Markov Model and the program HMMER were used to identify the YcaO cyclodehydratase (Pfam PF02624).51−53 The local genomic region (10 open reading frames on either side of the YcaO gene) was analyzed manually for the presence of a “lantibiotic dehydratase” gene and a putative precursor peptide. Only strains with the presence of all three genes were taken forward for reactivity-based screening.
Acknowledgments
We are grateful to W. van der Donk and D. Eastburn (University of Illinois at Urbana–Champaign, UIUC) for donating strains, in addition to M. Burke, S. Blanke, and P. Orlean (UIUC) for miscellaneous reagents. We thank L. Zhu for NMR assistance. This work was supported in part by a NIH Director’s New Innovator Award Program (DP2 OD008463 to D.A.M.), the David and Lucile Packard Fellowship for Science and Engineering (to D.A.M.), the Robert C. and Carolyn J. Springborn Endowment (to J.I.T.), and the American Society for Biochemistry and Molecular Biology Undergraduate Research Award (to K.S.). J.R.D. was supported by a fellowship from the Institute for Genomic Biology. The Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the National Center for Research Resources, National Institutes of Health (S10 RR027109 A).
Supporting Information Available
Additional experimental procedures, structural characterization details, and supporting figures as mentioned in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Newman D. J.; Cragg G. M. (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J. Nat. Prod. 75, 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. (2013) Platforms for antibiotic discovery. Nat. Rev. Drug Discovery 12, 371–387. [DOI] [PubMed] [Google Scholar]
- Fischbach M. A.; Walsh C. T. (2009) Antibiotics for emerging pathogens. Science 325, 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane C. D.; Mitchell D. A. (2014) Lessons learned from the transformation of natural product discovery to a genome-driven endeavor. J. Ind. Microbiol. Biotechnol. 41, 315–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velásquez J. E.; van der Donk W. A. (2011) Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 15, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J.; Ju K.-S.; Yu X.; Velásquez J. E.; Mukherjee S.; Lee J.; Zhao C.; Evans B. S.; Doroghazi J. R.; Metcalf W. W.; van der Donk W. A. (2014) Use of a phosphonate methyltransferase in the identification of the fosfazinomycin biosynthetic gene cluster. Angew. Chem., Int. Ed. 53, 1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odendaal A. Y.; Trader D. J.; Carlson E. E. (2011) Chemoselective enrichment for natural products discovery. Chem. Sci. 2, 760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D. D.; Wu C.-H.; Moree W. J.; Lamsa A.; Medema M. H.; Zhao X.; Gavilan R. G.; Aparicio M.; Atencio L.; Jackson C.; Ballesteros J.; Sanchez J.; Watrous J. D.; Phelan V. V.; van de Wiel C.; Kersten R. D.; Mehnaz S.; De Mot R.; Shank E. A.; Charusanti P.; Nagarajan H.; Duggan B. M.; Moore B. S.; Bandeira N.; Palsson B. Ø.; Pogliano K.; Gutiérrez M.; Dorrestein P. C. (2013) MS/MS networking guided analysis of molecule and gene cluster families. Proc. Natl. Acad. Sci. U.S.A. 110, E2611–E2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P.; Ma M.; Rateb M. E.; Shaaban K. A.; Yu Z.; Huang S. X.; Zhao L. X.; Zhu X.; Yan Y.; Peterson R. M.; Lohman J. R.; Yang D.; Yin M.; Rudolf J. D.; Jiang Y.; Duan Y.; Shen B. (2014) Biosynthetic potential-based strain prioritization for natural product discovery: a showcase for diterpenoid-producing actinomycetes. J. Nat. Prod. 77, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gersch M.; Kreuzer J.; Sieber S. A. (2012) Electrophilic natural products and their biological targets. Nat. Prod. Rep. 29, 659–682. [DOI] [PubMed] [Google Scholar]
- Bagley M. C.; Dale J. W.; Merritt E. A.; Xiong X. (2005) Thiopeptide antibiotics. Chem. Rev. 105, 685–714. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Yu Y.; Velasquez J. E.; van der Donk W. A. (2012) Evolution of lanthipeptide synthetases. Proc. Natl. Acad. Sci. U.S.A. 109, 18361–18366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.; Zhang Q.; van der Donk W. A. (2013) Insights into the evolution of lanthipeptide biosynthesis. Protein Sci. 22, 1478–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J.; Bibb M. (2010) Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc. Natl. Acad. Sci. U.S.A. 107, 16297–16302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesen J.; Bibb M. J. (2011) Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350. J. Bacteriol. 193, 2510–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiyama K.; Otoguro K.; Segawa T.; Shiomi K.; Yang H.; Takahashi Y.; Hayashi M.; Otani T.; Omura S. (1993) A new antibiotic, cypemycin. Taxonomy, fermentation, isolation and biological characteristics. J. Antibiot. (Tokyo) 46, 1666–1671. [DOI] [PubMed] [Google Scholar]
- Bonauer C.; Walenzyk T.; König B. (2006) α,β-Dehydroamino acids. Synthesis 2006, 1–20. [Google Scholar]
- Myers C. L.; Hang P. C.; Ng G.; Yuen J.; Honek J. F. (2010) Semi-synthetic analogues of thiostrepton delimit the critical nature of tail region modifications in the control of protein biosynthesis and antibacterial activity. Bioorg. Med. Chem. 18, 4231–4237. [DOI] [PubMed] [Google Scholar]
- Schoof S.; Baumann S.; Ellinger B.; Arndt H.-D. (2009) A fluorescent probe for the 70 S- ribosomal GTPase-associated center. ChemBioChem. 10, 242–245. [DOI] [PubMed] [Google Scholar]
- Schoof S.; Pradel G.; Aminake M. N.; Ellinger B.; Baumann S.; Potowski M.; Najajreh Y.; Kirschner M.; Arndt H.-D. (2010) Antiplasmodial thiostrepton derivatives: proteasome inhibitors with a dual mode of action. Angew. Chem., Int. Ed. 49, 3317–3321. [DOI] [PubMed] [Google Scholar]
- Wells L.; Vosseller K.; Cole R. N.; Cronshaw J. M.; Matunis M. J.; Hart G. W. (2002) Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post- translational modifications. Mol. Cell. Proteomics 1, 791–804. [DOI] [PubMed] [Google Scholar]
- Tseng H.-C.; Ovaa H.; Wei N. J. C.; Ploegh H.; Tsai L.-H. (2005) Phosphoproteomic analysis with a solid-phase capture-release-tag approach. Chem. Biol. 12, 769–777. [DOI] [PubMed] [Google Scholar]
- Li J.; Girard G.; Florea B. I.; Geurink P. P.; Li N.; van der Marel G. A.; Overhand M.; Overkleeft H. S.; van Wezel G. P. (2012) Identification and isolation of lantibiotics from culture: a bioorthogonal chemistry approach. Org. Biomol. Chem. 10, 8677–8683. [DOI] [PubMed] [Google Scholar]
- Challis G. L. (2008) Genome mining for novel natural product discovery. J. Med. Chem. 51, 2618–2628. [DOI] [PubMed] [Google Scholar]
- Bentley S. D.; Chater K. F.; Cerdeno-Tarraga A. M.; Challis G. L.; Thomson N. R.; James K. D.; Harris D. E.; Quail M. A.; Kieser H.; Harper D.; Bateman A.; Brown S.; Chandra G.; Chen C. W.; Collins M.; Cronin A.; Fraser A.; Goble A.; Hidalgo J.; Hornsby T.; Howarth S.; Huang C. H.; Kieser T.; Larke L.; Murphy L.; Oliver K.; O’Neil S.; Rabbinowitsch E.; Rajandream M. A.; Rutherford K.; Rutter S.; Seeger K.; Saunders D.; Sharp S.; Squares R.; Squares S.; Taylor K.; Warren T.; Wietzorrek A.; Woodward J.; Barrell B. G.; Parkhill J.; Hopwood D. A. (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417, 141–147. [DOI] [PubMed] [Google Scholar]
- Watve M. G.; Tickoo R.; Jog M. M.; Bhole B. D. (2001) How many antibiotics are produced by the genus Streptomyces?. Arch. Microbiol. 176, 386–390. [DOI] [PubMed] [Google Scholar]
- Scherlach K.; Hertweck C. (2009) Triggering cryptic natural product biosynthesis in microorganisms. Org. Biomol Chem. 7, 1753–1760. [DOI] [PubMed] [Google Scholar]
- Donovick R.; Pagano J. F.; Stout H. A.; Weinstein M. J. (1955) Thiostrepton, a new antibiotic. I. In vitro studies. Antibiot. Annu. 3, 554–559. [PubMed] [Google Scholar]
- Hensens O. D.; Albers-Schonberg G.; Anderson B. F. (1983) The solution conformation of the peptide antibiotic thiostrepton: a 1H NMR study. J. Antibiot. (Tokyo) 36, 799–813. [DOI] [PubMed] [Google Scholar]
- Ranu B. C.; Mandal T. (2007) Water-promoted highly selective anti-Markovnikov addition of thiols to unactivated alkenes. Synlett 925–928. [Google Scholar]
- Arnison P. G.; Bibb M. J.; Bierbaum G.; Bowers A. A.; Bugni T. S.; Bulaj G.; Camarero J. A.; Campopiano D. J.; Challis G. L.; Clardy J.; Cotter P. D.; Craik D. J.; Dawson M.; Dittmann E.; Donadio S.; Dorrestein P. C.; Entian K. D.; Fischbach M. A.; Garavelli J. S.; Goransson U.; Gruber C. W.; Haft D. H.; Hemscheidt T. K.; Hertweck C.; Hill C.; Horswill A. R.; Jaspars M.; Kelly W. L.; Klinman J. P.; Kuipers O. P.; Link A. J.; Liu W.; Marahiel M. A.; Mitchell D. A.; Moll G. N.; Moore B. S.; Muller R.; Nair S. K.; Nes I. F.; Norris G. E.; Olivera B. M.; Onaka H.; Patchett M. L.; Piel J.; Reaney M. J.; Rebuffat S.; Ross R. P.; Sahl H. G.; Schmidt E. W.; Selsted M. E.; Severinov K.; Shen B.; Sivonen K.; Smith L.; Stein T.; Süssmuth R. D.; Tagg J. R.; Tang G. L.; Truman A. W.; Vederas J. C.; Walsh C. T.; Walton J. D.; Wenzel S. C.; Willey J. M.; van der Donk W. A. (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N.; Tang W.; Goto Y.; Nair S. K.; van der Donk W. A. (2012) Lantibiotics from Geobacillus thermodenitrificans. Proc. Natl. Acad. Sci. U.S.A. 109, 5241–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N.; Oman T. J.; Andrew Wang T.-S.; De Gonzalo C. V.; Walker S.; van der Donk W. A. (2014) Mode of action and structure-activity relationship studies of geobacillin I. J. Antibiot. (Tokyo) 67, 133–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.; Qu X.; He X.; Duan L.; Wu G.; Bi D.; Deng Z.; Liu W.; Ou H.-Y. (2012) ThioFinder: a web-based tool for the identification of thiopeptide gene clusters in DNA sequences. PLoS One 7, e45878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Zhao H.; Liu J. (1982) Studies on the agricultural antibiotics 5102-II. Isolation and characterization of antibiotic 5102-II. Acta Microbiol. Sin. 22, 145–150. [Google Scholar]
- Wang J.; Yu Y.; Tang K.; Liu W.; He X.; Huang X.; Deng Z. (2010) Identification and analysis of the biosynthetic gene cluster encoding the thiopeptide antibiotic cyclothiazomycin in Streptomyces hygroscopicus 10–22. Appl. Environ. Microbiol. 76, 2335–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby J. O.; Nard N. J.; Mitchell D. A. (2011) Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 15, 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki M.; Ohtsuka T.; Yamada M.; Ohba Y.; Yoshizaki H.; Yasuno H.; Sano T.; Watanabe J.; Yokose K.; Seto H. (1991) Cyclothiazomycin, a novel polythiazole- containing peptide with renin inhibitory activity. Taxonomy, fermentation, isolation and physico-chemical characterization. J. Antibiot. (Tokyo) 44, 582–588. [DOI] [PubMed] [Google Scholar]
- Hashimoto M.; Murakami T.; Funahashi K.; Tokunaga T.; Nihei K.; Okuno T.; Kimura T.; Naoki H.; Himeno H. (2006) An RNA polymerase inhibitor, cyclothiazomycin B1, and its isomer. Bioorg. Med. Chem. 14, 8259–8270. [DOI] [PubMed] [Google Scholar]
- Aoki M.; Ohtsuka T.; Itezono Y.; Yokose K.; Furihata K.; Seto H. (1991) Structure of cyclothiazomycin, a unique polythiazole-containing peptide with renin inhibitory activity. Part 1. Chemistry and partial structures of cyclothiazomycin. Tetrahedron Lett. 32, 217–220. [Google Scholar]
- Aoki M.; Ohtsuka T.; Itezono Y.; Yokose K.; Furihata K.; Seto H. (1991) Structure of cyclothiazomycin, a unique polythiazole-containing peptide with renin inhibitory activity. Part 2. Total structure. Tetrahedron Lett. 32, 221–224. [Google Scholar]
- Dunbar K. L.; Mitchell D. A. (2013) Insights into the mechanism of peptide cyclodehydrations achieved through the chemoenzymatic generation of amide derivatives. J. Am. Chem. Soc. 135, 8692–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar K. L.; Melby J. O.; Mitchell D. A. (2012) YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat. Chem. Biol. 8, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melby J. O.; Li X.; Mitchell D. A. (2014) Orchestration of enzymatic processing by thiazole/oxazole-modified microcin dehydrogenases. Biochemistry 53, 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers A. A.; Walsh C. T.; Acker M. G. (2010) Genetic interception and structural characterization of thiopeptide cyclization precursors from Bacillus cereus. J. Am. Chem. Soc. 132, 12182–12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Horn P. B.; Backstrom A. D.; Stewart V.; Begley T. P. (1993) Structural genes for thiamine biosynthetic enzymes (thiCEFGH) in Escherichia coli K-12. J. Bacteriol. 175, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P.; Wu H.; Chen X. L.; Deng Z.; Bai L.; Pang X. (2014) Regulation of the biosynthesis of thiopeptide antibiotic cyclothiazomycin by the transcriptional regulator SHJG8833 in Streptomyces hygroscopicus 5008. Microbiology 10.1099/mic.0.076901-0. [DOI] [PubMed] [Google Scholar]
- Mizuhara N.; Kuroda M.; Ogita A.; Tanaka T.; Usuki Y.; Fujita K. (2011) Antifungal thiopeptide cyclothiazomycin B1 exhibits growth inhibition accompanying morphological changes via binding to fungal cell wall chitin. Bioorg. Med. Chem. 19, 5300–5310. [DOI] [PubMed] [Google Scholar]
- Just-Baringo X.; Albericio F.; Álvarez M. (2014) Thiopeptide antibiotics: retrospective and recent advances. Mar. Drugs 12, 317–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doroghazi J. R.; Metcalf W. W. (2013) Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics 14, 611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M.; Coggill P. C.; Eberhardt R. Y.; Mistry J.; Tate J.; Boursnell C.; Pang N.; Forslund K.; Ceric G.; Clements J.; Heger A.; Holm L.; Sonnhammer E. L.; Eddy S. R.; Bateman A.; Finn R. D. (2012) The Pfam protein families database. Nucleic Acids Res. 40, D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy S. R. (1998) Profile hidden Markov models. Bioinformatics 14, 755–763. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.