Abstract
Facioscapulohumeral dystrophy (FSHD) is caused by decreased epigenetic repression of the D4Z4 macrosatellite array and recent studies have shown that this results in the expression of low levels of the DUX4 mRNA in skeletal muscle. Several other mechanisms have been suggested for FSHD pathophysiology and it remains unknown whether DUX4 expression can account for most of the molecular changes seen in FSHD. Since DUX4 is a transcription factor, we used RNA-seq to measure gene expression in muscle cells transduced with DUX4, and in muscle cells and biopsies from control and FSHD individuals. We show that DUX4 target gene expression is the major molecular signature in FSHD muscle together with a gene expression signature consistent with an immune cell infiltration. In addition, one unaffected individual without a known FSHD-causing mutation showed the expression of DUX4 target genes. This individual has a sibling with FSHD and also without a known FSHD-causing mutation, suggesting the presence of an unidentified modifier locus for DUX4 expression and FSHD. These findings demonstrate that the expression of DUX4 accounts for the majority of the gene expression changes in FSHD skeletal muscle together with an immune cell infiltration.
INTRODUCTION
Facioscapulohumeral dystrophy (FSHD) is a human muscular dystrophy that initially affects the muscles of the face and upper extremities, but can progress to affect most skeletal muscles (1). The most common genetic cause of FSHD (FSHD1) is the deletion of a subset of D4Z4 macrosatellite repeats in the subtelomeric region of chromosome 4; whereas the less common form of FSHD (FSHD2) is caused, in the majority of cases, by a mutation in the SMCHD1 gene on chromosome 18 (2,3). The mutations for FSHD1 and FSHD2 both result in decreased epigenetic repression of the D4Z4 repeat in somatic tissue and mis-expression of a retrogene, DUX4, contained within each D4Z4 repeat on chromosome 4, as well as within nearly identical D4Z4 repeats in the subtelomeric region of chromosome 10 (4). Genetics strongly implicates the expression of DUX4 as necessary for FSHD because decreased D4Z4 epigenetic repression, either due to contraction of the repeats or to a mutation in SMCHD1, results in FSHD only in individuals with a specific FSHD-permissive haplotype that contains a poly-adenylation site for the DUX4 mRNA in the region adjacent to the D4Z4 repeat (3).
Despite the overwhelming genetic evidence that DUX4 mRNA expression is necessary for FSHD, its primary role has been questioned because of the extremely low abundance of the DUX4 mRNA in affected FSHD muscle cells and biopsies. However, the low mRNA abundance represents a variegated expression pattern with relatively high expression of DUX4 in a small number of nuclei at a single time point, possibly being expressed in short bursts in different muscle nuclei over time (5).
DUX4 is normally expressed in the male testis. Antibody detection and in situ hybridization indicate that cells in the seminiferous tubules are expressing DUX4, most likely the spermatogonia and primary spermatocytes, although additional studies using dual detection of lineage markers remain to be performed (5).
DUX4 is a double-homeobox transcription factor. When mis-expressed in primary human muscle cells in culture, DUX4 binds to a double-homeodomain motif and activates the expression of a broad set of genes, many involved in stem and germ cell biology (6). Some of the DUX4 targets have been previously identified as Cancer Testis Antigens, genes whose expression is normally restricted to the immune-privileged germline that induce an immune response when mis-expressed in cancer cells. Initial studies showed that a selected set of six DUX4 targets were detected by RT–PCR in FSHD muscle cultures and biopsies, but not in control muscle, demonstrating that the low levels of DUX4 mRNA expression in FSHD muscle was sufficient to activate its downstream program of gene expression (6).
In contrast, expression array studies of skeletal muscle biopsies did not identify DUX4 target genes as specifically mis-expressed in FSHD (7). However, because of the low and variegated expression of DUX4 in FSHD muscle, it is perhaps understandable that the targets of DUX4, also expressed at low levels, might not be easily detected in discovery-oriented studies using expression arrays. To determine whether a broader set of DUX4-regulated genes could be identified as mis-expressed in FSHD muscle, and whether other gene signatures that were not related to DUX4 could be identified in FSHD, we performed RNA-seq on both cultured muscle and muscle biopsies from control, FSHD1 and FSHD2 individuals. Our results identify DUX4-regulated genes as the major difference between control and FSHD, both muscle cells and biopsies, and that many of the genes not regulated by DUX4 that are associated with FSHD biopsies appear to be related to immune cell infiltration. These results suggest that DUX4 is the major determinant of abnormal gene expression in FSHD, together with an immune cell infiltrate. In addition, similar to other studies, we found that some individuals in FSHD families can express DUX4 and DUX4 target genes without the clinical manifestation of FSHD.
RESULTS
DUX4 robustly induces a core set of genes in skeletal muscle cells
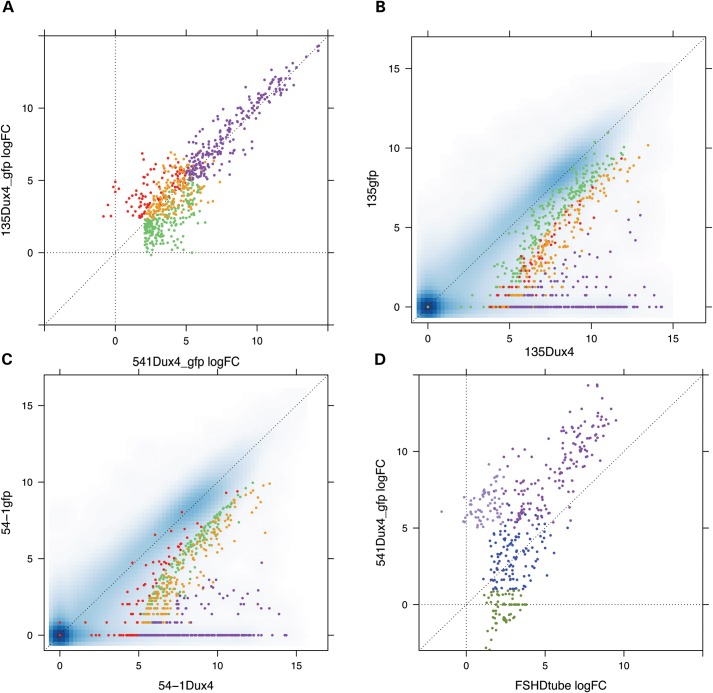
To identify genes regulated by DUX4 in skeletal muscle cells, we transduced a muscle biopsy-derived primary myoblast cell culture (MB135) from a control individual and an immortalized myoblast cell line (MB541) (6,8), both with a D4Z4 repeat in the unaffected range, with a lentiviral vector expressing DUX4 or GFP as a control. These control muscle cells do not express endogenous DUX4 and the lenti-viral transduction achieved DUX4 expression in most of the cells at a level roughly similar to that detected in the rare DUX4-expressing cell in FSHD cultures, based on immunodetection. Because the expression of DUX4 induces apoptosis after 48 h, RNA was harvested at 24 h (MB135) and 48 h (MB541). RNA-seq identified 507 DUX4-up-regulated genes in MB135 and 643 in MB541 with 416 genes shared by the two cell types using stringent statistical criteria (FDR <0.05, moderated log-fold-change >1 [approximate linear fold change >2.71 see Materials and Methods)]. The fold-change of DUX4-regulated genes in the two cell types was highly concordant (Fig. 1A), including most genes that did not meet the statistical threshold in both populations, indicating that up-regulated genes specific to an individual cell type are mostly marginal cases near the cut-off for statistical significance rather than cell-type-specific targets of DUX4. The most robust DUX4 targets, defined as the 213 up-regulated genes with a moderated log-fold-change >5 (linear fold-change greater than 75) (Fig. 1A, purple data points and Supplementary Material, Table S1), were generally not expressed in the absence of DUX4 and were robustly induced by DUX4 in both muscle cell cultures (Fig. 1B and C).
Figure 1.
Dux4 induced genes mis-expressed in FSHD muscle cells. (A) log-fold change of DUX4 up-regulated genes in MB135 (y-axis) and MB541 (x-axis) cultured myoblast cells transduced with lentiviral DUX4 compared with a control lenti-GFP (purple, genes upregulated by DUX4 > 5 log-fold change in both MB135 and MB541; orange, up-regulated by DUX4 in both cell types but <5 log-fold; red, DUX4 up-regulated only in MB135 cells; green, DUX4 up-regulated only in MB541cells). (B and C) Pairwise comparison of gene expression in DUX4-transduced (x-axes) and the control GFP-transduced (y-axes) MB541 (B) and MB135(C) cells. Gene counts were transformed as described in Materials and Methods. Genes are colored as in (A). (D) Comparison of the log-fold change of genes up-regulated by DUX4 in MB541 cells (y-axis) to the log-fold change of genes up-regulated in FSHD myotubes compared to control myotubes (x-axis) (dark purple, genes increased in FSHD compared with control cells that were up-regulated by DUX4 >5 log-fold in MB541; blue, increased in FSHD cells and up-regulated by DUX4 in MB51 with log-FC between one and five; olive, increased in FSHD cells and not called increased by DUX4 in MB541 (log-FC <1); light purple, not increased in FSHD cells compared with controls but up-regulated by DUX4 in MB541 cells).
In contrast to DUX4 up-regulated genes, there were very few genes down-regulated by DUX4. Compared with the lenti-GFP control, 28 down-regulated genes were common to both cell types. However, only one gene (CSF3) was also down-regulated relative to a no-virus control. Most of remaining 27 genes were involved in the innate immune response, consistent with our prior demonstration that DUX4 represses the innate immune response induced by lenti-viral transduction (6,9), a process partly mediated by up-regulation of DEFB103, a defensin peptide previously shown to block the innate immune response (9).
DUX4-regulated genes are mis-expressed in FSHD muscle cells
To determine whether DUX4 targets, or other genes, are mis-expressed in FSHD muscle cells, we compared gene expression in primary muscle cultures from five FSHD individuals (three FSHD2 and two FSHD1) and three unaffected control individuals. In undifferentiated myoblasts, 90 genes showed increased expression in FSHD cells compared with controls and over one-half of these (51 genes) were among the subset of 213 robust DUX4-regulated genes identified in the transduction experiments. In differentiated myotube cultures, 348 genes were differentially up-regulated in FSHD cells compared with controls, 158 of which were among the 213 most robust DUX4-regulated genes (Fig. 1D, purple dots) and an additional 118 were induced 2.71-fold or more by DUX4 transduction in myoblasts but did not meet the more rigorous criteria for a DUX4 target (Fig. 1D, blue dots); whereas only 72 (∼20%) differentially expressed genes were not identified as regulated by DUX4 (Fig. 1D, olive dots and Supplementary Material, Table S2), and only 18 of these met the statistical threshold in both FSHD myoblasts and myotubes. Therefore, the majority of gene expression changes in cultured FSHD muscle cells can be attributed to the expression of DUX4 and genes regulated by DUX4. The increase in the number of DUX4 target genes detected in differentiated muscle compared with myoblasts likely represents the increase in DUX4 mRNA and protein that occurs during FSHD muscle differentiation (8,10), which was also reflected in increased RNA-seq reads for DUX4 in differentiated FSHD muscle cells (data not shown).
DUX4 target genes are expressed in FSHD biopsy samples
To determine whether DUX4-regulated genes distinguish control from FSHD muscle biopsies, we performed RNA-Seq on 24 quadriceps needle biopsy samples (nine controls, nine FSHD1 and six FSHD2) (see Materials and Methods and Table 1 for the complete sample list and associated clinical data). Only 38 genes were consistently up-regulated across all FSHD samples compared with all of the control samples [P-value <0.05, moderated log-fold-change >1 and Wilcoxon-rank sum P-value <0.05 (see Materials and Methods)], and 28 of these (74%) were among the robust 213 DUX4-regulated genes identified in the transduction experiments, indicating that the most of the FSHD-specific gene expression in muscle biopsies was determined by DUX4.
Table 1.
Sample list and associated clinical information
| NMD # | Gender | Age | Muscle Bx'd | CSSb | Corr.CSSe | Path.Scorec | D4Z4: Kba | NMD# | Gender | Age | Muscle Bx'd |
|---|---|---|---|---|---|---|---|---|---|---|---|
| FSHD1 | Control | ||||||||||
| F1 | M | 47 | L QUAD | 0/10 | 0 | 2 | 26 | C1 | M | 37 | L QUAD |
| F2 | F | 45 | L QUAD | 5/10 | 111 | NHSd | 18 | C2 | M | 44 | L QUAD |
| F3 | F | 55 | R QUAD | 6/10 | 109 | 4 | 15 | C3 | M | 62 | L QUAD |
| F4 | M | 33 | R QUAD | 6/10 | 122 | 5 | 24 | C4 | M | 48 | L QUAD |
| F5 | M | 52 | R QUAD | 6/10 | 151 | NHS | 22 | C5 | M | 58 | L QUAD |
| F6 | F | 30 | L QUAD | 3/10 | 100 | 3 | 23 | C6 | F | 69 | R ANT TIB |
| F7 | F | 61 | R QUAD | 6/10 | 99 | 6 | 16 | C7 | F | 41 | L QUAD |
| F8 | F | 26 | R QUAD | 7/10 | 269 | 5 | 15 | C8 | F | 53 | L QUAD |
| F9 | M | 48 | L QUAD | 3/10 | 63 | 4 | 17 | C9 | F | 32 | R QUAD |
| NMD # | Gender | Age | Muscle Bx'd | CSS | Corr.CSSe | Path.Score | SMCHD1 mutation | FseI %f | |||
| Blood | Blasts | ||||||||||
| FSHD2 | |||||||||||
| F10 | M | 42 | R QUAD | 5/10 | 119 | 2 | YES | 12 | ** | ||
| F11 | M | 34 | L QUAD | 6/10 | 176 | 3 | YES | 16 | 2 | ||
| F12 | M | 26 | R QUAD | 6/10 | 231 | 4 | YES | 7 | 3 | ||
| F13 | M | 50 | L QUAD | 5/10 | 100 | 5 | NO | 13 | ** | ||
| F14 | F | 56 | R QUAD | 7/10 | 125 | 3 | YES | 25 | 7 | ||
| F15 | M | 59 | L QUAD | 2/10 | 34 | 2 | NO | 15 | 0 | ||
aAll refer to the size of the FSHD-permissive 4qA161 allele.
bCSS: clinical severity score: 0 asymptomatic-10 wheelchairbound.
cPath score: 0 = no pathology – 12 = severe pathology (none had inflammation).
dNHS, no histologic sample collected.
eAge corrected CSS = (CSS/age at examination) × 1000.
fFseI %: percentage CpG methylaion of D4Z4 as measured by the FseI restriction endonuclease.
**Blast FseI same as blood.
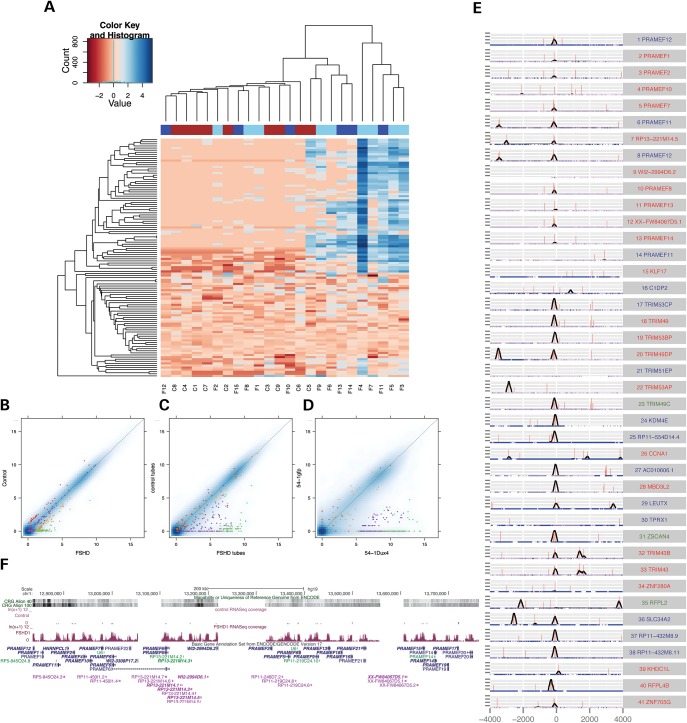
We then examined the expression of the DUX4-regulated genes across all biopsy samples (Fig. 2A), using 114 of the 213 DUX4-regulated genes that met a threshold of average expression across all samples (transformed expression level >0.5, see Materials and Methods), i.e. excluding the genes identified in the transduction experiments that were not detected as expressed in the biopsy samples. Nine of the fifteen FSHD samples had elevated expression levels of DUX4 targets compared with controls, whereas the other six FSHD biopsy samples had comparable expression of DUX4 targets as the controls. We also noted that control sample C5 had low but detectable expression of DUX4 targets and this control clustered with the FSHD biopsy samples that also expressed DUX4 targets.
Figure 2.
Expression of Dux4 targets in FSHD biopsy samples. (A) Heatmap of the expression of the most robust 114 DUX4 targets across all control and FSHD skeletal muscle biopsy samples. The 114 genes were selected as the subset of 213 robust DUX4-regulated genes that met a threshold of average expression across all of the samples (transformed expression level >0.5, see Materials and Methods). The expression levels were transformed and centered by subtracting the median for each gene and the color coding indicates the log-fold-change relative to the median. The columns are colored coded as: red, control; light blue, FSHD1; dark blue, FSHD2. (B–D) Pairwise comparisons of gene expression levels in (B) FSHD and control biopsy samples, (C) FSHD and control cultured myotubes and (D) MB541 transduced with DUX4 or GFP as a control. Colors indicate: green, DUX4 targets called up-regulated in FSHD biopsy samples; purple, DUX4 targets not called up-regulated in FSHD biopsy samples; orange, up-regulated in FSHD biopsies but not DUX4 targets; red, up-regulated in control biopsy samples. (E) DUX4-binding sites within ± 4 kb of TSSs of a set of candidate biomarker genes. The red vertical bar corresponds to the presence of a consensus motif for the DUX4 binding site, and height of the bar indicate the strength of the motif based on the PWM score. The black line indicates the DUX4 ChIP-Seq coverage in DUX4-transduced muscle cells. The blue gradient along the x-axis indicates the sequence uniqueness based on the ENCODE CRG Align 40 track. Gene names colored in red have annotated starting exons, those colored in green have internal exons activated by DUX4 as starting exons, and those colored in blue have novel starting exons activated by DUX4 in intergenic or intronic regions. The zero position on the y-axis represents the predicted TSS based on the RNA-seq. (F) The PRAMEF locus. The gene names in bold type correspond to members of the 67 candidate biomarker genes expressed in FSHD biopsies. The uniqueness tracks and RNA-seq tracks for one FSHD and one control myotube sample are also shown.
The segregation of FSHD samples into DUX4-target-positive and DUX4-target-negative samples likely reflects the biopsy of a muscle that is affected late in FSHD (the quadriceps) and a limited amount of sampled tissue with a small needle biopsy (see Discussion). The control sample C5 is a member of a complex FSHD family (Supplementary Material, Fig. S1). He is a clinically unaffected individual with 13 D4Z4 repeats on an FSHD-permissive haplotype, which is more than the standard threshold of 10 repeats for FSHD and would not be expected to express DUX4. His FSHD2-affected sibling (F15, also included in this study) has the same 13 repeat FSHD-permissive allele and shows the typical CpG hypomethylation at D4Z4 but without an identified pathological mutation in SMCHD1, suggesting that an as yet unknown modifier locus is segregating in this family and might be present in C5 (see Discussion).
To determine the most robust FSHD-DUX4 candidate biomarker genes in the biopsy samples, we removed C5 from subsequent analyses based on the assumption that C5 might represent a DUX4-expressing unaffected individual in a family with an unknown FSHD-causing mutation, and restricted our comparison with the remaining control samples. Of the 118 genes expressed significantly higher in DUX4-target-positive FSHD-versus-control samples, 67 were among the tissue-culture identified 213 robust DUX4-regulated genes identified in the cell transduction experiments (Supplementary Material, Table S3) and an additional 13 were boundary cases of DUX4-regulated genes that did not make the strict cut-off for the robust DUX4 targets. Therefore ∼69% of the genes associated with FSHD are regulated by DUX4. The 67 robust DUX4 target genes were among the most up-regulated genes in DUX4-target-positive FSHD biopsy samples, FSHD myotubes and DUX4-transduced cells (Fig. 2B–D, green dots) relative to their respective controls, and the expression of all these genes increased during myogenesis in cultured FSHD muscle cells but not in control cells (Supplementary Material, Fig. S2A and B). Most of these genes were not expressed in the corresponding control samples, making them good candidates for FSHD biomarkers.
We also compared gene expression between the DUX4-target-negative FSHD samples and the controls (excluding sample C5) and did not find any differentially expressed genes using similar statistical thresholds. Therefore, the DUX4-target-positive FSHD samples show gene expression differences compared with the controls, whereas the DUX4-target-negative FSHD samples have similar gene expression to controls, again indicating that the DUX4 target genes are the main discriminator between FSHD and control samples.
DUX4 target genes and candidate biomarkers
To determine whether these candidate FSHD biomarkers might be direct targets of DUX4, we examined the presence of DUX4-binding sites adjacent to the transcription start site as determined by the analysis of the RNA-seq from the DUX4-transduced myoblasts. Many of the biomarker candidate genes belong to highly related gene families and repetitive regions and their TSSs are not well annotated. In addition, we observed novel (unannotated based on GENCODE v19) TSSs at some candidate biomarker loci. Therefore, to identify the functional TSS for the analysis of DUX4 binding at DUX4-regulated genes, we developed a method to predict the TSS based on RNA-seq reads (see Materials and Methods for details) and screened the 67 candidate biomarker genes with this method to identify the functional TSS using a very stringent cut-off. We identified 41 TSSs for the 67 biomarker genes (Fig. 2E and Supplementary Material, Table S4), which included 22 annotated and 19 novel, TSSs, including three TSSs initiating from annotated middle exons (TRIM49C, ZSCAN4, and RFPL2). For the analysis of DUX4 ChIP-Seq data, we included reads that mapped to multiple locations to identify DUX4 binding sites within the repetitive regions. Out of the 41 TSSs, 32 had at least one DUX4 ChIP-seq peak within ± 500 bp window, most of which clustered within 200 bp upstream of TSS. All except one of the nearest peaks for these 32 sites contained at least one DUX4-binding-site motif, and 21 contained at least two motifs. Therefore, ∼75% of these candidate biomarkers have DUX4 binding near their TSS and are likely to be directly regulated by DUX4. Some TSSs from repetitive gene families, such as PRAMEF10, TRIM51EP, did not have DUX4 ChIP-seq peaks near their TSS and it is possible that the RNA-Seq reads mapped to these genes are from other homologous regions. Other genes, such as TPRX1, ZNF280A and CCNA1 are likely to be regulated by distal enhancers, or indirectly by DUX4.
It was striking that most biomarker candidates are members of highly homologous gene families that are clustered spatially on the chromosomes, including PRAMEF (preferentially expressed in melanoma), TRIM (tripartite motif-containing), MBDL (methyl-CpG binding protein-like), ZSCAN (zinc finger and SCAN domain containing) and RFPL (ret-finger protein-like) families (Supplementary Material, Fig. S3). The most dramatic example is the PRAMEF locus (Fig. 2F), which spans an approximately one megabase region and includes 28 candidate biomarker candidates grouped into several different RNA-Seq clusters.
From this pool of candidate biomarkers, we identified four genes as a core set of candidate biomarkers (the genome structure and RNA-seq reads for these genes are shown in Supplementary Material, Fig. S4). The candidate with the strongest statistical support is LEUTX, (leucine 20 homeobox). DUX4 activates a novel TSS of LEUTX that is spliced into the second annotated exon that contains the beginning of the open reading frame. In the PRAMEF locus, PRAMEF2 has the strongest statistical support with almost no expression in the control samples (except C5). TRIM43 is the strongest candidate among TRIM family members. KHDC1L, an isoform of KHDC1, and a nearby but non-overlapping pseudo gene KHDC1P1, are also good candidate biomarkers with DUX4 binding sites within their promoters, whereas the overlapping KHDC1 is not discriminative (see Supplementary Material, Table S4 for first exon sequences).
Association of DUX4-target gene expression with clinical severity
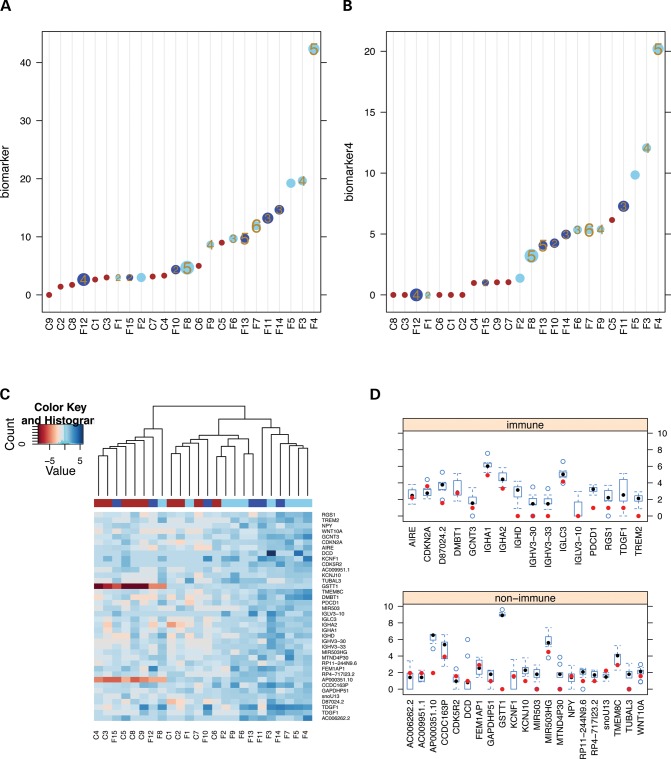
To associate expression of the candidate biomarkers with clinical data, we took the total number of reads that mapped to any of the 67 candidate biomarker genes in each sample (scaled and transformed by the square-root) and plotted this measure of biomarker gene expression with representation of the pathology score and the corrected clinical severity score (CSS) (11–13). Individuals with high pathology and CSS scores generally had higher biomarker expression (Fig. 3A). The discrimination between FSHD and control was slightly improved using only reads over the four selected biomarkers (LEUTX, PRAMEF2, TRIM43 and KHDC1L) (Fig. 3B).
Figure 3.
(A and B) Association of DUX4-target gene expression with clinical severity. (A) The y-axis is the total number of reads that mapped to any of the 67 candidate biomarker genes in each sample (scaled and square-root transformed). The biopsy samples are color coded as: red, control; light blue, FSHD1; dark blue, FSHD2. The size of the circle corresponds to the corrected clinical severity score (CSS) and the numbers within the circles correspond to the pathology score, with larger size indicating a higher severity. The samples with missing numbers did not have pathology scores. (B) Similar to A but using the total reads mapped to the selected four biomarkers: LEUTX, PRAMEF2, TRIM43 and KHDC1L. (C and D) Genes expressed at higher levels in FSHD samples that are not DUX4 targets. (C) Heatmap of genes expressed more highly in FSHD samples but that are not induced (logFC <1) by DUX4-transduction of human muscle cells with presentation and coding as described in Figure 2A. (D) Boxplot of the expression distribution for the genes in (C) based on RNA-seq reads from the FSHD samples that were DUX4-target-positive, stratified as immune and non-immune genes. The red dots correspond to the expression of each gene in control sample C5. Note that some loci have copy number variation in the human population, such as GSTT1 that is not present in some individuals. Boxplot: vertical bounds of rectangle represent the 25th and 75th percentiles, the whisker extends to the extreme value (minimum or maximum) bounded by 1.5 times the IQR (25th and 75th inter-quartile range), and the filled circles are the median and the open circles are the outliers.
We assessed whether the discrimination of FSHD samples from controls might be improved by the addition of additional biomarker candidate genes to the four selected candidate biomarkers (see Supplementary Material, Fig. S5). None of the other candidate biomarker genes improved the discrimination of FSHD and control samples and the four FSHD samples with very low expression of DUX4 targets (F1, F12, F15, F2) remained boundary cases that were not clearly separable from the control samples.
Immune genes expressed in FSHD biopsies
Although 80 of the 118 genes associated with the DUX4-target-positive FSHD samples were DUX4-regulated genes (67 robust and 13 boundary cases), 38 genes were not identified as DUX4 regulated. The expression of these 38 genes across all samples was plotted in a clustered heat map (Fig. 3C). As anticipated, the DUX4-target-positive FSHD samples cluster together, however, the control C5 sample that had DUX4 target expression does not show a higher elevation of the 38 non-DUX4-target genes and no longer clusters with the FSHD samples. Sixteen of these FSHD-associated non-DUX4-target genes are expressed in cells of the immune system (based on Gene Ontology annotation of ‘immune system process’ and HUGO definitions of immunoglobulins gene family), including members of immunoglobulin clusters IGHA, IGHD, IGLC and IGLV, and plotting their distribution in DUX4-target-positive FSHD biopsies showed that the C5 sample from the asymptomatic family member had lower expression of immune-associated genes compared with the FSHD-affected samples (Fig. 3D). Therefore, a major component of the non-DUX4-target gene changes likely represents the presence of immune cells in FSHD muscle biopsies, and the asymptomatic control individual C5 that expresses some DUX4 target genes does not demonstrate expression changes indicating an immune infiltration. In addition, the FSHD biopsy samples with absent or low expression of DUX4 target genes also showed lower expression of the sixteen immune-associated genes (Supplementary Material, Fig. S6), indicating that the immune genes were mostly associated with muscle biopsies expressing DUX4 target genes.
FSHD1 and FSHD2 have similar gene expression profiles
Comparing gene expression between the FSHD1 and FSHD2 biopsy samples, we detected only three genes expressed more highly in FSHD1 than FSHD2, and six genes expressed more highly in FSHD2 than FSHD1 (Supplementary Material, Table S5); however, small sample sizes limited statistical power. Interestingly, five of the six FSHD2-specific genes were members of homologous gene clusters in repeat arrays. PCDHB2 is a member of the protocadherin cluster and several other members (PCDHA2, PCDHA3 and PCDHA8) were also preferentially expressed in FSHD2 patients, although they did not meet the statistical stringency. TP53TG3C is a member of TP53TG3 gene family that has two adjacent copies and two more copies within 600 kb. Similarly, RP11–3N2.13 and RP11-3N2.1 are adjacent and homologous non-coding genes, while RP11-760D2.5 is nearby.
We also compared FSHD1 and FSHD2 differentiated muscle cells in tissue culture, although this analysis had even less power because of the sample size. The largest category of differentially expressed genes were genes that are increased during control muscle differentiation, suggesting that the FSHD2 cultures had better muscle differentiation compared to FSHD1 cultures (data not shown). We then examined if the genes differentially expressed between FSHD1 and FSHD2 biopsy samples showed similar changes between cultured FSHD1 and FSHD2 myotubes. FSHD1 biopsy-specific genes did not show higher expression in FSHD1 cultured myotubes, while FSHD2 biopsy-specific genes tended to be expressed at higher levels in cultured FSHD2 myotubes (Supplementary Material, Table S5), suggesting that a set of genes enriched in gene clusters or repetitive regions might be up-regulated in FSHD2 but not in FSHD1.
DISCUSSION
Our results indicate that the majority of gene expression changes associated with FSHD, either in cultured muscle cells or in muscle biopsy material, can be attributed to DUX4 expression. Of the 118 genes identified as significantly up-regulated in FSHD biopsy samples, 80 (68%) were regulated by DUX4. An additional sixteen of the 118 genes were consistent with an immune infiltration in the FSHD muscle biopsies. Taken together, over 80% of the genes dysregulated in FSHD muscle biopsies could be attributed to DUX4 targets and an immune infiltration. Therefore, DUX4-regulated genes and an associated immune cell infiltration accounts for the majority of the gene expression changes associated with FSHD muscle in this study. If there are other genes critical for FSHD that act independently of DUX4, such as has been suggested for FRG1, ANT1, FAT1 and others (14–16), these genes have a relatively minor transcriptional impact in FSHD skeletal muscle.
Although we have reliably detected DUX4 expression and DUX4-target expression in muscle cultures derived from FSHD quadriceps muscle biopsies, and other reports have reliably demonstrated DUX4 expression in muscle cultures derived from FSHD deltoid and biceps muscles (17), six of the fifteen FSHD muscle biopsies did not show significantly elevated expression of the set of the 213 most robust DUX4-regulated genes, and four did not show a higher expression of the selected set of four DUX4 candidate biomarkers (see Fig. 3B). Based on our current understanding of FSHD pathophysiology, there are two reasonable explanations for the lack of a perfect association between DUX4 target gene expression and FSHD. First, a hallmark of FSHD is the asymmetric and regional progression of the disease and recent MRI studies have documented disease-associated change restricted to specific muscle groups (18–20). The quadriceps muscle that was biopsied for this study is usually affected later in the disease and it is possible that this muscle was not yet affected in the FSHD individuals without DUX4 target gene expression. Future studies using MRI characteristics of muscle involvement in FSHD should determine whether the expression of DUX4 and DUX4-target gene expression is correlated with MRI documentation of disease activity. Second, the needle biopsy samples a very small region of the muscle. Although very little is known regarding DUX4 expression in skeletal muscle of FSHD individuals, the tissue culture studies demonstrate a variegated expression pattern with very few nuclei expressing DUX4 (5,17). Therefore, sampling a small amount of tissue might also result in a set of samples without DUX4 or DUX4 target gene expression. Supporting this interpretation, cultured muscle cells derived from the F12 muscle biopsy showed a strong expression of DUX4 target genes (see Supplementary Material, Fig. S2A), whereas the RNA-seq directly on the F12 biopsy sample did not identify the expression of DUX4-target genes (see Fig. 1A). Whatever the reason for some of the FSHD biopsies not showing the expression of DUX4 target genes, it is important to note that they did not show any significant gene expression differences compared with the control biopsies. Therefore, there was little evidence of a non-DUX4 process in these samples.
A prior study reported the expression of the full-length DUX4 mRNA in muscle biopsies from a small number of genetically unaffected relatives of FSHD individuals (17), although generally at lower levels compared with the FSHD individuals. The subject C5 in this study appears to be a similar finding. This individual has 13 D4Z4 repeats on an FSHD-permissive allele, which is more than the 10 repeats generally considered the upper limit for FSHD1. However, the family shows evidence for the independent segregation of a modifier locus, since one brother has FSHD1 with six D4Z4 repeats on an FSHD-permissive allele and one brother has FSHD2 with 13 repeats and low levels of D4Z4 methylation but without a pathological mutation in SMCHD1 identified by exome sequencing and the father also showed hypomethylation but without an FSHD-permissive allele and without FSHD (2) (see Supplementary Material, Fig. S1). Individual C5 has higher D4Z4 methylation than the brother that is mildly affected by FSHD2 and both carry the same 13 repeat FSHD-permissive allele. Because the mutation causing FSHD2 has not been identified in this family, it is possible that the same modifier locus is present in both brothers and slightly more penetrant in the affected individual, or that more than one modifier locus might be segregating in this family, each with a partial reduction in D4Z4 repression.
It is also interesting that C5 showed DUX4-target gene expression in the skeletal muscle biopsy without FSHD symptomology, further indicating that the expression of DUX4 and its downstream gene network are not sufficient for disease. This could simply be due to lower levels of expression that do not reach a threshold for the pathophysiological mechanisms, or represent an unknown biological or stochastic protection. The relative absence of immune cell gene expression in the biopsy from this individual is intriguing in this regard, as it would be consistent with an immune-mediated mechanism of disease. However, it could also represent the absence of a secondary immune response in the absence of muscle disease from another mechanism and further studies remain necessary.
The DUX4 target genes expressed in FSHD muscle cells and biopsies were the genes most highly induced by DUX4-transduced cultured muscle cells. We have previously shown that the extremely low levels of the DUX4 mRNA in FSHD muscle cultures represents relatively high expression in a small number of cells at any time point (5), suggesting that a subset of cells loses repression of DUX4 and that results in a burst of DUX4 expression in a limited subpopulation of nuclei. The observation that only the most highly expressed DUX4 genes are reliably detected across FSHD muscle biopsies is consistent with a ‘tip-of-the-iceberg’ problem because the more moderately expressed targets are obscured by the background noise from the larger number of non-expressing nuclei. We identified a core set of four DUX4-regulated genes based on gene-family representation, and the fold and significance of induction. These four candidate biomarkers (LEUTX, PRAMEF2, TRIM43 and KHDC1L) show slightly improved discrimination between control and FSHD biopsies (see Fig. 3) and it will be interesting to test them in a larger clinical series.
It is interesting that previous expression array studies comparing FSHD to control muscle biopsies did not identify the same discrete group of DUX4-regulated genes (7,21–23). For example, none of the 15 candidate biomarker genes identified in the most extensive and recent study of FSHD muscle biopsies is one of the robust DUX4 target genes (7). One of the reasons for this might be that the DUX4 target genes are not well represented on expression arrays. For example, LEUTX is a primate-specific homeobox gene that has not been studied or included in most arrays and KHDC1L is a transcript embedded in the KHDC1 gene, which itself is not a good biomarker for FSHD. Others are members of larger highly related gene families (see Supplementary Material, Fig. S3) that might obscure gene-specific signals through cross-hybridization of similar sequences to the tiled probes on an array. Finally, the low levels of expression and the fact that not all biopsied FSHD muscles showed DUX4 target gene expression decreases the power to identify these genes in a purely discovery-based approach using array technology. Nonetheless, a set of FSHD muscle biopsies cluster together based on the expression of the DUX4 target genes identified in tissue culture (see Fig. 2A), whereas the FSHD biopsy samples in our study do not cluster together based on the expression level of the 15 candidate biomarkers for the prior array-based study (7) (Supplementary Material, Fig. S7). Although future validation studies are necessary, these results suggest that these DUX4 target genes will be useful as biomarkers for FSHD.
The comparison of FSHD1 and FSHD2 samples did not show any reliable FSHD1-specific changes, but did show a small number of genes specific to FSHD2. These tended to be genes that were in multiple copies or clustered in highly related families, such as the protocadherin clusters (24,25). Four of the six FSHD2 individuals biopsied for this study have a mutation in SMCHD1 (see Table 1), and SMCHD1 has a role in repeat-mediated epigenetic repression as well as in the regulation of protocadherin clusters. Therefore, the dysregulation of this set of genes in FSHD2 samples is consistent with loss of SMCHD1 activity.
In summary, we have shown that DUX4 target gene expression is the major molecular signature in FSHD muscle. Although the expression of DUX4 target genes is the major molecular characteristic of FSHD, some FSHD biopsies did not show expression of these genes, possibly reflecting the involvement of subsets of muscles in FSHD individuals and the limited sample size of a needle biopsy. In addition, an unaffected family member with an FSHD-permissive allele in the short-normal range expressed DUX4 target genes, indicating that expressing DUX4 target genes is not sufficient to guarantee disease penetrance. The facts that this individual appeared to lack an immune cell infiltration, based on low expression of immune cell genes, and had both an FSHD1 sibling and an FSHD2 sibling without an SMCHD1 mutation, suggests a possible role of the immune response and other modifier loci beyond SMCHD1.
MATERIALS AND METHODS
Transduction of muscle cells cultures with DUX4
The transduction of the 54-1 and MB135 human myoblast lines was previously described (6). RNA was harvested after transduction by lenti-pgk-DUX4 or lenti-pgk-GFP and non-transduced cells as controls after 48 h (54-1) or 24 h (MB135).
Muscle cell cultures
Muscle cells derived from two FSHD1 and three FSHD2 were cultured and RNA was harvested from both myoblasts and myotubes. These cultures were described previously (26). As previously described, primary myoblast cell lines were received from the University of Rochester biorepository (http://www.urmc.rochester.edu/fields-center) and were cultured in DMEM/F-10 media (Gibco) in the presence of 20% heat-inactivated fetal bovine serum (Gibco), 1% penicillin/streptomycin (Gibco). Media was supplemented with 10 ng/ml rhFGF (Promega) and 1 μM dexamethasone (SIGMA). Myoblasts were fused at 80% confluence in DMEM/F-12 Glutamax media containing 2% KnockOut serum replacement formulation (Gibco) for 36 h.
Muscle biopsy
All muscle biopsies were obtained at the University of Rochester Medical Center under an IRB-approved protocol from clinically and genetically confirmed FSHD individuals and controls that included family members without clinical or genetic diagnostic features associated with FSHD or other muscular dystrophies. Muscle biopsy samples were obtained under local anesthesia by the needle biopsy technique utilizing the modified Bergstrom needle as previously described (27,28). A muscle sample, adjacent to the sample used for RNA extraction, was oriented and mounted and then frozen in isopentane cooled in liquid nitrogen for histologic evaluation. A twelve point scale (0 = normal muscle and 12 = end-stage muscle) was used for histopathological grading of the sample based on: degree of muscle fiber variability, extent of central nucleation, presence of muscle fiber necrosis/regeneration and extent of interstitial fibrosis. (http://www.urmc.rochester.edu/fields-center/protocols/PathologicSeverityScoring.cfm)
RNA-seq library preparation and sequencing
Library preparation and sequencing was carried out by the FHCRC Genomics Shared Resource. Sequencing libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina) according to the manufacturer's instructions, poly(A) selected, and subjected to Illumina sequencing using standard protocols to generate 100 bp single-end reads. Library size distributions were validated using an Agilent 2100 Bioanalyzer. Additional library quality control, blending of pooled indexed libraries and cluster optimization were performed using the QPCR NGS Library Quantization Kit (Agilent Technologies). Sequencing occurred on an Illumina HiSeq 2000 using a single-read, 100 base read length (SR100) sequencing strategy. RNA-seq datasets have been submitted to GEO (accession number, GSE56787).
RNA-seq data processing and analysis
Image analysis and base calling were performed with the Illumina's Real Time Analysis v1.17.20 software. Files were demultiplexed of indexed reads and generated in FASTQ format using the Illumina's CASAVA v1.8.2 software. Reads were removed that did not pass Illumina's base call quality threshold. Reads were aligned to human genome ensemble assembly GRCh37, using TopHat 2.0.8 (Trapnell et al., 2009). We collected read counts for all genes using Bioconductor Rsamtools package. To include reads that mapped to multiple homologs in the genome, each hit of a read with multiple hits received a fractional count equal to one over the number of hits. Differential expressed genes were detected using Bioconductor package DESeq 1.14. Based on recommendation of DESeq package, to compare two groups of samples with fewer than six replicates for each condition, we used ‘estimateDispersions’ function using the ‘fit-only’ sharing mode, so that dispersions were estimated based on local regression against mean values. For comparison of groups involving more than six replicates for each condition, we used ‘maximum’ sharing mode, which were the maximum of fitted values and per-gene estimates. To visualize gene expression clustering and to compute fold change differences, we applied to asinh transformation to gene counts of each sample scaled by the corresponding size-factor. The adjusted log-fold-change is computed as the difference of the average transformed values under the two conditions in comparison. The log-fold-change computed this way has more regulated behavior at values close to or at zero, and approximates log-fold-change for larger values. This approach is very similar to the variance-stabilization transformation method recommended by the most recent version of DESEQ. To make sure that differentially expressed genes were not driven by few extreme outliers, we applied the non-parametric Wilcoxon-rank sum test to samples in comparison as another filter. The P-value thresholds were selected based on the number of tested samples.
Novel exon prediction
We developed a new method for detecting novel exons using RNA-seq datasets. The method exploits un-annotated splicing junctions derived from RNA-Seq reads to infer the boundaries of novel exons. The region bounded by two novel junctions at both ends with the same strand and decent coverage between them, or the region with a novel junction only at one end, and tails off to background is determined as a novel exon. The strands of predicted splicing junctions are determined by matching the sequences at both ends to the motif derived from the splicing donors/acceptors of known introns. The strands of predicted exons are determined by the strands of the connecting splicing junctions. We also examine whether the annotated exons are supported by RNA-Seq reads, i.e. whether the exons have corresponding splicing junctions, and if the entire exons have decent read coverage without internal splicing junctions that truncate the exons. Finally, we use splicing junctions to assemble the predicted novel exons into genes, and determined whether they are exons of known genes or not.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Institutes of Health (NINDS P01NS5069539 to S.V.M., R.T. and S.J.T.; NIAMS R01AR045203, to S.J.T.; and NCRR UL1RR024160, to R.T.), Friends of FSH Research, to S.J.T.; Prinses Beatrix Spierfonds, to S.V.M.; Stichting FSHD, to S.V.M.; FSH Society, to S.V.M. and Geraldi Norton Foundation and the Eklund Family, to R.T. and S.V.M.
Conflict of Interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the individuals who participated in this study to provide the necessary biological samples.
REFERENCES
- 1.Pandya S., King W.M., Tawil R. Facioscapulohumeral dystrophy. Phys. Ther. 2008;88:105–113. doi: 10.2522/ptj.20070104. [DOI] [PubMed] [Google Scholar]
- 2.Lemmers R.J., Tawil R., Petek L.M., Balog J., Block G.J., Santen G.W., Amell A.M., van der Vliet P.J., Almomani R., Straasheijm K.R., et al. Digenic inheritance of an SMCHD1 mutation and an FSHD-permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat. Genet. 2012;44:1370–1374. doi: 10.1038/ng.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lemmers R.J., van der Vliet P.J., Klooster R., Sacconi S., Camano P., Dauwerse J.G., Snider L., Straasheijm K.R., van Ommen G.J., Padberg G.W., et al. A unifying genetic model for facioscapulohumeral muscular dystrophy. Science. 2010;329:1650–1653. doi: 10.1126/science.1189044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Maarel S.M., Miller D.G., Tawil R., Filippova G.N., Tapscott S.J. Facioscapulohumeral muscular dystrophy: consequences of chromatin relaxation. Curr. Opin. Neurol. 2012;25:614–620. doi: 10.1097/WCO.0b013e328357f22d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Snider L., Geng L.N., Lemmers R.J., Kyba M., Ware C.B., Nelson A.M., Tawil R., Filippova G.N., van der Maarel S.M., Tapscott S.J., et al. Facioscapulohumeral dystrophy: incomplete suppression of a retrotransposed gene. PLoS Genet. 2010;6:e1001181. doi: 10.1371/journal.pgen.1001181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geng L.N., Yao Z., Snider L., Fong A.P., Cech J.N., Young J.M., van der Maarel S.M., Ruzzo W.L., Gentleman R.C., Tawil R., et al. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev. Cell. 2012;22:38–51. doi: 10.1016/j.devcel.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahimov F., King O.D., Leung D.G., Bibat G.M., Emerson C.P., Jr, Kunkel L.M., Wagner K.R. Transcriptional profiling in facioscapulohumeral muscular dystrophy to identify candidate biomarkers. Proc. Natl Acad. Sci. USA. 2012;109:16234–16239. doi: 10.1073/pnas.1209508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krom Y.D., Dumonceaux J., Mamchaoui K., den Hamer B., Mariot V., Negroni E., Geng L.N., Martin N., Tawil R., Tapscott S.J., et al. Generation of isogenic D4Z4 contracted and noncontracted immortal muscle cell clones from a mosaic patient: a cellular model for FSHD. Am. J. Pathol. 2012;181:1387–1401. doi: 10.1016/j.ajpath.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Semple F., Webb S., Li H.N., Patel H.B., Perretti M., Jackson I.J., Gray M., Davidson D.J., Dorin J.R. Human beta-defensin 3 has immunosuppressive activity in vitro and in vivo. Eur. J. Immunol. 2010;40:1073–1078. doi: 10.1002/eji.200940041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block G.J., Narayanan D., Amell A.M., Petek L.M., Davidson K.C., Bird T.D., Tawil R., Moon R.T., Miller D.G. Wnt/beta-catenin signaling suppresses DUX4 expression and prevents apoptosis of FSHD muscle cells. Hum. Mol. Genet. 2013;22:4661–4672. doi: 10.1093/hmg/ddt314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balog J., Thijssen P.E., de Greef J.C., Shah B., van Engelen B.G., Yokomori K., Tapscott S.J., Tawil R., van der Maarel S.M. Correlation analysis of clinical parameters with epigenetic modifications in the DUX4 promoter in FSHD. Epigenetics. 2012;7:579–584. doi: 10.4161/epi.20001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ricci E., Galluzzi G., Deidda G., Cacurri S., Colantoni L., Merico B., Piazzo N., Servidei S., Vigneti E., Pasceri V., et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol. 1999;45:751–757. doi: 10.1002/1531-8249(199906)45:6<751::aid-ana9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 13.van Overveld P.G., Enthoven L., Ricci E., Rossi M., Felicetti L., Jeanpierre M., Winokur S.T., Frants R.R., Padberg G.W., van der Maarel S.M. Variable hypomethylation of D4Z4 in facioscapulohumeral muscular dystrophy. Ann. Neurol. 2005;58:569–576. doi: 10.1002/ana.20625. [DOI] [PubMed] [Google Scholar]
- 14.Gabellini D., D'Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M.A., Bottinelli R., et al. Facioscapulohumeral muscular dystrophy in mice overexpressing FRG1. Nature. 2006;439:973–977. doi: 10.1038/nature04422. [DOI] [PubMed] [Google Scholar]
- 15.Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A. Increased levels of adenine nucleotide translocator 1 protein and response to oxidative stress are early events in facioscapulohumeral muscular dystrophy muscle. J. Mol. Med. 2005;83:216–224. doi: 10.1007/s00109-004-0583-7. [DOI] [PubMed] [Google Scholar]
- 16.Caruso N., Herberth B., Bartoli M., Puppo F., Dumonceaux J., Zimmermann A., Denadai S., Lebosse M., Roche S., Geng L., et al. Deregulation of the protocadherin gene FAT1 alters muscle shapes: implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 2013;9:e1003550. doi: 10.1371/journal.pgen.1003550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones T.I., Chen J.C., Rahimov F., Homma S., Arashiro P., Beermann M.L., King O.D., Miller J.B., Kunkel L.M., Emerson C.P., Jr, et al. Facioscapulohumeral muscular dystrophy family studies of DUX4 expression: evidence for disease modifiers and a quantitative model of pathogenesis. Hum. Mol. Genet. 2012;21:4419–4430. doi: 10.1093/hmg/dds284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frisullo G., Frusciante R., Nociti V., Tasca G., Renna R., Iorio R., Patanella A.K., Iannaccone E., Marti A., Rossi M., et al. CD8(+) T cells in facioscapulohumeral muscular dystrophy patients with inflammatory features at muscle MRI. J. Clin. Immunol. 2011;31:155–166. doi: 10.1007/s10875-010-9474-6. [DOI] [PubMed] [Google Scholar]
- 19.Friedman S.D., Poliachik S.L., Carter G.T., Budech C.B., Bird T.D., Shaw D.W. The magnetic resonance imaging spectrum of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2012;45:500–506. doi: 10.1002/mus.22342. [DOI] [PubMed] [Google Scholar]
- 20.Friedman S.D., Poliachik S.L., Otto R.K., Carter G.T., Budech C.B., Bird T.D., Miller D.G., Shaw D.W. Longitudinal features of stir bright signal in FSHD. Muscle Nerve. 2013;49:257–260. doi: 10.1002/mus.23911. [DOI] [PubMed] [Google Scholar]
- 21.Osborne R.J., Welle S., Venance S.L., Thornton C.A., Tawil R. Expression profile of FSHD supports a link between retinal vasculopathy and muscular dystrophy. Neurology. 2007;68:569–577. doi: 10.1212/01.wnl.0000251269.31442.d9. [DOI] [PubMed] [Google Scholar]
- 22.Winokur S.T., Chen Y.W., Masny P.S., Martin J.H., Ehmsen J.T., Tapscott S.J., van der Maarel S.M., Hayashi Y., Flanigan K.M. Expression profiling of FSHD muscle supports a defect in specific stages of myogenic differentiation. Hum. Mol. Genet. 2003;12:2895–2907. doi: 10.1093/hmg/ddg327. [DOI] [PubMed] [Google Scholar]
- 23.Arashiro P., Eisenberg I., Kho A.T., Cerqueira A.M., Canovas M., Silva H.C., Pavanello R.C., Verjovski-Almeida S., Kunkel L.M., Zatz M. Transcriptional regulation differs in affected facioscapulohumeral muscular dystrophy patients compared to asymptomatic related carriers. Proc. Natl Acad. Sci. USA. 2009;106:6220–6225. doi: 10.1073/pnas.0901573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gendrel A.V., Tang Y.A., Suzuki M., Godwin J., Nesterova T.B., Greally J.M., Heard E., Brockdorff N. Epigenetic functions of smchd1 repress gene clusters on the inactive X chromosome and on autosomes. Mol. Cell. Biol. 2013;33:3150–3165. doi: 10.1128/MCB.00145-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mould A.W., Pang Z., Pakusch M., Tonks I.D., Stark M., Carrie D., Mukhopadhyay P., Seidel A., Ellis J.J., Deakin J., et al. Smchd1 regulates a subset of autosomal genes subject to monoallelic expression in addition to being critical for X inactivation. Epigenetics Chromatin. 2013;6:19. doi: 10.1186/1756-8935-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young J.M., Whiddon J.L., Yao Z., Kasinathan B., Snider L., Geng L.N., Balog J., Tawil R., van der Maarel S.M., Tapscott S.J. DUX4 binding to retroelements creates promoters that are active in FSHD muscle and testis. PLoS Genet. 2013;9:e1003947. doi: 10.1371/journal.pgen.1003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Welle S., Brooks A.I., Delehanty J.M., Needler N., Bhatt K., Shah B., Thornton C.A. Skeletal muscle gene expression profiles in 20–29 year old and 65–71 year old women. Exp. Gerontol. 2004;39:369–377. doi: 10.1016/j.exger.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 28.Welle S., Bhatt K., Thornton C.A. High-abundance mRNAs in human muscle: comparison between young and old. J. Appl. Physiol. 2000;89:297–304. doi: 10.1152/jappl.2000.89.1.297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.