Abstract
Keratoconus, a common inherited ocular disorder resulting in progressive corneal thinning, is the leading indication for corneal transplantation in the developed world. Genome-wide association studies have identified common SNPs 100 kb upstream of ZNF469 strongly associated with corneal thickness. Homozygous mutations in ZNF469 and PR domain-containing protein 5 (PRDM5) genes result in brittle cornea syndrome (BCS) Types 1 and 2, respectively. BCS is an autosomal recessive generalized connective tissue disorder associated with extreme corneal thinning and a high risk of corneal rupture. Some individuals with heterozygous PRDM5 mutations demonstrate a carrier ocular phenotype, which includes a mildly reduced corneal thickness, keratoconus and blue sclera. We hypothesized that heterozygous variants in PRDM5 and ZNF469 predispose to the development of isolated keratoconus. We found a significant enrichment of potentially pathologic heterozygous alleles in ZNF469 associated with the development of keratoconus (P = 0.00102) resulting in a relative risk of 12.0. This enrichment of rare potentially pathogenic alleles in ZNF469 in 12.5% of keratoconus patients represents a significant mutational load and highlights ZNF469 as the most significant genetic factor responsible for keratoconus identified to date.
INTRODUCTION
Keratoconus (MIM 148300), a common bilateral, progressive corneal thinning disorder (1), is the leading indication for corneal transplantation in the developed world, accounting for 25% of the 2500 corneal transplants performed annually in the UK and a similar proportion of the 32 000 grafts performed annually in the USA (2). Keratoconus usually arises in the teenage years and presents a significant health burden in work-age adults. The minimum incidence is 1 in 2000, but it is much more common in some ethnic groups (1,3). There is strong evidence for a heritable component in the development of keratoconus (4,5). Most studies describe autosomal dominant inheritance, with incomplete penetrance or variable expressivity (4). However, in a genetic modelling study in a multi-ethnicity population a major recessive genetic defect was the most parsimonious (6), although no recessive loci for keratoconus have been described to date.
The progressive corneal thinning associated with keratoconus [mean central corneal thickness (CCT) 450–500 µm] (7) results in myopia and irregular corneal astigmatism. In healthy humans, CCT is a normally distributed quantitative trait with a mean of 536 ± 31 µm (8), which has an estimated heritability up to 95% (9). Genome-wide association studies (GWAS) in the healthy European and Asian populations have identified CCT-associated loci, with common SNPs upstream of zinc finger 469 (ZNF469 [MIM 612078]) the most strongly associated with CCT (10–14). Mutations in three genes (ZNF469, COL5A1 and COL8A2) close or within these identified loci are responsible for rare Mendelian conditions that affect the corneal structure: brittle corneal syndrome, Ehlers–Danlos syndrome and posterior polymorphous corneal dystrophy, respectively (10–12).
Brittle cornea syndrome (BCS) is an autosomal recessive generalized connective tissue disorder associated with extreme corneal thinning (220–450 µm) and a high risk of corneal rupture (15,16). Homozygous mutations in ZNF469 and PR domain-containing protein 5 [PRDM5 (MIM 614161)] genes result in BCS Type 1 [BCS1 (MIM 229200)] (17) and BCS Type 2 [BCS2 (MIM 614170)] (15), respectively. Some individuals with heterozygous PRDM5 mutations demonstrate a carrier ocular phenotype which includes a mildly reduced CCT (480–505 µm), keratoconus and blue sclera (15). In one family with BCS2, there was a relationship between the severity and age of onset of keratoconus and PRDM5 mutational status. Family members with a homozygous PRDM5 mutation (deletion of exons 9–14) developed early and severe keratoconus, whereas one heterozygous family member developed keratoconus which was clinically milder with a later onset (15). The relationship between the degree of CCT reduction and the presence of homozygous or heterozygous mutations in PRDM5 suggested a dosage effect (15). Although none of the CCT-associated loci have been mapped to PRDM5, a common SNP (rs10518367) which is 70 kb upstream of PRDM5 has been associated with CCT in the European population at the significance level of P = 8.9 × 10−5.
Given that rare ZNF469 and PRDM5 homozygous mutations result in the extreme corneal thinning disorder (BCS), and that there was evidence of a carrier ocular phenotype (keratoconus and corneal thinning) in some individuals with PRDM5 heterozygous mutations, and that common SNPs 100 kb upstream of ZNF469 are strongly associated with CCT, we undertook Sanger sequencing of both genes in patients with isolated keratoconus; a common ocular disease characterized by progressive corneal thinning and ectasia.
RESULTS
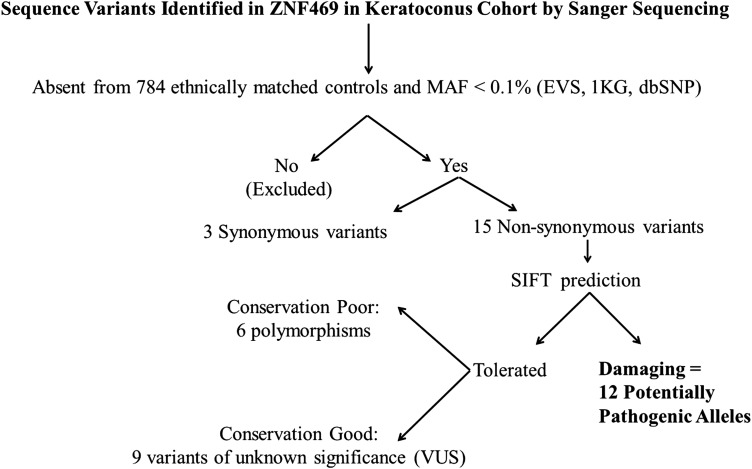
Heterozygous PRDM5 mutations in carrier individuals from BCS2 families can result in keratoconus (15), and so the entire coding region and intron–exon junctions of PRDM5 (exons 1–16) were sequenced in an initial 96 unrelated European patients with keratoconus. This analysis failed to identify any pathogenic variants, although eight known SNPs were detected (rs146268537, rs74320998, rs343192, rs17051264, rs34666716, rs12499000, rs75893420 and rs55774575). We therefore proceeded to Sanger sequence ZNF469 in the original cohort increased with additional keratoconus cases (total number of cases = 112) of unrelated European patients with keratoconus from three study centres (Belfast, Leeds and Lausanne). Sequence variants detected by Sanger sequencing were classified as potentially pathogenic alleles by filtering using (i) ethnically matched population-specific control data from 784 individuals (outlined in the Materials and Methods); (ii) the data from dbSNP (Build 137), the May 2012 release of the 1000 Genomes (1KG) Project and the Exome Variant Server (EVS), NHLBI Exome Sequencing Project (ESP), with no allele having a minor allele frequency (MAF) >0.1% and (iii) classified as damaging using the Sorting Intolerant from Tolerant (SIFT) programme as outlined in the Materials and Methods and Figure 2.
Figure 2.
Hierarchical flow diagram of filtering process performed on sequence variants identified in ZNF469 in keratoconus cohort by Sanger sequencing.
From these stringently filtered sequencing data, 12 potentially pathogenic non-synonymous heterozygous alleles were detected in the keratoconus cohort (Table 1) and two in-frame deletions: c.2904_2909delGTCGGG; p.Ser969_Gly970del and c.9011_9025delTTCCCGGGAACACCC; p.Leu3004_Thr3008del. In the keratoconus cohort following filtering, there remained 15 non-synonymous classified as tolerated by SIFT which were absent from control data and had an MAF <0.1% (Table 2). On the basis of poor conservation, six of these variants were classified as polymorphisms leaving nine variants of unknown significance. We detected 34 non-synonymous and 33 synonymous variants that were observed in both cases and controls, had an MAF ≥0.1% or were common variants, and were deemed non-pathogenic (Supplementary Material, Table S1). Overall, this study identified 34 novel variants (29 non-synonymous and 5 synonymous) that have been submitted to the NCBI dbSNP (Supplementary Material, Table S2). The severity of keratoconus was graded using the Amsler–Krumeich classification (18,19), and 10 individuals had grade Stage III or above indicating severe disease (illustrated in Fig. 1). Stages III and IV usually require surgical approaches for visual rehabilitation, and three individuals required corneal transplantation.
Table 1.
Potentially pathologic ZNF 469 alleles identified in keratoconus and control subjects
| Nucleotide changea | Amino acid changea | Present in 1KG data [MAF (%)] | Present in EVS data [MAF (%)] | rs number | SIFT prediction (SIFT score)b | Amsler–Krumeich classificationc |
Corneal transplantation | |
|---|---|---|---|---|---|---|---|---|
| Right eye | Left eye | |||||||
| Keratoconus cohort | ||||||||
| c.290C > T | p.Pro97Leu | No | No | rs273585617 | Damaging (0) | Stage III | Stage II | No |
| c.337G > A | p.Glu113Lys | No | No | NA | Damaging (0) | Stage II | Stage II | No |
| c.2063C > A | p.Thr688Asn | No | No | NA | Damaging (0) | Stage IV | Stage III | No |
| c.2699C > G | p.Pro900Arg | No | No | rs273585618 | Damaging (0.02) | Stage III | Stage II | Yes; right eye |
| c.2699C > T | p.Pro900Leu | No | No | rs273585618 | Damaging (0) | Stage II | Stage II | No |
| c.2904_2909del GTCGGG | p.Ser969_Gly970 del (in-frame deletion) | No | No | NA | NA | Stage IV | Stage III | Yes; right eye |
| c.3119A > C | p.Lys1040Thr | No | No | rs273585619 | Damaging (0) | Stage III | Stage III | Yes; right eye |
| c.4363G > T | p.Ala1455Ser | No | No | rs116532825 | Damaging (0.02) | Stage I | Stage III | Yes; left eye |
| c.5464C > A | p.Pro1822Thr | Yes (NA) | No | rs74032866 | Damaging (0.04) | Stage I | Stage I | No |
| c.6095C > A | p.Ser2032Tyr | No | No | rs273585623 | Damaging (0.05) | Stage II | Stage III | No |
| c.8912G > T | p.Gly2971Val | No | No | rs273585625 | Damaging (0.04) | Stage III | Stage II | No |
| c.9011_9025del TTCCCGGGAACACCC | p.Leu3004_Thr3008 del (in-frame deletion) | No | No | NA | NA | Stage III | Stage II | No |
| c.9047C > T | p.Thr3016Met | No | No | rs273585626 | Damaging (0.02) | Stage II | Stage I | No |
| c.11615C > T | p.Pro3872Leu | No | No | rs273585630 | Damaging (0.03) | Stage II | Stage III | No |
| Normal controls | ||||||||
| c.1701G > T | p.Gln567His | No | No | NA | Damaging | NA | NA | NA |
NA, not available or applicable.
aZNF469 Ensembl transcript ENST00000437464 or NCBI NM_001127464.1 (Build GRCh37/hg19).
bPositions with normalized probabilities of <0.05 are predicted to be damaging, those ≥0.05 are predicted to be tolerated.
Table 2.
ZNF 469 sequence variants identified in keratoconus and control subjects
| Nucleotide change | Amino acid change | rs number | Present in 1KG data [MAF (%)] | Present in EVS data [MAF (%)] | SIFT prediction | Classificationa |
|---|---|---|---|---|---|---|
| Keratoconus cohort | ||||||
| Non-synonymous variants | ||||||
| c.77G > C | p.Ser26Thr | rs273585616 | No | No | Tolerated | Variant of unknown significance (VUS) |
| c.1627G > A | p.Gly543Ser | NA | No | No | Tolerated | Polymorphism (Ser common) |
| c.2297G > A | p.Arg766Gln | rs144492145 | Yes (0.05) | No | Tolerated | Polymorphism |
| c.3236G > A | p.Arg1079Gln | NA | No | No | Tolerated | Polymorphism (Gln in marmoset) |
| c.4394C > T | p.Pro1465Leu | rs369382753 | No | Yes (0.02) | Tolerated | Polymorphism (Leu in rat and mouse) |
| c.4826G > C | p.Arg1609Pro | rs273585621 | No | No | Tolerated | Polymorphism (Pro in dog) |
| c.5060G > A | p.Arg1687Lys | NA | No | No | Tolerated | VUS |
| c.5597A > T | p.Gln1866Leu | NA | No | No | Tolerated | VUS |
| c.6007G > A | p.Glu2003Lys | rs273585622 | No | No | Tolerated | Polymorphism (Lys in gorilla) |
| c.6725C > A | p.Ser2242Tyr | rs273585624 | No | No | Tolerated | VUS |
| c.7527G > C | p.Glu2509Asp | rs199519673 | No | Yes (0.090) | Tolerated | VUS |
| c.7747G > A | p.Glu2583Lys | NA | No | No | Tolerated | VUS |
| c.7847G > A | p.Arg2616Gln | NA | No | No | Tolerated | VUS |
| c.9835A > G | p.Thr3279Ala | rs273585627 | No | No | Tolerated | VUS |
| c.11101G > A | p.Gly3701Ser | rs273585629 | No | No | Tolerated | VUS |
| Synonymous variants | ||||||
| c.99G > A | p.Pro33Pro | rs273585631 | No | No | NA | VUS |
| c.720G > A | p.Glu240Glu | rs273585632 | No | No | NA | VUS |
| c.2478G > T | p.Pro826Pro | rs273585634 | No | No | NA | VUS |
| c.6453T > C | p.Asp2151Asp | NA | No | No | NA | VUS |
| c.10843C > T | p.Leu3615Leu | NA | No | No | NA | VUS |
| Normal control cohort | ||||||
| Non-synonymous variants | ||||||
| c.10115C > T | p.Pro3372Leu | NA | No | No | Tolerated | VUS |
| c.11252G > A | p.Arg3751Lys | NA | No | No | Tolerated | Polymorphism (Lys in dog) |
| Synonymous variants | ||||||
| c.30G > A | p.Pro10Pro | NA | No | No | NA | VUS |
| c.4281C > A | p.Leu1427Leu | NA | No | No | NA | VUS |
aIf predicted to be tolerated using SIFT the conservation of the residue was assessed and if poorly conserved the variant was classified as a polymorphism
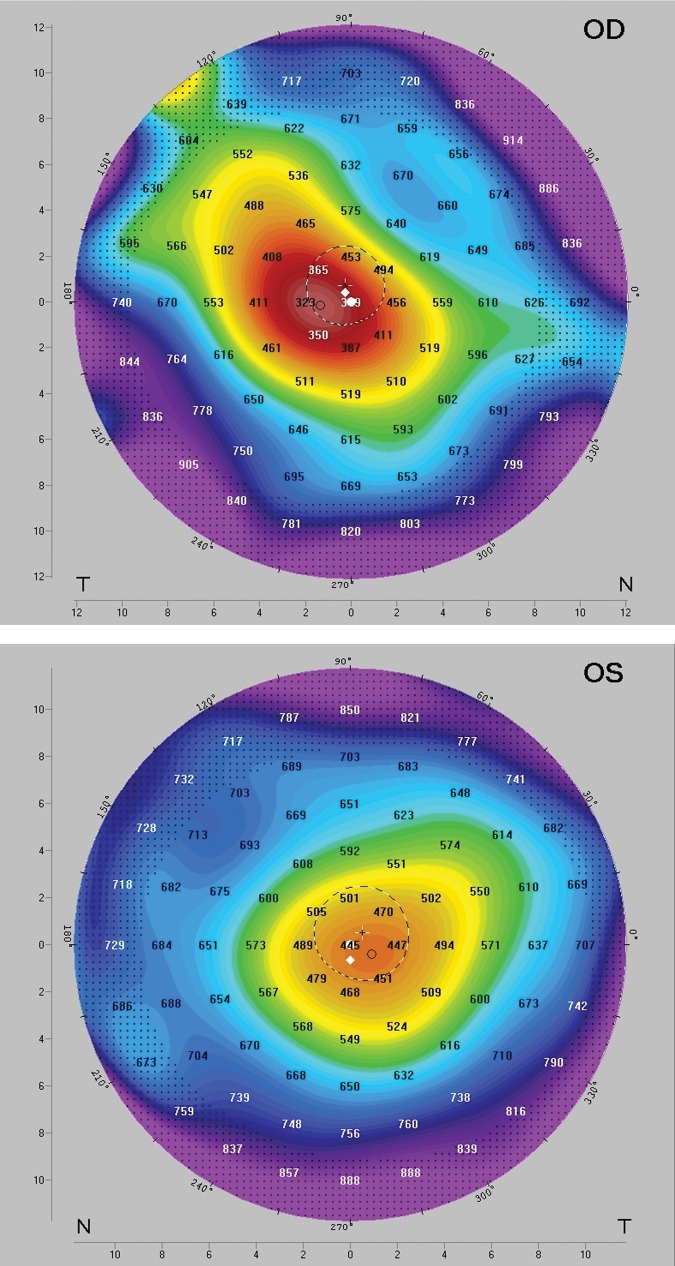
Figure 1.
Corneal topography of the 28-year-old European patient from the UK with a c.3119A > C (p.Lys1040Thr) ZNF469 pathogenic allele (Table 1) using Pentacam corneal topography; OD indicates right eye and OS left eye. The topography shows the anterior corneal steepening associated with keratoconus with a large cone centrally in the right eye and paracentrally in the left eye associated with corneal thinning underlying the cones; minimum corneal thickness of 318 µm (OD) and 438 µm (OS). The keratoconus is Stage III in both eyes (Amsler–Krumeich classification) with a best corrected Snellen acuity of 6/36 right and 6/24 left. The patient subsequently underwent a deep anterior lamellar keratoplasty (corneal transplant) in the right eye.
As there was the possibility that the alleles identified in the keratoconus subjects represented chance events, we Sanger sequenced the complete coding sequence of ZNF469 (13 203 bp) in 96 unaffected and unrelated European control samples (192 chromosomes) using the same experimental stringency as the case sequencing, and detected one non-synonymous heterozygous allele (c.1701G > T; p.Gln567His) deemed potentially pathogenic given our filtering criteria (Table 1). There was a statistically significant enrichment of potentially pathogenic ZNF469 alleles in the keratoconus subjects (14 variants) compared with the 96 European controls (one variant); P = 0.00102 [odds ratio (OR) 13.6, relative risk (RR) 12.0]. The allele frequency differences make it impossible for the rare ZNF469 alleles to be in linkage disequilibrium with the common variant signal that is within a 53 kb linkage disequi librium block 117 kb away from the 5′ end of ZNF46, and this has been replicated in diverse ancestries groups. The common variant, although strongly associated with corneal thickness, is not strongly associated with keratoconus {OR 1.25 [95% confidence interval (CI) 1.11–1.40]} (13). Further functional studies and assays are required to confirm the pathogenicity of all alleles absent from ethnically matched controls and the population control data (dbSNP, EVS, 1KG).
DISCUSSION
Mutations in four genes, VSX1 (MIM 605020) (20,21), SOD1 (MIM 147450) (21,22), MIR184 (MIM 613146) (23) and ZEB1 (MIM 189909) (24), have been implicated in the pathogenesis of keratoconus in a minority of cases (<4%) (21,25). Two GWAS have been conducted in keratoconus cohorts which identified an SNP rs4954218, located near the RAB3GAP1 gene (MIM 602536) (26), and polymorphisms in HGF (MIM 142409) (27), associated with keratoconus susceptibility, but neither study reported genome-wide significant association. The identification of alleles that are predicted to be potentially pathogenic in 12.5% of keratoconus patients (14/112) makes ZNF469 the most significant genetic factor responsible for keratoconus identified to date. The small sizes of unrelated keratoconus case cohorts favour a candidate genes approach so that while the P-value obtained for evidence of a burden of ZNF469 rare damaging variants in the keratoconus cases is statistically significant, it would not have been in a genome-wide context.
BCS is a rare recessive connective tissue disorder associated with consanguinity, with most BCS patients originating from countries in the Middle East and North Africa (16). ZNF469, the gene for BCS1, was originally mapped to chromosome 16q24 in a single large Palestinian family and a homozygous frameshift mutation, (c.9527delG) predicted to result in a pre-mature termination codon (p.Gln3178ArgfsX23), was subsequently reported (17). Five further homozygous ZNF469 mutations have been reported in the literature: a founder mutation in five Tunisian patients (c.5934delA) predicted to result in a pre-mature termination codon (p.Gly1983AlafsX16), p.Gln1392X (Syrian origin) (28), p.Phe717SerfsX14 (15), p.Gln1757X (15) and one homozygous missense mutation (p.Cys3339Tyr) in a consanguineous Norwegian family (29). Homozygous mutations in PRDM5 result in BCS2, and in some families PRDM5 heterozygous gene carriers display an ocular carrier state that includes a mildly reduced CCT (480–505 µm), keratoconus and blue sclera (15). Families harbouring homozygous and heterozygous PRDM5 mutations show a gene dosage relationship in terms of the degree of CCT reduction and the severity and age of onset of keratoconus (15). There is no data available of CCT measurement or corneal topography for heterozygous carriers of ZNF469 mutations in BCS1 families. Our data for ZNF469 in the keratoconus population mirrors that seen in the ocular phenotype of PRDM5 heterozygous carriers. Heterozygous ZNF469 pathological alleles result in progressive corneal thinning and ectasia producing the keratoconus phenotype. The majority of potentially pathogenic alleles in the keratoconus cohort were missense variants likely to have a less deleterious effect on protein function than the ZNF469 truncating mutations commonly associated with BCS1. This further supports a gene dosage phenomenon wherein homozygous, severely deleterious ZNF469 mutations result in an early-onset severe and visually devastating ocular phenotype (extreme corneal thinning, ectasia and spontaneous rupture) (15), whereas heterozygous, missense ZNF469 mutations result in corneal thinning, ectasia and keratoconus. Further functional studies and cell-based assays are required to interrogate the molecular pathology and mutational mechanisms associated with these potentially pathogenic ZNF469 alleles.
ZNF469 is a 3925 amino acid evolutionarily poorly conserved C2H2 zinc finger (C2H2-ZNF) protein of unknown function (30). C2H2-ZNF genes constitute the largest class of transcription factors in humans making up ∼2% of all the human genes and represent the second largest gene family in humans (30). The first identified members of the C2H2-ZNF family were Xenopus TFIIIA and Drosophila Kruppel, and thus genes of this family are often called ZNF genes of the TFIIIA or Kruppel type (30–32). Most C2H2-ZNF genes code for transcription factors which can bind DNA, RNA, DNA–RNA hybrids and proteins (32). The physiological role of ZNF469 is not well established, but there is evidence that ZNF469 regulates extra-cellular matrix development and maintenance (15). ZNF469 shows 30% sequence similarity to the helical parts of COL1A2 (MIM 120160), COL1A1 (MIM 120150) and COL4A1 (MIM 120130), all of which are highly expressed in the cornea (17). The cornea is composed of 70% collagen, mostly collagen Type I, and there is evidence of a dysregulation of collagen homeostasis in the keratoconic cornea (33,34). Corneal thinning has been reported in osteogenesis imperfect (35), which results from mutations in COL1A1 or COL1A2.
We have identified an enrichment of potentially pathogenic alleles in ZNF469 in patients with keratoconus. Further work is required to determine the functional impact of these variants, and the pathways regulated by ZNF469 which are involved in the development of keratoconus. Identifying genes responsible for keratoconus may also provide insights into the genetic basis for the normal variation in CCT. Decreased CCT has been proposed as a risk factor for primary open-angle glaucoma [POAG (MIM 137760)], the leading cause of irreversible blindness worldwide affecting >60 million people (36). Individual patients with a thin cornea have a substantially increased risk for developing POAG (37,38), and glaucoma patients with a thin CCT have an increased severity and more rapid progression of visual field loss (39). The genetic basis of CCT may provide insights into the development of glaucoma. Common SNPs near ZNF469 are the strongest CCT-associated loci, although the functional role of these SNPs is not known (10–13). The role ZNF469 plays in the development of POAG, and maintenance of CCT in normal subjects has not been determined. Combining resequencing with GWAS has yielded success in identifying rare disease-associated variants (40,41). Our study establishes the significant role ZNF469 plays in the development of keratoconus.
MATERIALS AND METHODS
All studies adhered to the tenets of the Declaration of Helsinki and were approved by the relevant institutions with all participants giving written informed consent.
Patients
Clinically affected keratoconus patients of European ethnicity were recruited as part of ongoing studies from Belfast (Belfast Health and Social Care Trust, UK), Leeds (St. James's University Hospital, Leeds, UK) and Lausanne (Jules-Gonin Eye Hospital, Lausanne and Institute for Research in Ophthalmology, Sion, Switzerland); and genomic DNA was extracted from peripheral blood leukocytes using commercial kits. The diagnosis of keratoconus was performed by an experienced ophthalmologist based on well-established clinical signs on slit-lamp biomicroscopy and cycloplegic retinoscopy; and a confirmatory videokeratographic map obtained using the Topographic Modelling System-1 (Computed Anatomy Inc., NY, USA), Orbscan II (Bausch & Lomb, Salt Lake City, UT, USA) or the Pentacam (Oculus, Wetzlar, Germany) (20,27). Slit-lamp biomicroscopy was used to identify the key features of keratoconus including stromal corneal thinning, Vogt's striae and Fleischer rings in affected individuals. The oil droplet sign and scissoring of the red reflex were assessed by retinoscopy performed with a fully dilated pupil. Patients were considered as having keratoconus if they had at least one clinical sign of the disease in conjunction with a confirmatory videokeratography map (20,27). The severity of keratoconus was graded using the Amsler–Krumeich classification (18,19):
| Amsler–Krumeich classification | |
| Stage I | Eccentric corneal steppening Myopia and/or astigmatism <5.00D Mean central K readings <48.00D |
| Stage II | Myopia and/or astigmatism 5.00–8.00D Mean central K readings <53.00D Absence of scarring Minimal corneal thickness >400 µm |
| Stage III | Myopia and/or astigmatism 8.00–10.00D Mean central K readings >53.00D Absence of scarring Minimal corneal thickness 300–400 µm |
| Stage IV | Refraction not measurable Mean central K readings >55.00D Central corneal scarring Minimal corneal thickness 200 µm |
Ethnically matched population-specific control data
All affected and control individuals were of European ethnicity, and population-specific control data were obtained from three sources: (i) a total of 96 unrelated individuals (192 chromosomes) without ocular disease (aged 60 and over) from the Northern Irish population (UK) underwent Sanger sequencing; (ii) exome data from 275 non-glaucomatous individuals from the Manchester population (UK), which are effectively ethnically identical to the Leeds population and (iii) normative control data for 413 normal individuals from Lausanne (Swiss population; European) were obtained from the CoLaus study (http://www.colaus.ch/) (42).
DNA sequencing and statistical analysis
Polymerase chain reaction (PCR) primers for amplification of the 16 exons and flanking intron sequences of PRDM5 were designed using Primer3 (v. 0.4.0) software (http://frodo.wi.mit.edu/primer3/) (43) and are listed in Supplementary Material, Table S3. PCR and Sanger sequencing of ZNF469 was undertaken with primers identical to those previously used by Christensen et al. (29) (personal communication) with adapted conditions (Supplementary Material, Table S4). Sequencing results were analysed manually using the sequence analysis software SeqScape 2.1.1 (Applied Biosystems, USA). Identified sequence variants were described according to the guidelines published by the Human Genome Variation Society. Variants were annotated in accordance with Ensembl transcript ENST00000437464 or NCBI NM_001127464.1 (Build GRCh37/hg19). The sequence variants were passed through a series of filtering steps shown in Figure 2.
If the sequence variants were present in the ethnically matched population-specific control data, they were excluded. The remaining sequence variants were required to have an MAF of <0.1% in the data from dbSNP (Build 137), the May 2012 release of the 1KG Project and the EVS and NHLBI ESP. Following this, the remaining non-synonymous alleles were filtered using SIFT which can identify if an amino acid substitution influences protein function resulting in a phenotypic change; classified as damaging or tolerated (44). SIFT can distinguish between functionally neutral and deleterious amino acid changes in mutagenesis studies and on human polymorphisms (45,46). There are alternative prediction tools that use a combination of methods based on sequence homology, protein structure information and physicochemical properties of amino acids for prediction (44). Given that ZNF469 is a poorly characterized protein with no verified structural homologues, the SIFT algorithm was applied as SIFT computes a combined score derived from the distribution of amino acid residues observed at a given position in the sequence alignment and the estimated unobserved frequencies of amino acid distribution calculated from a Dirichlet mixture and does not rely on structural or physiochemical information (44). The conservation of the affected amino acid across species was analysed using Homologene (http://www.ncbi.nlm.nih.gov/homologene/) and multiple sequence alignment with ClustalX (http://www.clustal.org/) visualized with GeneDoc software (http://www.nrbsc.org/gfx/genedoc/).
The collapsing method (47) was used to compare the frequency of remaining potentially pathogenic alleles between case and control subjects with the level of significance set to P< 0.05. This method involves collapsing genotypes across variants and applying a univariate test which is powerful for analysing rare variants (47). Specifically, each individual was assigned an indicator variable that takes the value one if the subject carries at least one potentially pathogenic variant and zero otherwise. Whether the proportions of individuals with index variable one differ significantly in cases and controls were tested using a Fisher exact test on the corresponding contingency table of indicator variable counts. The estimated OR, RR, 95% CI and Fisher's exact P-value were calculated using JavaStat (http://statpages.org/ctab2x2.html).
Web Resources
1000 Genomes Project: http://browser.1000genomes.org/index.html.
Exome Variant Server, NHLBI Exome Sequencing Project, Seattle, WA: http://eversusgs.washington.edu/EVS/.
NCBI dbSNP: http://www.ncbi.nlm.nih.gov/snp.
Fisher's exact test calculations: http://statpages.org/ctab2x2.html.
Sorting Intolerant from Tolerant (SIFT: http://sift.jcvi.org/.
Online Mendelian Inheritance in Man (OMIM): http://www.omim.org/.
Homologene: http://www.ncbi.nlm.nih.gov/homologene/.
ClustalX: http://www.clustal.org/.
GeneDoc software: http://www.nrbsc.org/gfx/genedoc/.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Northern Ireland Research and Development Office RRG (grant number 4.46 to C.E.W.); Fight for Sight (grant number 1787 to C.E.W. and J.L.) and the NIHR Manchester Biomedical Research Centre (L.F.P., F.D.M. and G.C.B.). J.L. was a Fight for Sight (UK) PhD student. We are grateful for access to exome sequence data from the CoLaus cohort, which was sequenced as part of a partnership between the Wellcome Trust Sanger Institute, the CoLaus principal investigators and the Quantitative Sciences Department of GlaxoSmithKline.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to the patients who participated in this study. We thank Eyvind Rødahl from the University of Bergen, Norway for providing the sequence of the ZNF469 primers. Special thanks to Drs S. George, D.G. Frazer, U. Donnelly and J.E. Moore (Belfast Health and Social Care Trust, UK) for assistance with DNA collection and recruitment.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rabinowitz Y.S. Keratoconus. Surv. Ophthalmol. 1998;42:297–319. doi: 10.1016/s0039-6257(97)00119-7. [DOI] [PubMed] [Google Scholar]
- 2.Rahman I., Carley F., Hillarby C., Brahma A., Tullo A.B. Penetrating keratoplasty: indications, outcomes, and complications. Eye (Lond.) 2009;23:1288–1294. doi: 10.1038/eye.2008.305. [DOI] [PubMed] [Google Scholar]
- 3.Pearson A.R., Soneji B., Sarvananthan N., Sandford-Smith J.H. Does ethnic origin influence the incidence or severity of keratoconus? Eye (Lond.) 2000;14:625–628. doi: 10.1038/eye.2000.154. [DOI] [PubMed] [Google Scholar]
- 4.Edwards M., McGhee C.N., Dean S. The genetics of keratoconus. Clin. Exp. Ophthalmol. 2001;29:345–351. doi: 10.1046/j.1442-9071.2001.d01-16.x. [DOI] [PubMed] [Google Scholar]
- 5.Tuft S.J., Hassan H., George S., Frazer D.G., Willoughby C.E., Liskova P. Keratoconus in 18 pairs of twins. Acta Ophthalmol. 2012;90:e482–e486. doi: 10.1111/j.1755-3768.2012.02448.x. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Rabinowitz Y.S., Rotter J.I., Yang H. Genetic epidemiological study of keratoconus: evidence for major gene determination. Am. J. Med. Genet. 2000;93:403–409. [PubMed] [Google Scholar]
- 7.Fontes B.M., Ambrosio R., Jr, Jardim D., Velarde G.C., Nose W. Corneal biomechanical metrics and anterior segment parameters in mild keratoconus. Ophthalmology. 2010;117:673–679. doi: 10.1016/j.ophtha.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 8.Doughty M.J., Zaman M.L. Human corneal thickness and its impact on intraocular pressure measures: a review and meta-analysis approach. Surv. Ophthalmol. 2000;44:367–408. doi: 10.1016/s0039-6257(00)00110-7. [DOI] [PubMed] [Google Scholar]
- 9.Dimasi D.P., Burdon K.P., Craig J.E. The genetics of central corneal thickness. Br. J. Ophthalmol. 2010;94:971–976. doi: 10.1136/bjo.2009.162735. [DOI] [PubMed] [Google Scholar]
- 10.Vitart V., Bencic G., Hayward C., Skunca Herman J., Huffman J., Campbell S., Bucan K., Navarro P., Gunjaca G., Marin J., et al. New loci associated with central cornea thickness include COL5A1, AKAP13 and AVGR8. Hum. Mol. Genet. 2010;19:4304–4311. doi: 10.1093/hmg/ddq349. [DOI] [PubMed] [Google Scholar]
- 11.Vithana E.N., Aung T., Khor C.C., Cornes B.K., Tay W.T., Sim X., Lavanya R., Wu R., Zheng Y., Hibberd M.L., et al. Collagen-related genes influence the glaucoma risk factor, central corneal thickness. Hum. Mol. Genet. 2011;20:649–658. doi: 10.1093/hmg/ddq511. [DOI] [PubMed] [Google Scholar]
- 12.Lu Y., Dimasi D.P., Hysi P.G., Hewitt A.W., Burdon K.P., Toh T., Ruddle J.B., Li Y.J., Mitchell P., Healey P.R., et al. Common genetic variants near the brittle cornea syndrome locus ZNF469 influence the blinding disease risk factor central corneal thickness. PLoS Genet. 2010;6:e1000947. doi: 10.1371/journal.pgen.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu Y., Vitart V., Burdon K.P., Khor C.C., Bykhovskaya Y., Mirshahi A., Hewitt A.W., Koehn D., Hysi P.G., Ramdas W.D., et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat. Genet. 2013;45:155–163. doi: 10.1038/ng.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoehn R., Zeller T., Verhoeven V.J., Grus F., Adler M., Wolfs R.C., Uitterlinden A.G., Castagne R., Schillert A., Klaver C.C., et al. Population-based meta-analysis in Caucasians confirms association with COL5A1 and ZNF469 but not COL8A2 with central corneal thickness. Hum. Genet. 2012;131:1783–1793. doi: 10.1007/s00439-012-1201-3. [DOI] [PubMed] [Google Scholar]
- 15.Burkitt Wright E.M., Spencer H.L., Daly S.B., Manson F.D., Zeef L.A., Urquhart J., Zoppi N., Bonshek R., Tosounidis I., Mohan M., et al. Mutations in PRDM5 in brittle cornea syndrome identify a pathway regulating extracellular matrix development and maintenance. Am. J. Hum. Genet. 2011;88:767–777. doi: 10.1016/j.ajhg.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Al-Hussain H., Zeisberger S.M., Huber P.R., Giunta C., Steinmann B. Brittle cornea syndrome and its delineation from the kyphoscoliotic type of Ehlers-Danlos syndrome (EDS VI): report on 23 patients and review of the literature. Am. J. Med. Genet. A. 2004;124A:28–34. doi: 10.1002/ajmg.a.20326. [DOI] [PubMed] [Google Scholar]
- 17.Abu A., Frydman M., Marek D., Pras E., Nir U., Reznik-Wolf H. Deleterious mutations in the Zinc-Finger 469 gene cause brittle cornea syndrome. Am. J. Hum. Genet. 2008;82:1217–1222. doi: 10.1016/j.ajhg.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krumeich J.H., Daniel J., Knulle A. Live-epikeratophakia for keratoconus. J. Cataract. Refract. Surg. 1998;24:456–463. doi: 10.1016/s0886-3350(98)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Choi J.A., Kim M.S. Progression of keratoconus by longitudinal assessment with corneal topography. Invest. Ophthalmol. Vis. Sci. 2012;53:927–935. doi: 10.1167/iovs.11-8118. [DOI] [PubMed] [Google Scholar]
- 20.Dash D.P., George S., O’Prey D., Burns D., Nabili S., Donnelly U., Hughes A.E., Silvestri G., Jackson J., Frazer D., et al. Mutational screening of VSX1 in keratoconus patients from the European population. Eye (Lond.) 2010;24:1085–1092. doi: 10.1038/eye.2009.217. [DOI] [PubMed] [Google Scholar]
- 21.De Bonis P., Laborante A., Pizzicoli C., Stallone R., Barbano R., Longo C., Mazzilli E., Zelante L., Bisceglia L. Mutational screening of VSX1, SPARC, SOD1, LOX, and TIMP3 in keratoconus. Mol. Vis. 2011;17:2482–2494. [PMC free article] [PubMed] [Google Scholar]
- 22.Udar N., Atilano S.R., Brown D.J., Holguin B., Small K., Nesburn A.B., Kenney M.C. SOD1: a candidate gene for keratoconus. Invest. Ophthalmol. Vis. Sci. 2006;47:3345–3351. doi: 10.1167/iovs.05-1500. [DOI] [PubMed] [Google Scholar]
- 23.Hughes A.E., Bradley D.T., Campbell M., Lechner J., Dash D.P., Simpson D.A., Willoughby C.E. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am. J. Hum. Genet. 2011;89:628–633. doi: 10.1016/j.ajhg.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lechner J., Dash D.P., Muszynska D., Hosseini M., Segev F., George S., Frazer D.G., Moore J.E., Kaye S.B., Young T., et al. Mutational spectrum of the ZEB1 gene in corneal dystrophies supports a genotype-phenotype correlation. Invest. Ophthalmol. Vis. Sci. 2013;54: 3215–3223. doi: 10.1167/iovs.13-11781. [DOI] [PubMed] [Google Scholar]
- 25.Stabuc-Silih M., Strazisar M., Hawlina M., Glavac D. Absence of pathogenic mutations in VSX1 and SOD1 genes in patients with keratoconus. Cornea. 2010;29:172–176. doi: 10.1097/ICO.0b013e3181aebf7a. [DOI] [PubMed] [Google Scholar]
- 26.Li X., Bykhovskaya Y., Haritunians T., Siscovick D., Aldave A., Szczotka-Flynn L., Iyengar S.K., Rotter J.I., Taylor K.D., Rabinowitz Y.S. A genome-wide association study identifies a potential novel gene locus for keratoconus, one of the commonest causes for corneal transplantation in developed countries. Hum. Mol. Genet. 2012;21:421–429. doi: 10.1093/hmg/ddr460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burdon K.P., Macgregor S., Bykhovskaya Y., Javadiyan S., Li X., Laurie K.J., Muszynska D., Lindsay R., Lechner J., Haritunians T., et al. Association of polymorphisms in the hepatocyte growth factor gene promoter with keratoconus. Invest. Ophthalmol. Vis. Sci. 2011;52:8514–8519. doi: 10.1167/iovs.11-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khan A.O., Aldahmesh M.A., Mohamed J.N., Alkuraya F.S. Blue sclera with and without corneal fragility (brittle cornea syndrome) in a consanguineous family harboring ZNF469 mutation (p.E1392X) Arch. Ophthalmol. 2010;128:1376–1379. doi: 10.1001/archophthalmol.2010.238. [DOI] [PubMed] [Google Scholar]
- 29.Christensen A.E., Knappskog P.M., Midtbo M., Gjesdal C.G., Mengel-From J., Morling N., Rodahl E., Boman H. Brittle cornea syndrome associated with a missense mutation in the zinc-finger 469 gene. Invest. Ophthalmol. Vis. Sci. 2010;51:47–52. doi: 10.1167/iovs.09-4251. [DOI] [PubMed] [Google Scholar]
- 30.Tadepally H.D., Burger G., Aubry M. Evolution of C2H2-zinc finger genes and subfamilies in mammals: species-specific duplication and loss of clusters, genes and effector domains. BMC Evol. Biol. 2008;8:176. doi: 10.1186/1471-2148-8-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuh R., Aicher W., Gaul U., Cote S., Preiss A., Maier D., Seifert E., Nauber U., Schroder C., Kemler R., et al. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. [DOI] [PubMed] [Google Scholar]
- 32.Laity J.H., Lee B.M., Wright P.E. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 33.Critchfield J.W., Calandra A.J., Nesburn A.B., Kenney M.C. Keratoconus: I. Biochemical studies . Exp. Eye. Res. 1988;46:953–963. doi: 10.1016/s0014-4835(88)80047-2. [DOI] [PubMed] [Google Scholar]
- 34.Kenney M.C., Nesburn A.B., Burgeson R.E., Butkowski R.J., Ljubimov A.V. Abnormalities of the extracellular matrix in keratoconus corneas. Cornea. 1997;16:345–351. [PubMed] [Google Scholar]
- 35.Evereklioglu C., Madenci E., Bayazit Y.A., Yilmaz K., Balat A., Bekir N.A. Central corneal thickness is lower in osteogenesis imperfecta and negatively correlates with the presence of blue sclera. Ophthalmic. Physiol. Opt. 2002;22:511–515. doi: 10.1046/j.1475-1313.2002.00062.x. [DOI] [PubMed] [Google Scholar]
- 36.Quigley H.A., Broman A.T. The number of people with glaucoma worldwide in 2010 and 2020. Br. J. Ophthalmol. 2006;90:262–267. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gordon M.O., Beiser J.A., Brandt J.D., Heuer D.K., Higginbotham E.J., Johnson C.A., Keltner J.L., Miller J.P., Parrish R.K., 2nd, Wilson M.R., et al. The ocular hypertension treatment study: baseline factors that predict the onset of primary open-angle glaucoma. Arch. Ophthalmol. 2002;120:714–720. doi: 10.1001/archopht.120.6.714. discussion 829–730. [DOI] [PubMed] [Google Scholar]
- 38.Dueker D.K., Singh K., Lin S.C., Fechtner R.D., Minckler D.S., Samples J.R., Schuman J.S. Corneal thickness measurement in the management of primary open-angle glaucoma: a report by the American Academy of Ophthalmology. Ophthalmology. 2007;114:1779–1787. doi: 10.1016/j.ophtha.2007.04.068. [DOI] [PubMed] [Google Scholar]
- 39.Leske M.C., Heijl A., Hyman L., Bengtsson B., Dong L., Yang Z. Predictors of long-term progression in the early manifest glaucoma trial. Ophthalmology. 2007;114:1965–1972. doi: 10.1016/j.ophtha.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Johansen C.T., Wang J., Lanktree M.B., Cao H., McIntyre A.D., Ban M.R., Martins R.A., Kennedy B.A., Hassell R.G., Visser M.E., et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat. Genet. 2010;42:684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gloyn A.L., McCarthy M.I. Variation across the allele frequency spectrum. Nat. Genet. 2010;42:648–650. doi: 10.1038/ng0810-648. [DOI] [PubMed] [Google Scholar]
- 42.Firmann M., Mayor V., Vidal P.M., Bochud M., Pecoud A., Hayoz D., Paccaud F., Preisig M., Song K.S., Yuan X., et al. The CoLaus study: a population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc. Disord. 2008;8:6. doi: 10.1186/1471-2261-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 44.Kumar P., Henikoff S., Ng P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 45.Ng P.C., Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ng P.C., Henikoff S. Accounting for human polymorphisms predicted to affect protein function. Genome Res. 2002;12:436–446. doi: 10.1101/gr.212802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li B., Leal S.M. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am. J. Hum. Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.