Abstract
Fibrodysplasia ossificans progressiva (FOP) is a disabling genetic disorder of progressive heterotopic ossification (HO). Here, we report a patient with an ultra-rare point mutation [c.619C>G, p.Q207E] located in a codon adjacent to the most common FOP mutation [c.617G>A, p.R206H] of Activin A Receptor, type 1 (ACVR1) and that affects the same intracellular amino acid position in the GS activation domain as the engineered constitutively active (c.a.) variant p.Q207D. It was predicted that both mutations at residue 207 have similar functional effects by introducing a negative charge. Transgenic p.Q207D-c.a. mice have served as a model for FOP HO in several in vivo studies. However, we found that the engineered ACVR1Q207D−c.a. is significantly more active than the classic FOP mutation ACVR1R206H when overexpressed in chicken limbs and in differentiation assays of chondrogenesis, osteogenesis and myogenesis. Importantly, our studies reveal that the ACVR1Q207E resembles the classic FOP receptor in these assays, not the engineered ACVR1Q207D−c.a.. Notably, reporter gene assays revealed that both naturally occurring FOP receptors (ACVR1R206H and ACVR1Q207E) were activated by BMP7 and were sensitive to deletion of the ligand binding domain, whereas the engineered ACVR1Q207D−c.a. exhibited ligand independent activity. We performed an in silico analysis and propose a structural model for p.Q207D-c.a. that irreversibly relocates the GS domain into an activating position, where it becomes ligand independent. We conclude that the engineered p.Q207D-c.a. mutation has severe limitations as a model for FOP, whereas the naturally occurring mutations p.R206H and p.Q207E facilitate receptor activation, albeit in a reversible manner.
INTRODUCTION
Fibrodysplasia ossificans progressiva (FOP, MIM #135100) is a rare autosomal dominant genetic disorder. Ectopic bone forms progressively within soft tissues throughout the life to an extent that is unique in diseases of heterotopic ossification (HO) (1,2). FOP patients are born without evidence of ectopic bone but can be diagnosed by congenital great toe malformation. This feature, together with postnatal progressive HO that appears in a characteristic anatomic pattern, defines the classic FOP phenotype. Additional common but variable FOP features are frequently found in patients with a classic phenotype (3), including malformations of the thumb (in about 50% of all FOP patients) or the cervical spine (>80%), conductive hearing impairment (>50%), short and broad femoral necks (>70%) and tibial osteochondromas (>90%). Patients possessing a classic phenotype typically carry an p.R206H mutation in the cytoplasmic GS domain of the cell surface receptor Activin A receptor type I (ACVR1; alias ALK2) that leads to overactivation of the bone morphogenetic protein (BMP) signalling pathway (3,4). Subgroups of the FOP phenotypic spectrum were proposed based on additional (FOP plus) or atypical (FOP variant) features (3). The FOP variant group was associated with non-R206H mutations within the ACVR1 receptor, whereas the FOP plus group predominantly carries the classical p.R206H mutation and additional features are thought to be caused by other genetic and/or environmental influences (3).
ACVR1 is one of the four known BMP type I receptors (ACVRL1, ACVR1, BMPR1A, BMPR1B) that binds ligands of the TGFβ superfamily, such as BMP6 and BMP7. BMPs are secreted, multifunctional growth factors that were initially discovered due to their bone-inductive property but are now recognized to be key regulators of embryonic development, tissue homeostasis and regeneration of several organs and deregulation of the BMP signalling pathway is associated with a number of different human diseases (reviewed in 5).
Upon ligand binding, the BMP type I receptors are activated via transphosphorylation by BMP type II receptors (ACVR1A, ACVR2B, BMPR2). The type I receptor then transduces the signal by activating downstream cascades either in the SMAD pathway or the MAPK pathway, subsequently leading to induction or repression of target gene transcription (6,7).
The FOP mutations p.Q207E and p.R206H introduce an amino acid substitution in the GS domain of ACVR1, which plays a central role in the receptor transactivation by type II receptors and is highly conserved among all known type I receptors. The GS domain consists of two helices that are interconnected by a flexible loop which forms a tightly folded inhibitory wedge that is buried between the αC-helix and the N-lobe of the kinase β-sheets. This inactive receptor conformation sterically hinders access to the ATP binding pocket and prevents movement of the receptor into its active conformation (7), which is stabilized by binding of FK506 binding protein 12 (FKBP12) to the unphosphorylated GS domain (8).
Phosphorylation of serine and threonine residues within the GS domain induces a conformational change of the intracellular domain transforming the receptor conformation into an active state that releases FKBP12 and promotes SMAD binding and phosphorylation (9,10). An engineered ACVR1 p.Q207D mutation generates a constitutively activated (c.a.) receptor, which will be henceforth referred to as p.Q207D-c.a. (11–13). This variant was designed based on in vitro analyses of the closely related TGFBR1 (transforming growth factor β receptor 1; alias: ALK5) receptor which revealed that replacement of the last threonine amino acid in the GS domain with a negatively charged aspartic acid is sufficient to induce constitutive activation of TGFβ signalling (14). Since both p.Q207D and p.Q207E replace the neutral amino acid glutamine 207 by a negatively charged residue (aspartic acid in p.Q207D and glutamic acid in p.Q207E), these mutations were thought to have similar functional effects on BMP signalling and the conditional Acvr1Q207D−c.a.-overexpressing transgenic mice has served as an in vivo model for FOP (15,16).
Here, we analysed the rare, naturally occurring p.Q207E and the common p.R206H mutation and identified drastic functional differences when compared with the engineered p.Q207D-c.a. mutation.
RESULTS
The ACVR1 p.Q207E mutation causes a classic FOP clinical phenotype
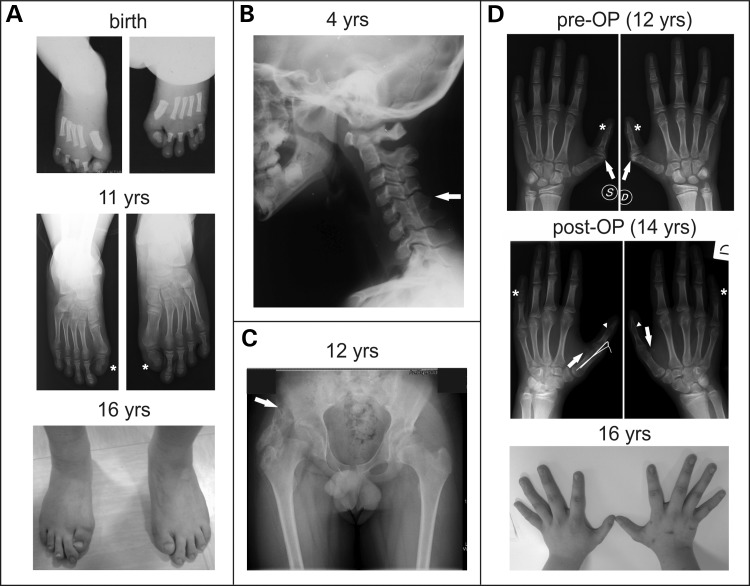
We report a male patient who was diagnosed with FOP at the age of 12. Bilateral malformations of the great toes and thumbs were recognized at birth, but not correlated with FOP at that time. Great toes showed monophalangism, were shortened and displayed a notable valgus deviation (Fig. 1A). Auxological parameters at birth and neuro-psychological development were described as clinically inconspiciuous apart from a documented slight delay in speech development. Neurological examination revealed no abnormalities except for the presence of a bilateral mild conductive hearing impairment. Fusion of cervical vertebrae C5–C6 was radiographically noticed at 4 years of age (Fig. 1B). The first HO occurred at the age of 12 located at the right hip after a mild traumatic event (Fig. 1C) and was followed by a 1 year quiescent phase, when no additional bone was formed.
Figure 1.
ACVR1 missense mutation p.Q207E is associated with a classic FOP phenotype. Clinical phenotype of a FOP patient carrying the p.Q207E mutation in ACVR1. (A) Radiographs and clinical picture of feet showing bilateral hallux valgus deformation and monophlangism (asterisk in middle panel) of great toes, which is characteristic for classically affected FOP patients. (B) Radiograph of the neck demonstrating orthotopic fusion of the facet joints between cervical vertebrae C5–C6 (indicated by arrow). (C) Radiograph of the hip indicating a large area of HO, which occurred 6 months after mild trauma at the age of 12 years, connecting the femoral neck with the hip (indicated by arrow). (D) Radiographs and clinical picture of hands showing bilateral dimorphism of the thumbs. X-ray analysis revealed abnormality of articular cartilage of the metacarpal–phalangeal (M.P.) joint (upper panel, arrow) leading to a clinical finding of camptodactyly which interfered with function (upper panel, asterisk). Fusion of M.P. joint of the thumb in neutral position resulted in a much wider opening of the hand which significantly improved function (middle panel, arrowhead). Additionally, mild bilateral abnormality of sesamoid bone shape in metacarpal bone was observed (arrow, middle panel). Bilateral malformation of the middle phalanges was found in the 5th digit with shortening of the overall size and broadening of the diaphysis (middle panel, asterisk).
Due to increasing disability in both thumbs, the patient underwent surgery to restore normal physiology and motion range. X-ray investigation prior to surgery revealed bilateral abnormality of interphalangeal joint formation between metacarpal and both phalanges. Malformation of the articular cartilage in the epiphysis of proximal phalanx resulted in a slightly broader joint space (Fig. 1D, arrow in upper panel), whereas irregularity in the distal portion of the proximal phalanx and the proximal portion of the distal phalanx led to a narrower interphalangeal joint (Fig. 1D, asterisk, upper panel). The shape of the sesamoid bone in the metacarpal epiphyses appeared to be slightly broader than normal (Fig. 1D, arrow, middle panel). In addition, middle phalanges of the fifth digits were found to be bilateral malformed with shortening of the overall size and thickening of the diaphysis (Fig. 1D, asterisks, middle panel).
Surgical metacarpal–phalangeal arthrodesis of the right thumb was performed according to up-dated guidelines of anaesthesiological management of FOP patients (17). Surgery was successful without provoking new HO formation and the left thumb was operated on 2 years later.
Based on these clinical findings, the overall phenotype of this patient can be classified as classic FOP. Although most patients with classic FOP have an ACVR1 c.617G>A (p.R206H) mutation, genetic analysis of this patient and his parents revealed a de novo heterozygous missense mutation in the ACVR1 gene at c.619C>G (RefSeq NM_001111067.2) which is predicted to cause a substitution of the neutral amino acid glutamine at codon 207 with the negatively charged glutamate (p.Q207E).
The ACVR1 mutation p.Q207E is located within the highly conserved GS domain and affects the same residue as the engineered constitutively active ACVR1 mutation p.Q207D
The human ACVR1 receptor consists of 509 amino acids and is subdivided into four functional units comprising a ligand binding domain (LBD), a single-pass transmembrane domain, a GS domain and a kinase domain. Alignment of the GS domain amino acid sequence among the seven known human TGFβ/BMP type I receptors shows high conservation within the GS motif (Fig. 2A). Protein alignment of ACVR1 also revealed a sequence conservation of the GS domain between human, mouse and chicken (Fig. 2A). The most frequent FOP-causing point mutation p.R206H is located at the C-terminal end of the GS domain within the α2-helix. The terminal glutamine (Q) residue of the GS box is replaced by aspartic acid in the constitutively active Q207D-c.a. (14). The FOP mutation p.Q207E alters the same amino acid and also introduces a negative charge by substitution of the glutamine (Q) with glutamic acid (E).
Figure 2.

Schematic representation of FOP mutations p.Q207E and p.R206H located within the GS domain of ACVR1. (A) Left: Schematic of ACVR1 indicating functional domains. LBD, ligand binding domain; TM, transmembrane domain; GS, glycine–serine rich domain; KD, kinase domain. Right: Amino acid alignment of the ACVR1 GS domain from different species (Hs: Homo sapiens, Mm: Mus musculus, Gg: Gallus gallus) and type I receptors of the TGFβ superfamily. Positions of the FOP-associated mutations p.R206H and p.Q207E are indicated at the top of the protein alignment. (B and C) In silico binding study identifies amino acids that are involved in intramolecular complex formation as well as binding to the inhibitor FKBP12. Alanine mutations of residues within the ACVR1(KD):ACVR1(GS) or the ACVR1(GS, KD):FKBP12 interface reduce (ΔΔG > 1) or facilitate binding (ΔΔG < 1). Changes of total free energy (ΔΔG) are colour coded and mapped on the contact surfaces (PDB ID: 3H9R). Side chains of R206 and Q207 contribute to FKBP12 but not to GS binding. (B) The GS domain residues L180 and V204 form a hydrophobic cluster with I266 and W227 of the kinase domain, while S194 and R258 make several main chain connections to maintain GS binding. R206 forms hydrogen bonds with D269 and M270 (data not shown), whereas Q207 forms a hydrogen bond with W227, but while W227 and D269 are essential hot spots for GS domain binding, both residues (R206 and Q207) are only marginally involved in the interaction of the GS domain with the kinase domain. (C) ACVR1(GS, KD):FKBP12 complex formation requires four residues of FKBP12 (R43, F47, P89 and F100) and two residues of ACVR1 (F198 and L199). R206 and Q207 do interact with FKBP12, but only slightly contribute to this interaction.
To identify residues that are important for interactions between the GS domain and the kinase domain or for FKBP12 binding to ACVR1, we performed an alanine scan (18). Changes in free energy (ΔΔG) were mapped on the contact surfaces of the kinase and GS domain of ACVR1 (Fig. 2B) as well as on the intracellular contact surface of the ACVR1 GS and kinase domain (ACVR1(GS, KD)) and FKBP12 (Fig. 2C).
Our results support that L180 and V204 in the GS domain form a hydrophobic cluster with I266 and W227 of the kinase domain, while S194 in the GS domain and R258 in the kinase domain make several main chain connections to stabilize an inactive receptor conformation.
The ACVR1(GS, KD):FKBP12 complex engages four residues of FKBP12 (R43, F47, P89 and F100) and two residues of ACVR1 (F198 and L199). Interestingly, ACVR1 side chains of R206 and Q207 facilitate FKBP12 binding to the GS domain (Fig. 2C). These residues are directly coupled and amino acid changes would affect the side chain orientation of the neighbouring residue.
We calculated the changes of free binding energies (ΔΔG) induced by each individual receptor mutation in the crystal structure of ACVR1 (PDB ID:3H9R), alone or bound to FKBP12 (Table 1), and found that p.Q207E as well as p.Q207D show reduced binding capacity to FKBP12, which is caused by charge repulsion between both molecules. The mutation p.R206H, however, does not directly interfere with FKBP12 binding.
Table 1.
Structure and function relation of FOP mutations on FKBP12 binding
| ACVR1 variant | +FKBP12 | −FKBP12 | Effect | Activation by reduced FKBP12 binding |
|---|---|---|---|---|
| ΔΔG (kcal/M) | ||||
| p.R206H | 0.62 | 0.32 | Steric hindrance | 3-fold (19) |
| p.Q207D | 2.47 | −0.70 | Charge repulsion | No binding (10) |
| p.Q207E | 2.45 | 0.40 | Charge repulsion | No binding (10) |
The changes in free binding energy (ΔΔG) were calculated for ACVR1 (WT, p.R206H, p.Q207E, p.Q207D-c.a.) with or without FKBP12 by FOLD-X. Both, p.Q207D and p.Q207E, show an increase in ΔΔG on the complex formation with FKBP12, whereas p.R206H has only a marginal effect comparable to experimental findings (19).
In order to evaluate the biological activity of p.Q207E, we performed functional assays to compare this receptor to the classic FOP mutation p.R206H and the engineered p.Q207D-c.a. variant.
ACVR1Q207E induces ectopic cartilage and digit malformation during embryonic development in chicken
We used the chicken as an in vivo model to study the consequences of FOP mutations on embryonic limb development. ACVR1 variants were overexpressed in chicken limbs at day 1.5 and skeletal development was analysed by staining after 10 days. At this time point (HH37/38), most skeletal elements exist as cartilaginous anlagen with only femur, tibia and metatarsals showing primary ossification centres. The left, non-injected limb served as an internal control for normal development. A representative non-injected limb is shown where all phalanges (p1–p3) are visible (Fig. 3A). Viral overexpression of the ACVR1WT receptor had almost no influence on skeletal development. Out of 34, 32 injected embryos displayed no phenotypical differences between injected and control extremities during embryonic development (Fig. 3B). In contrast, overexpression of ACVR1Q207D−c.a. caused severe skeletal malformations in >90% of injected embryos with large ectopic cartilage condensations in all major limb joints (Fig. 3C). Digits as well as the knee were preferentially affected but the ankle joint and pelvic surface also developed extra skeletal cartilage (data not shown). Massive thickening of skeletal elements, e.g. femur, metatarsals or phalanges, was also evident. Fusions of adjacent elements were frequently found, especially in the toes (Fig. 3C). In addition, ectopic cartilage that was not connected to skeletal elements was formed within muscle tissue, after injecting the engineered mutation ACVR1Q207D−c.a. (data not shown). Thirty-four percent of all ACVR1Q207D−c.a. injected embryos developed a malformation of the phalanx distalis in one or more toes.
Figure 3.
FOP mutation ACVR1Q207E induces ectopic cartilage formation and joint fusion during chicken limb development. Skeletal stainings of hind limbs at day 10. (A) Left hind limbs were non-infected and served as internal controls. Right hind limb fields of HH10 embryos were injected with RCASBP retrovirus containing either (B) ACVR1WT, (C) ACVR1Q207D−c.a., (D) ACVR1R206H or (E) ACVR1Q207E. Upper panel: Dorsal view of the foot. Middle panel: Magnification of the second digit. Phalanges (p1–p3) in control and ACVR1WT digits are clearly separated by interphalangeal joints (arrows in control digit). Severe thickening of skeletal elements and monophalangism was frequently found in ACVR1Q207D−c.a. embryos. Both FOP mutations ACVR1R206H and ACVR1Q207E caused ectopic formation of cartilaginous elements in interphalangeal joints (arrowhead in ACVR1Q207E) or fusion of adjacent elements (asterisks) up to monophalangism. Nail dysplasia (arrow) was found to a variable degree in ACVR1Q207D−c.a. and both FOP variants. Lower panel: Frontal higher magnification view of the knee. The joint space between femur and tibia is clearly visible in the control and ACVR1WT limb. Ectopic cartilage masses surrounding the knee joint were found in ACVR1Q207D−c.a. embryos. Formation of ectopic cartilage within the joint space was also found in the FOP mutants (arrows). Scale bars correspond to 1 mm in length. Total numbers of injected embryos are depicted in the lower right corner. Mt, metatarsus; p, phalanx.
The classic ACVR1R206H mutation induced ectopic cartilage formation in 87% of injected embryos that, in contrast to ACVR1Q207D−c.a.-mediated effects, was restricted to the knee (60%) and interphalangeal joints (∼72%). We observed interphalangeal joint fusions (Fig. 3D) and loose, cartilaginous nodules (Fig. 3E). Only a small fraction of the ACVR1R206H injected embryos (∼5%) developed a malformed phalanx distalis.
ACVR1Q207E induced ectopic cartilage formation in the knee (∼60%) and toe joints (75%) (Fig. 3E) to a comparable degree as found in ACVR1R206H. Interestingly, compared with ACVR1R206H, ACVR1Q207E induced a significant increase of phalanx distalis malformation (∼27%), similar to the rate in ACVR1Q207D−c.a. injected embryos.
Taken together, expression of the FOP mutations p.Q207E and p.R206H resulted in milder skeletal phenotype effects compared with the p.Q207D-c.a.
ACVR1Q207E inhibits muscle differentiation and enhances chondrogenesis and osteogenesis in vitro
Induction of HO in FOP patients occurs either spontaneously or in response to trauma and leads to necrosis of the affected skeletal muscle and subsequent replacement with ectopic bone via endochondral ossification, a process that includes a gradual replacement of a cartilaginous template by bone. We therefore aimed to investigate the influence of the FOP mutations (p.R206H, p.Q207E) on the differentiation of various cell types such as myoblasts, chondrocytes and osteoblasts in comparison with p.Q207D-c.a.
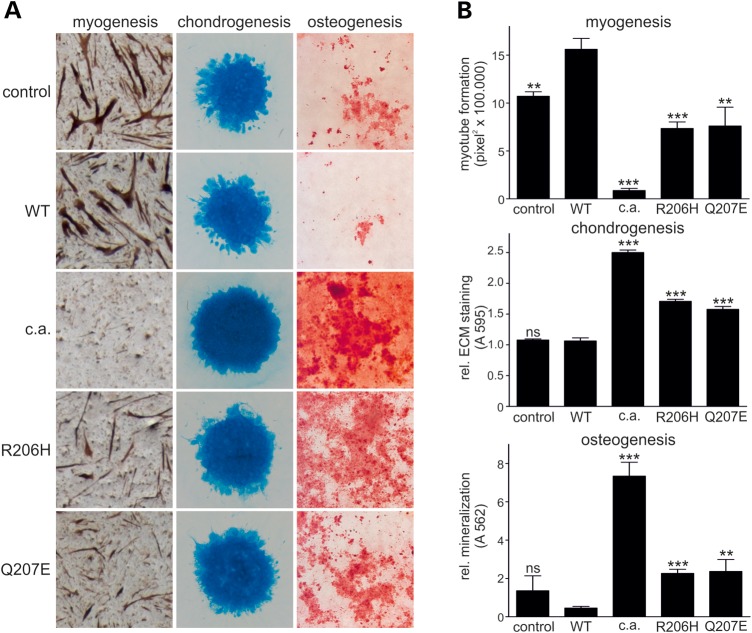
In order to analyse myogenic differentiation, we used the QM7 cell line (20). Retroviral overexpression of ACVR1WT caused increased myotube formation when compared with cells infected with an empty vector control virus (Fig. 4A). In contrast, ACVR1Q207D−c.a. completely prevented formation of multinucleated myotubes, as almost no myosin-staining was visible (Fig. 4A) and the remaining myosin-positive cells did not possess typical muscle cell shape. Both FOP mutations, ACVR1Q207E and ACVR1R206H, significantly inhibited the formation of myotubes, reducing the total number and size of muscle fibres, as determined by myosin-staining and histomorphometric analysis (Fig. 4B); however, the inhibitory effect was reduced compared with cells expressing ACVR1Q207D−c.a..
Figure 4.
FOP mutation ACVR1Q207E inhibits myogenesis and enhances chondrogenic and osteogenic differentiation in vitro. QM7 cells (for myogenesis), chicken micromass cultures (for chondrogenesis) or chicken BMSCs (for osteogenesis) were infected with either empty viral vector (control), ACVR1WT, ACVR1Q207D−c.a. or the indicated FOP mutation. Staining to detect myogenic, chondrogenic and osteogenic differentiation are depicted in (A) and respective quantifications are shown in (B). Myogenesis: Differentiation into myotubes was monitored 6 days after initiation of myoblast differentiation. Overexpression of ACVR1WT slightly enhanced myogenesis, whereas ACVR1Q207D−c.a. completely inhibited it. Myotube formation was significantly inhibited by both FOP mutations. Myotube formation is visualized by anti-myosin staining and quantified as pixel2 × 100.000 by histomorphometric analysis. Chondrogenesis: Micromass cultures were stained with Alcian blue to visualize cartilage formation after 7 days of induction. Cartilaginous matrix production was slightly enhanced by the FOP mutations while ACVR1Q207D−c.a.-infected micromass cultures produced excessive amounts of cartilage in comparison with control and ACVR1WT. Alcian blue staining of chicken micromass cultures was quantified by extraction and photometric quantification. Osteogenesis: BMSCs were stained with Alizarin red to visualize mineralization 7 days after initiation of differentiation into the osteogenic lineage. FOP mutations show significantly enhanced mineralization in BMSCs compared with ACVR1WT, but to a lower extent than ACVR1Q207D−c.a.. Alizarin red staining of BMSCs was quantified by extraction and photometric quantification. In (B), bars represent mean ± SD of quadruplicate samples from one representative of three independent experiments. The two-tailed student's t-test was performed. Significant differences in comparison with ACVR1WT are given as: **P < 0.01 or ***P < 0.001, ns not significant, n = 4.
To investigate the consequences of the FOP mutation p.Q207E on chondrogenic differentiation in vitro, we used the chicken micromass culture system (21). We infected embryonic limb bud-derived mesenchymal progenitor cells with a retrovirus (RCAS) expressing ACVR1WT, ACVR1Q207E, ACVR1R206H or ACVR1Q207D−c.a. and monitored cartilaginous matrix accumulation by Alcian blue staining and quantification (Fig. 4A and B). Expression of ACVR1WT had no significant impact on cartilage formation compared with empty vector control. Expression of ACVR1Q207E or ACVR1R206H led to a slight but statistically significant increase of matrix accumulation when compared with ACVR1WT, while cartilaginous matrix was strongly enhanced in cultures expressing ACVR1Q207D−c.a..
Next, we investigated the impact of the FOP-associated point mutations on osteogenic differentiation. Bone marrow stromal cells (BMSCs) were derived from 17-day-old chicken embryos. After initiation of osteogenesis, Alizarin red staining detected calcification (Fig. 4A). Overexpression of ACVR1WT had little influence on the onset and magnitude of extracellular matrix mineralization in comparison with the empty viral control. Elevated mineralization occurred with overexpression of the FOP mutations ACVR1R206H and ACVR1Q207E. Again, the effect of the constitutively active p.Q207D-c.a. variant was significantly stronger compared with either of the naturally occurring FOP mutations p.Q207E and p.R206H (Fig. 4A and B).
Prochondrogenic activity of ACVR1Q207E is inhibited by the BMP antagonist NOG
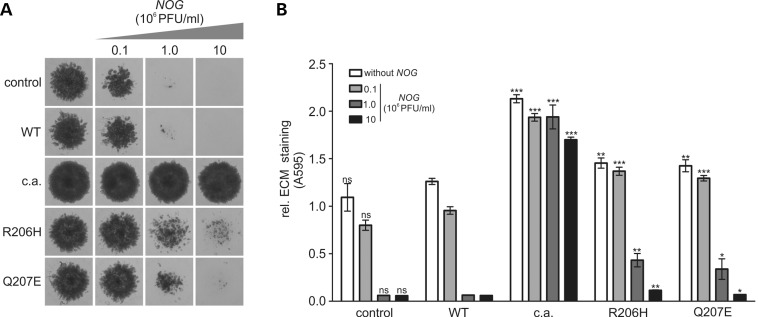
To test for a general ligand-dependent activation mechanism of FOP receptors, we co-infected micromass cultures with an ACVR1 variant (WT, p.Q207D-c.a., p.R206H or p.Q207E) (20 × 106 PFU/ml) and increasing concentrations of a retrovirus expressing the BMP antagonist NOG (0.1–10 × 106 PFU/ml). NOG is an extracellular antagonist that inhibits BMP signalling by preventing ligand binding (22). The formation of cartilaginous matrix in cultures infected with ACVR1WT receptor and control vector was reduced at the lowest NOG concentration and almost completely prevented at 1 × 106 PFU/ml (Fig. 5). Inhibition of chondrogenic matrix formation in cultures expressing FOP mutant receptors required higher concentration of NOG, with the chondrogenic effect of both FOP mutations almost completely inhibited using a NOG concentration of 10 × 106 PFU/ml. In contrast, cartilage formation in cultures expressing ACVR1Q207D−c.a. receptor was minimally altered with increasing NOG titres, indicating NOG resistance by this receptor.
Figure 5.
Prochondrogenic activity of ACVR1Q207E is suppressed by BMP antagonist NOG. (A) Chicken micromass cells were co-infected with RCAS virus encoding ACVR1WT, ACVR1Q207D−c.a. or indicated FOP mutation (viral titre 2 × 107 PFU/ml) and increasing titres of RCAS expressing NOG. Cultures were stained with Alcian blue after 7 days to visualize cartilage formation. While ACVR1Q207D−c.a-infected cultures produced extracellular cartilage matrix in the presence of NOG, control and ACVR1WT cultures were strongly inhibited by NOG. Cartilage matrix production of ACVR1Q207E and ACVR1R206H infected cultures was also inhibited with increasing NOG titres but were less sensitive compared with ACVR1WT. (B) Photometric Alcian blue quantification of chicken micromass cultures shown in (A) bars represent mean ± SD of quadruplicate samples from one representative of three experiments. Two-tailed Student's t-test was performed. Significant differences in comparison with ACVR1WT with the indicated NOG titre are given as: *P < 0.05; **P < 0.01; ***P < 0.001, ns not significant, n = 4.
ACVR1Q207E has enhanced basal activity, which is further amplified by BMP7 stimulation
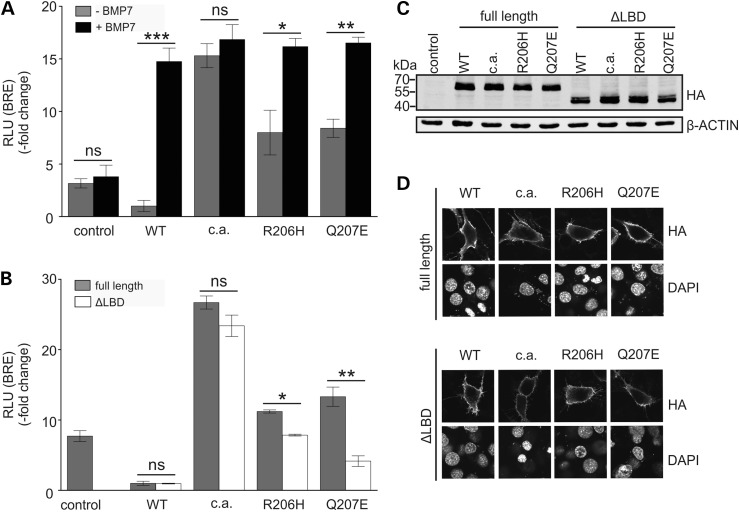
As NOG was able to inhibit activities of both FOP receptors in the chicken micromass assay, we tested if the FOP mutations have increased ligand sensitivity, an acquired ligand independency, or a combination of both. First, we tested the responsiveness of FOP receptors to BMP7, a known agonist of ACVR1 (23), to evaluate if GS domain mutations induce increased ligand sensitivity. We used a luciferase reporter construct containing a BMP-SMAD responsive element (BRE), which was co-transfected into NIH/3T3 cells together with the normalization vector pRL-TK and an Acvr1 receptor construct or an empty vector control (Fig. 6A). Overexpression of Acvr1WT caused mild inhibition of BMP signalling compared with the empty vector control. In contrast, ACVR1Q207E and ACVR1R206H induced increased activation of the BRE reporter to similar degrees while cells expressing Acvr1Q207D−c.a. activated the BRE-reporter even more strongly. Treatment with recombinant BMP7 induced luciferase activity of ACVR1WT and both FOP receptors to the level of ACVR1Q207D−c.a., which was not further increased upon stimulation with BMP7 (Fig. 6A).
Figire 6.
FOP mutation ACVR1Q207E is ligand sensitive. (A) NIH/3T3 cells were co-transfected with pGL3ti-BRE, pRL-TK and either full-length Acvr1WT, Acvr1Q207D−c.a., Acvr1R206H or Acvr1Q207E. The empty pCS2+ vector served as control. Cells were stimulated with 2 nm of recombinant BMP7. Activation of BMP signalling by Acvr1Q207E and Acvr1R206H in comparison with the WT was enhanced while the overexpression of Acvr1Q207D−c.a. more strongly activated the BRE reporter. Upon BMP7 stimulation, Acvr1WT as well as Acvr1R206H and Acvr1Q207E showed significantly enhanced Smad activity, while activity of Acvr1c.a. was not altered. For each experiment, relative luciferase activity (RLU) was normalized to the relative luciferase activity of cells expressing Acvr1WT. Bars represent mean ± SD of triplicate samples from one representative of three experiments. Two-tailed Student's t-test was performed. Significant differences are given as: *P < 0.05; **P < 0.01 or ***P < 0.001, ns not significant, n = 3. (B) NIH/3T3 cells were co-transfected with pGL3ti-BRE, pRL-TK and either full-length or ligand binding domain (ΔLBD) constructs of Acvr1WT, Acvr1Q207D−c.a., Acvr1R206H or Acvr1Q207E. The empty pCS2+ vector served as control. ΔLBD constructs of Acvr1R206H and Acvr1Q207E receptors showed significantly reduced activity compared with their respective full-length versions, while ΔLBD of Acvr1c.a. showed no reduction of activity. For each experiment, RLU was normalized to the relative luciferase activity of cells expressing Acvr1WT. Bars represent mean ± SD of triplicate samples from one representative of three experiments. Two-tailed Student's t-test was performed. Significant differences are given as: P < 0.05* or P < 0.01**, ns not significant. (C) NIH/3T3 cells were transfected with HA-tagged full-length and ΔLBD constructs of Acvr1WT, Acvr1Q207D−c.a., Acvr1R206H and Acvr1Q207E. Cell lysates were immunoblotted with anti-HA and anti-β-Actin antibodies. FOP mutations and LBD deletions show comparable expression levels. (D) Confocal microscopy images are shown of NIH/3T3 cells transfected with HA-tagged full-length and ΔLBD constructs of Acvr1WT, Acvr1Q207D−c.a., Acvr1R206H and Acvr1Q207E. Immunostaining against HA on non-permeabilized cells demonstrates that ACVR1 full-length and ΔLBD receptors are localized at the cell surface. Cell nuclei are stained with DAPI. The scale bar represents 20 µm.
To determine whether the activation of the BRE reporter by the ACVR1 FOP mutations is dependent on binding of BMP ligand to the receptor, we designed expression constructs lacking the extracellular ligand binding domain (ΔLBD). Luciferase activity of Acvr1WT and Acvr1Q207D−c.a. was not altered by the deletion of the LBD, whereas BRE activation by ΔLBD receptors of Acvr1Q207E and Acvr1R206H was significantly reduced compared with their respective full-length receptor constructs. In comparison with the full-length WT receptor, over-activation of BMP signalling by the ΔLBD FOP receptors still occurred indicating a partial ligand independency (Fig. 6B).
A shortened form of ACVR1 protein was synthesized by ΔLBD constructs leading to a size shift of the protein bands detected in the immunoblot (Fig. 6C). Apart from the difference in size, no difference in protein levels of the ACVR1 variants was observed.
ACVR1 is a transmembrane receptor mainly located at the cell surface. In order to evaluate, whether the FOP mutations or ΔLBD influence protein expression or localization of the receptor, N-terminal HA-tagged Acvr1 constructs were expressed in NIH/3T3 cells and analysed by immunoblotting and confocal immunofluorescence imaging. Immunofluorescence staining of HA-tagged ACVR1 variants showed that all of them translocated to the cell membrane. Deletion of the ACVR1 LBD therefore does not hinder membrane localization (Fig. 6D).
Together, these findings suggest different degrees of ligand independent activation of SMAD mediated BMP signalling by FOP mutations that can be further enhanced by ligand stimulation.
Loss of inhibitory receptor conformation in p.Q207D-c.a. is decisive for conferring constitutive activation
In previous structure-functional analyses, the mutations p.Q207E and p.Q207D-c.a. were predicted to have similar functional effects due to the replacement of Q207 with a negatively charged amino acid, which induces a charge repulsion of the inhibitor FKBP12 (3,10). However, all of our functional analyses revealed that ACVR1Q207E acts more like the classic FOP mutation ACVR1R206H, with both showing significantly milder activity than the ACVR1Q207D−c.a. mutation.
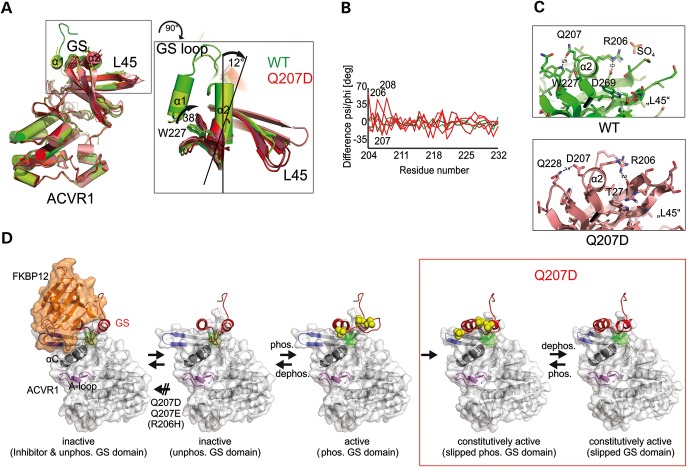
Available crystal structures of WT (PDB 3H9R; 4C02) and p.Q207D-c.a. (PDB 3MTF; 3Q4U; 3OOM, 4BGG, 4DYM) were aligned to detect alterations in receptor conformation induced by this mutation. The overall structure of the kinase domains of WT and p.Q207D-c.a. appear very similar (Fig. 7A). A difference dihedral angle plot was performed to describe conformational changes between WT and p.Q207D-c.a. crystal structures independent of an otherwise biasing structural alignment. Large deviations above mean values at single positions can indicate hinge regions that rotate adjacent domains or loops as rigid bodies towards each other. Here, the changes in psi and phi angles at 206–208 of p.Q207D structures (Fig. 7B, red), when compared with WT structure (Fig. 7B, green, PDB ID: 3H9R), define a hinge that laterally moves α2 apart from the kinase domain of ACVR1 (Fig. 7B). Interestingly, all five available p.Q207D-c.a. structures show a 12° shift of the α2 helix and a 38° shift of W227 (Fig. 7A), which prevent a hydrogen bond between Q207D and W227, that is located at the C-terminal end of the β2 strand (Fig. 7C).
Figure 7.
Model of constitutive activation of p.Q207D-c.a. by an irreversible loss of inhibitory GS domain conformation. (A) Alignment of available WT (PDB ID: 3H9R, 4C02) in green and p.Q207D-c.a. (pdb: 3MTF, 3Q4U, 3OOM, 4DYM, 4BGG) structures in red. Close up of aligned structures indicating a 12° shift of the α2 helix and a 38° shift of W227 between WT and p.Q207D-c.a. (B) A difference dihedral angle plot was calculated to detect conformational changes in WT and Q207D crystal structures. Changes of psi or phi angles of the peptide plane between WT (green: 4C02) and p.Q207D-c.a. mutant structures (red: 3MTF, 3Q4U) towards 3H9R above 35° shows a hinge comprising residues 206–208 that allow a rigid motion of the GS domain from the L45 SMAD sensitive loop and the remainder of the kinase domain. (C) Q207 forms a hydrogen bond with W227, whereas in p.Q207D-c.a. structures, Q207D is out of reach to W227 but stabilized by a new connection to Q228. In WT structures, R206 base stacks with R202 and binds to sulphate in pdb:3H9R, while in p.Q207D-c.a. structures a new H-bond forms to residue T271 of the L45 loop. (D) FKBP12 (orange) binds to ACVR1 and locks the GS domain (red) in its inactive position. ACVR1 is inactive when the GS loop blocks the motion of helix αC (dark grey) at an inhibitory region (green). p.Q207E and p.Q207D-c.a. prevent FKBP12 binding by charge repulsion. Phosphorylation (yellow) of the GS loop e.g. at S190, S192 and S194 will unlock the GS loop (residues 187–196) and free αC. The activity of the FOP mutation p.Q207E is regulated by phosphorylation and dephosphorylation with motion restricted to the GS loop, whereas the phosphorylation of the GS loop in mutation p.Q207D-c.a. causes a rigid motion (relocation) of the entire GS domain, which then becomes permanently unlocked and thereby causing constitutive activation of the receptor.
We propose that the shift of the α2 helix in the p.Q207D-c.a. structure will lead to an irreversible loss of the inhibitory GS loop conformation upon the first phosphorylation event and accordingly induce constitutive activation in this receptor (Fig. 7D). The ACVR1 p.Q207E structure predicts the same negative effect on FKBP12 binding as p.Q207D, but due to its longer side chain, Q207E can still form a hydrogen bond with W227 of the kinase domain. This decisive difference allows the Q207E receptor (in contrast to p.Q207D-c.a.) to still be regulated by phosphorylation and dephosphorylation of the GS domain, and explains its preserved ligand sensitivity and its overall milder effects in functional assays and in patients.
DISCUSSION
Here, we report a FOP patient who carries a heterozygous ACVR1 mutation at codon 207, p.Q207E. Compared with the ACVR1 p.R206H mutation which occurs in the majority of patients with FOP, this is only the second reported case of ACVR1 p.Q207E worldwide (3). The previously described patient was clinically classified as ‘FOP plus’ due to a history of severe growth retardation in addition to the classic FOP symptoms, raising the possibility of a distinct pathology caused by this mutation (3). In contrast, the patient described in our study presents only classic clinical characteristics with common variable FOP features and therefore is clinically evaluated as phenotypically equivalent to p.R206H patients. Thus p.Q207E appears to cause a classic FOP phenotype and the additional feature in the previously described patient may not be due to the ACVR1 mutation but rather to the patient's genetic background. However, limited patient numbers for extremely rare FOP mutations, such as p.Q207E, considerably complicate the establishment of reliable genotype–phenotype correlations.
It is presently unclear why p.R206H is the most prevalent of the 12 reported FOP mutations. The recurrent ACVR1 c.617G>A mutation that causes the p.R206H substitution alters a CpG dinucleotide, which has been recognized to be associated with higher mutation rates, while other FOP ACVR1 mutations do not (24). One possibility is that mutations at other positions within ACVR1 are not compatible with viability, as it might be the case for the p.Q207D mutation. Alternatively, an increased mutation frequency at this position might be due to a selective advantage conferred on the male germline cells by the respective mutation (24).
In the present study, we examined the functional differences among the two ACVR1 mutations p.Q207E and p.R206H that occurred as spontaneous, naturally occurring mutations in FOP patients and the engineered constitutively activating ACVR1 mutation of p.Q207D.
In vivo overexpression of either FOP mutation, ACVR1R206H or ACVR1Q207E, in the developing chicken limb induced skeletal malformations such as joint fusions and ectopic cartilage formation. This phenotypic effect indicates an enhanced BMP signalling pathway in the autopod and has also been linked to other human diseases, such as proximal symphalangism or multiple synostosis syndrome (25,26). Overexpression of the engineered ACVR1Q207D−c.a. receptor led to a dramatic expansion of cartilaginous elements, consistent with previously reported data (12), and indicates considerable differences in receptor activity between p.Q207D-c.a. and the FOP mutations.
Ectopic bone formation in FOP has been well described and was shown to develop through a series of catabolic and anabolic processes. Affected skeletal muscle first undergoes apoptosis before new bone is formed through endochondral ossification, a process that requires a chondrogenic template which is subsequently replaced by bone (27). In vitro differentiation assays revealed that ACVR1R206H and ACVR1Q207E indeed facilitate an amplified commitment of MSCs into the chondrogenic and osteogenic lineages at the expense of myogenic differentiation. Consistent with our observations of the skeletal development of chicken limbs, we found that activities of both FOP receptors were considerably less pronounced compared with ACVR1Q207D−c.a.. We further determined that co-expression of the BMP antagonist NOG abrogated cartilage formation in cells expressing either of the FOP receptors, while ACVR1Q207D−c.a. expressing cells were not inhibited by NOG, indicating that the FOP receptors mediate signalling through a ligand dependent activation mechanism.
We further investigated the ligand dependency of ACVR1Q207E and ACVR1R206H to determine whether they result of increased ligand independency or sensitivity or a combination of both. Using a BMP-SMAD-dependent reporter assay, we observed that the ACVR1Q207E mutation enhanced SMAD signalling similar to ACVR1R206H levels. Both FOP mutations were considerably less active compared with ACVR1Q207D−c.a. Treatment with BMP7, a known agonist of ACVR1 (23), further enhanced BMP signalling by both ACVR1Q207E and ACVR1R206H, verifying that the FOP receptors respond to ligand stimulation. To exclude effects through ligand binding to ACVR1 and NOG antagonism, we generated receptor constructs lacking the LBD. While receptor activation by both FOP receptors was significantly reduced compared with their respective full-length counterparts, the LBD-deleted receptors remained considerably above WT receptor activity, suggesting for the first time a partial ligand independent activation mechanism for ACVR1Q207E and confirming results found for ACVR1R206H (28). In contrast, no difference in BMP signalling activation was found for ACVR1Q207D−c.a. when the LBD was deleted.
Our experimental findings thus demonstrate that FOP mutations p.R206H and p.Q207E are functionally distinct from the constitutively active p.Q207D mutation.
To address what molecular mechanism causes constitutive activation specifically by mutation p.Q207D, we analysed the available two WT and five Q207D mutant structures (Figs 2 and 7). We calculated the change of free binding energies (ΔΔG) that each individual mutation induces into the structure of the ACVR1 GS domain (PDB ID:3H9R) with and without FKBP12 bound and found that Q207D directly reduces binding of FKBP12 by intermolecular charge repulsion of aspartic acid with E55 (Table 1). The same mechanism is seen in the p.Q207E variant, which is in accordance with the previous study of Chaikuad et al. (10). However, reduced binding of FKBP12 alone does not explain why Q207D is the most active variant. Activation of ACVR1 requires (i) the release of the inhibitor FKBP12 and (ii) phosphorylation of the GS domain prior to final phosphorylation of the activation loop (A-loop). The latter is a canonical activation step of kinases. Principally, a mutation that constitutively activates ACVR1 prevents both inhibitory processes, FKBP12 binding and GS domain deactivation. The precise mechanism of how the GS loop separates from the kinase domain and activates ACVR1 is not fully understood, yet. But it is remarkable that all available Q207D structures, albeit truncated to few residues of the GS domain, show a consistent lateral movement of α2 helix while retaining the hydrophobic cluster maintained by V204 and W227. The latter tryptophan is apparently the central organizer binding and positioning of the GS domain (Fig. 2). We propose that in WT ACVR1, phosphorylation lifts the GS loop but retains positions of α1 and α2 which are stabilized by a hydrophobic cluster surround W227. In WT structures, Q207 makes an H-bond to indol nitrogen of W227 (Fig. 7C) that stabilizes hydrophobicity and reduces solvent accessibility of W227 and thereby likely constrains lateral movement of the GS domain. Current Q207D structures propose that a loss of the single H-bond between Q207 to indol nitrogen (Fig. 7C) changes the hydrophobicity of W227 and weakens constraints to prevent lateral movements. The loss of the H-bond between Q207D and W227 leads to a slip of the entire GS domain into an arrested position, which will unlock the GS domain permanently and lead to a constitutive activation. In FOP mutation p.Q207E, an amino group is changed to a negatively charged carboxyl group, while the length of the side chain is not altered; this prevents binding to E55 of FKBP12 but maintains binding geometry to W227. The activation of ACVR1 by p.R206H is still not clear; the histidine does introduce a marginal steric hindrance in FKBP12 binding (Table 1); however, inhibitor binding affinity is only reduced 3-fold (19). The initial idea of a pH-dependent regulation of the R206H to D269 interaction could not be verified (19,29). On the other hand, the p.Q207D structures show new interactions of R206 to residues of loop L45 (Fig. 7B) that in turn is sensitive to SMAD proteins. While p.Q207E does constantly prevent FKBP12 binding, p.R206H might spatially and temporally change FKBP12 binding and subsequent phosphorylation of R-SMADs or feedback inhibition of e.g. SMAD 6/7 through altering loop L45.
Taken together, our results emphasize that p.Q207D has a distinctly different activation mode compared with the naturally occurring FOP mutations, which in contrast to p.Q207D seem to remain under regulatory control and might be susceptible to treatment, for example, by FKBP12 variants. Our results further imply that specific knock-in mouse models carrying homologous FOP mutations are better suited to perform preclinical tests for potential drugs to treat FOP.
MATERIALS AND METHODS
Clinical investigation and molecular analysis
All clinical investigations were conducted in accordance with the provision of the Declaration of Helsinki principles. Diagnosis of FOP was based on clinical investigation and verified by genetic analysis for which informed consent was obtained from the legal authorities. Genomic DNA was purified from blood and mutation screening in ACVR1 (RefSeq NM_001111067.2) was performed as previously described (4,30).
Alignment
Protein sequences comprising the highly conserved GS domain of the human TGFβ receptor type I family as well as chicken and mouse ACVR1 were aligned using CLUSTAL X (31) and shaded using CHROMA (32).
Free binding energy calculation
The structure of the ACVR1:FKBP12 complex (PDB ID: 3H9R) has been solved by X-ray crystallography (10). Hot spot residues at the interfaces between kinase and GS domain and between ACVR1 and inhibitor FKBP12 have been determined by calculating changes of individual and total free binding energies (ΔΔG) for ACVR1 mutants, including FOP-related mutations using FOLD-X (18). Structures were visualized using PyMol Molecular Graphics System, Schrödinger, LLC (http://www.pymol.org/).
Cloning of expression plasmids
The coding sequence of murine Acvr1 was cloned into the shuttle vector pSLAX-13. This construct was used as a template for in vitro mutagenesis to introduce the mutations p.Q207D, p.R206H and p.Q207E (sequences for mutagenic primers are listed in Supplementary Material, Table S1). These constructs were used for further site-directed mutagenesis to generate ΔLBD-Acvr1, in which the sequence for amino acids 35–99 encoding the LBD of the receptor is deleted. HA-tagged Acvr1 constructs were generated using site-directed mutagenesis to insert an N-terminal HA-tag. After mutagenesis, all coding sequences were subcloned into the expression vector pCS2+ by digestion with ClaI.
Retroviral constructs and virus production
Cloning of chicken coding sequences of ACVR1WT, ACVR1Q207D−c.a., ACVR1R206H into the retroviral vector RCASBP(A) was described previously (33). The p.Q207E mutation was introduced into the coding sequence of the chicken ACVR1 receptor by site-directed mutagenesis in the shuttle vector pSLAX-13. Insert was subcloned into RCASBP(A) after ClaI digestion. Chicken NOG in RCASBP(B) was published previously (34).
For viral production, each plasmid was transfected into DF1 cells. Supernatant containing viral particles was harvested on three consecutive days and ultracentrifuged as described previously to obtain high-titre concentrates (35). Titres of all viruses were determined as plaque forming units (PFU/ml) prior to any functional application.
In vivo manipulation of chicken limb buds and skeletal preparations
Fertilized eggs were incubated for 36 h to obtain embryos at Hamburger and Hamilton (HH) stage 10. Viral titres were adjusted to 2 × 108 PFU per ml and injected into the right hind limb field as described previously (36). The non-injected left hind limb served as an internal control. Each viral construct (ACVR1WT; ACVR1Q207D−c.a.; ACVR1R206H and ACVR1Q207E) was used in two independent injection rounds. Embryos were sacrificed 10 days after fertilization and dehydrated over night in 70% ethanol. Skeletal staining and preparation were performed as described previously (37). Skeletons were stored in 80% glycerol.
Cultivation of QM7 cells and muscle differentiation assay
The quail myogenic cell line QM7 (ATCC) was cultured in growth medium [Medium 199 GlutaMAX (Invitrogen), 10% tryptose phosphate broth, 10% FBS (Biochrom), 2% chicken serum (Gibco), pen/strep (Lonza)], which was replaced daily. Cells were infected with empty RCASBP(A) (control) or an ACVR1 receptor construct at a titre of higher than 1 × 108 PFU/ml and grown for 3 days to ensure complete viral infection before seeding into 24-well plates with equal cell density of 2.5 × 104 cells per well. To induce myogenesis, cells were transferred to differentiation medium [Medium 199 with GlutaMAX (Invitrogen), 10% tryptose phosphate broth, 0.5% FBS (Biochrom), 0.2% chicken serum (Gibco), pen/strep (Lonza)] for 6 days. Myotubes were stained using anti-myosin heavy chain antibody (MF20, Developmental Studies Hybridoma Bank) and Vectastain ABC system (Vector Laboratories) according to the manufacturer's instruction. Muscle staining was quantified by histomorphometric analysis using the Autmess AxioVision™ 4.6 software (Carl Zeiss).
Preparation and cultivation of micromass cultures
Fertilized eggs were incubated for 4.5 days to obtain HH 24–25 chicken embryos. Limb buds were dissected and enzymatically digested to obtain a single cell suspension as described previously (33). Cells were infected with empty RCASBP(A) (control) or an ACVR1 receptor construct at a titre of 2 × 107 PFU/ml or coinfected with NOG-RCASBP(B) (0.1–10 × 106 PFU/ml) and seeded as a drop culture containing 2 × 105 cells. Cells were allowed to attach to the plate surface at 39°C before adding growth medium [DMEM:F12 (Biochrom), 10% FBS (FBS superior Biochrom), 0.2% chicken serum (Gibco), 2 mm L-Gln (Lonza) and pen/strep (Lonza)]. Cultures were fixed in Kahle's fixative and stained with 0.05% Alcian blue (Sigma-Aldrich) dissolved in 0.1 m HCl to visualize cartilage-specific matrix. Excess dye was removed by consecutive washing steps with water and cultures were dried and subsequently documented. Quantification of incorporated dye was determined by extraction with 6 m guanidine hydrochloride, followed by photometric measurement at A595 nm.
Preparation and cultivation of avian bone marrow stromal stem cells
Fertilized eggs were incubated for 17 days to obtain stage HH43 chicken embryos. Embryos were sacrificed and the tibiotarsus isolated. The bone marrow was extracted by flushing the bones with DPBS using a 0.45 mm syringe. The cell suspension was plated into a cell culture flask and cells were allowed to adhere to the plate surface for 1 h. After removing non-adherent blood cells by carefully washing the culture with DPBS, cells were cultured in growth medium [DMEM low glucose (Lonza), 10% FBS superior (Biochrom), 2 mm L-Gln (Lonza), pen/strep (Lonza)] in a humidified atmosphere of 5% CO2 and 39°C. Medium was replaced three times a week. Cultures were allowed to reach 80–95% confluency before trypsinizing. The cell suspension was then adjusted to 2.5 × 104 cells/well, seeded into a 24-well format and infected with indicated viruses at a concentration of 5 × 104 PFU/ml. Osteogenic differentiation was initiated when cultures reached 100% confluency and growth medium was replaced with osteogenic differentiation medium [DMEM low glucose (Lonza), 10% FBS superior (Biochrom), 2 mm L-Gln (Lonza), pen/strep (Lonza), 60 µg/ml ascorbic acid and 2.5 mm β-glycerophosphate]. Mineralization was visualized by staining calcified deposition with 0.5% Alizarin red S solution (pH 4.0) and quantified after extraction with 10% cetylpyridinium chloride by photometric measurements at A562 nm.
Luciferase reporter gene assay
NIH/3T3 cells (ATCC) were seeded in 96-well plates at a density of 1 × 104 cells per well and cultured for 24 h in growth medium [DMEM high glucose (Lonza), 10% FBS superior (Biochrom), 2 mm L-Gln (Lonza), pen/strep (Lonza)]. Cells were transfected with the BMP responsive firefly luciferase reporter pGL3ti-BRE (38), the renilla luciferase normalization vector pRL-TK (Promega) and pCS2+ plasmids containing the murine coding sequences of Acvr1WT, Acvr1Q207D−c.a., Acvr1R206H and Acvr1Q207E. Cells were transfected with Lipofectamine 2000 (Invitrogen). Twenty hours after transfection, indicated cells were stimulated with 2 nm of recombinant BMP7 (RnD). Forty-eight hours after transfection, cells were lysed in potassium phosphate buffer (9 mm potassium dihydrogen phosphate, 91 mm dipotassium phosphate, 0.2% Triton X-100) and dual luciferase activity was determined as described previously (39) using Mithras LB 940 (Berthold Detection Systems).
Immunocytochemistry
NIH/3T3 cells were seeded on cover glasses (Marienfeld GmbH & CoKG) in 24-well plates and transfected with HA-tagged Acvr1 constructs in pCS2+ using Lipofectamine 2000 (InvitrogenTM, Life Technologies). Twenty-four hours after transfection, cells were starved for 1 h in serum free culture medium and subsequently fixed for 15 min at room temperature in 4% paraformaldehyde in PBS. To block non-specific antibody binding sites, cells were incubated in 10% FBS/PBS over night at 4°C. Subsequently, cells were incubated with rabbit anti-HA antibody (H6908, Sigma-Aldrich) at a 1:100 dilution in 10% FBS/PBS for 1 h at room temperature. After incubation with anti-rabbit Alexa Fluor 488 antibody (A11008, Molecular Probes® Life Technologies) diluted 1:2000 in 10% FBS/PBS and counterstaining with DAPI (InvitrogenTM, Life Technologies), cover glasses were mounted on microscope slides (‘SuperFrost®Plus’, Menzel GmbH & CoKG) using Fluoromount-G (Southern Biotech). Confocal fluorescence images were recorded at 63-fold magnification using Zeiss Axio Imager.M2 equipped with a LSM700 confocal module (Carl Zeiss).
Immunoblot
NIH/3T3 cells were seeded in six-well plates at a density of 2 × 105 cells/well and transfected with HA-tagged Acvr1 constructs in pCS2+ using Lipofectamine 2000 (InvitrogenTM, Life Technologies). Cells were harvested 24 h after transfection and lysed in lysis buffer [50 mm HEPES, 10 mm EDTA 10% glycerin, 1% Triton X-100, 50 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 2 mm sodium fluoride, 5 mm tetrasodium pyrophosphate, 1× Complete Mini Protease Inhibitor (Roche)].
A total of 45 µg protein was diluted in reducing sample buffer (20 mm Tris-buffer, pH 7.5, 125 mm β-mercaptoethanol, 10% glycerol, 1% SDS, 0.01% Bromphenol blue) and separated on a 12% SDS–PAGE, transferred onto nitrocellulose membrane (Santa Cruz) and probed with rabbit anti-HA (H6908, Sigma-Aldrich) and mouse monoclonal anti-β-Actin antibody (A5441, Sigma-Aldrich). After incubation with secondary antibodies, IRDye goat anti-rabbit 800CW and IRDye goat anti-mouse 680 (LI-COR) signals were detected using the Odyssey detection system.
Calculation of dihedral angle differences
Dihedral angles, psi and phi from Ramachandran plot of wild-type (PDB ID: 4C02) and Q207D mutant crystal structures (PDB ID:3MTF, 3Q4U) have been subtracted from corresponding values of WT ACVR1 structure (PDB ID: 3H9R) (40).
Modelling of p.Q207D-c.a
The partial GS domain comprising α2 helix (residues 204–207) of the truncated p.Q207D-c.a. structure (PDB ID: 3MTF) was completed by structurally aligning the corresponding α2 helix of the full GS domain from ACVR1: FKBP12 complex (PDB ID: 3H9R) using Swiss-PdbViewer (http://www.expasy.org/spdbv/). Dephosphorylated and phosphorylated states of the GS domain have been modelled using loopdb and by constraining coordinates of helices α1 and α2. Structures were visualized using PyMol Molecular Graphics System, Schrödinger, LLC (http://www.pymol.org/).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by a research grant from ‘The Center for Research in Fibrodysplasia Ossificans Progressiva’ from the University of Pennsylvania, by the German FOP e. V. and by the International Fibrodysplasia Ossificans Progressiva Association, the Center for Research in FOP and Related Disorders, the Ian Cali Endowment for FOP Research, the Whitney Weldon Endowment for FOP Research, the Isaac & Rose Nassau Professorship of Orthopaedic Molecular Medicine (to F.S.K.), the Cali-Weldon Research Professorship in FOP (to E.M.S.), the Rita Allen Foundation, the Penn Center for Musculoskeletal Disorders (NIH P30-AR050950), and the National Institutes of Health (NIH R01-AR41916). Contributions were made possible by Deutsche Forschungsgemeinschaft funding through the Berlin-Brandenburg School for Regenerative Therapies (GSC 203 to A.D., K.S.).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank the patient's family for their collaboration and contribution to this project, Maria Walther and Mareen Schmidt-von Kegler for excellent technical assistance and Lutz Schomburg for critical remarks on the article. The plasmid pGl3ti-BRE was kindly provided by Peter ten Dijke. The MF20 antibody developed by Donald A. Fischman, M.D. was obtained from the ‘Developmental Studies Hybridoma Bank’ developed under the auspices of the NICHD and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA 52242.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Thickman D., Bonakdar-pour A., Clancy M., Van Orden J., Steel H. Fibrodysplasia ossificans progressiva. AJR Am. J. Roentgenol. 1982;139:935–941. doi: 10.2214/ajr.139.5.935. [DOI] [PubMed] [Google Scholar]
- 2.Connor J.M., Evans D.A. Fibrodysplasia ossificans progressiva. The clinical features and natural history of 34 patients. J. Bone Joint Surg. Br. 1982;64:76–83. doi: 10.1302/0301-620X.64B1.7068725. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan F.S., Xu M., Seemann P., Connor J.M., Glaser D.L., Carroll L., Delai P., Fastnacht-Urban E., Forman S.J., Gillessen-Kaesbach G., et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum. Mutat. 2009;30:379–390. doi: 10.1002/humu.20868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shore E.M., Xu M., Feldman G.J., Fenstermacher D.A., Cho T.J., Choi I.H., Connor J.M., Delai P., Glaser D.L., LeMerrer M., et al. A recurrent mutation in the BMP type I receptor ACVR1 causes inherited and sporadic fibrodysplasia ossificans progressiva. Nat. Genet. 2006;38:525–527. doi: 10.1038/ng1783. [DOI] [PubMed] [Google Scholar]
- 5.Gordon K.J., Blobe G.C. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim. Biophys. Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 7.Huse M., Muir T.W., Xu L., Chen Y.G., Kuriyan J., Massague J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol. Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 8.Huse M., Chen Y.G., Massague J., Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 9.Huse M., Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 10.Chaikuad A., Alfano I., Kerr G., Sanvitale C.E., Boergermann J.H., Triffitt J.T., von Delft F., Knapp S., Knaus P., Bullock A.N. Structure of the bone morphogenetic protein receptor ALK2 and implications for fibrodysplasia ossificans progressiva. J. Biol. Chem. 2012;287:36990–36998. doi: 10.1074/jbc.M112.365932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki A., Kaneko E., Ueno N., Hemmati-Brivanlou A. Regulation of epidermal induction by BMP2 and BMP7 signaling. Dev. Biol. 1997;189:112–122. doi: 10.1006/dbio.1997.8652. [DOI] [PubMed] [Google Scholar]
- 12.Zhang D., Schwarz E.M., Rosier R.N., Zuscik M.J., Puzas J.E., O'Keefe R.J. ALK2 functions as a BMP type I receptor and induces Indian hedgehog in chondrocytes during skeletal development. J. Bone Miner. Res. 2003;18:1593–1604. doi: 10.1359/jbmr.2003.18.9.1593. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda T., Scott G., Komatsu Y., Araya R., Kawano M., Ray M.K., Yamada M., Mishina Y. Generation of a mouse with conditionally activated signaling through the BMP receptor, ALK2. Genesis. 2006;44:159–167. doi: 10.1002/dvg.20201. [DOI] [PubMed] [Google Scholar]
- 14.Wieser R., Wrana J.L., Massague J. GS domain mutations that constitutively activate T beta R-I, the downstream signaling component in the TGF-beta receptor complex. EMBO J. 1995;14:2199–2208. doi: 10.1002/j.1460-2075.1995.tb07214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimono K., Tung W.E., Macolino C., Chi A.H., Didizian J.H., Mundy C., Chandraratna R.A., Mishina Y., Enomoto-Iwamoto M., Pacifici M., et al. Potent inhibition of heterotopic ossification by nuclear retinoic acid receptor-gamma agonists. Nat. Med. 2011;17:454–460. doi: 10.1038/nm.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu P.B., Deng D.Y., Lai C.S., Hong C.C., Cuny G.D., Bouxsein M.L., Hong D.W., McManus P.M., Katagiri T., Sachidanandan C., et al. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan F.S., Shore E.M., Pignolo R.J. The medical management of fibrodysplasia ossificans progressiva: current treatment considerations. Clin. Proc. Intl Clin. Consort. FOP. 2011;4:1–100. [Google Scholar]
- 18.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 19.Groppe J.C., Wu J., Shore E.M., Kaplan F.S. In vitro analyses of the dysregulated R206H ALK2 kinase-FKBP12 interaction associated with heterotopic ossification in FOP. Cells Tissues Organs. 2011;194:291–295. doi: 10.1159/000324230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antin P.B., Ordahl C.P. Isolation and characterization of an avian myogenic cell line. Dev. Biol. 1991;143:111–121. doi: 10.1016/0012-1606(91)90058-b. [DOI] [PubMed] [Google Scholar]
- 21.Mello M.A., Tuan R.S. High density micromass cultures of embryonic limb bud mesenchymal cells: an in vitro model of endochondral skeletal development. In Vitro Cell Dev. Biol. Anim. 1999;35:262–269. doi: 10.1007/s11626-999-0070-0. [DOI] [PubMed] [Google Scholar]
- 22.Groppe J., Greenwald J., Wiater E., Rodriguez-Leon J., Economides A.N., Kwiatkowski W., Affolter M., Vale W.W., Izpisua Belmonte J.C., Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- 23.Macias-Silva M., Hoodless P.A., Tang S.J., Buchwald M., Wrana J.L. Specific activation of Smad1 signaling pathways by the BMP7 type I receptor, ALK2. J. Biol. Chem. 1998;273:25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 24.Arnheim N., Calabrese P. Understanding what determines the frequency and pattern of human germline mutations. Nat. Rev. Genet. 2009;10:478–488. doi: 10.1038/nrg2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gong Y., Krakow D., Marcelino J., Wilkin D., Chitayat D., Babul-Hirji R., Hudgins L., Cremers C.W., Cremers F.P., Brunner H.G., et al. Heterozygous mutations in the gene encoding noggin affect human joint morphogenesis. Nat. Genet. 1999;21:302–304. doi: 10.1038/6821. [DOI] [PubMed] [Google Scholar]
- 26.Seemann P., Schwappacher R., Kjaer K.W., Krakow D., Lehmann K., Dawson K., Stricker S., Pohl J., Ploger F., Staub E., et al. Activating and deactivating mutations in the receptor interaction site of GDF5 cause symphalangism or brachydactyly type A2. J. Clin. Invest. 2005;115:2373–2381. doi: 10.1172/JCI25118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pignolo R.J., Suda R., Kaplan F.S. The fibrodysplasia ossificans progressiva lesion. Clin. Rev. Bone Miner. Metab. 2005;3:195–200. [Google Scholar]
- 28.Le V.Q., Wharton K.A. Hyperactive BMP signaling induced by ALK2(R206H) requires type II receptor function in a Drosophila model for classic fibrodysplasia ossificans progressiva. Dev. Dyn. 2012;241:200–214. doi: 10.1002/dvdy.22779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groppe J.C., Shore E.M., Kaplan F.S. Functional modeling of the ACVR1 (R206H) mutation in FOP. Clin. Orthop. Relat. Res. 2007;462:87–92. doi: 10.1097/BLO.0b013e318126c049. [DOI] [PubMed] [Google Scholar]
- 30.Bocciardi R., Bordo D., Di Duca M., Di Rocco M., Ravazzolo R. Mutational analysis of the ACVR1 gene in Italian patients affected with fibrodysplasia ossificans progressiva: confirmations and advancements. Eur. J. Hum. Genet. 2009;17:311–318. doi: 10.1038/ejhg.2008.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodstadt L., Ponting C.P. CHROMA: consensus-based colouring of multiple alignments for publication. Bioinformatics. 2001;17:845–846. doi: 10.1093/bioinformatics/17.9.845. [DOI] [PubMed] [Google Scholar]
- 33.Shen Q., Little S.C., Xu M., Haupt J., Ast C., Katagiri T., Mundlos S., Seemann P., Kaplan F.S., Mullins M.C., et al. The fibrodysplasia ossificans progressiva R206H ACVR1 mutation activates BMP-independent chondrogenesis and zebrafish embryo ventralization. J. Clin. Invest. 2009;119:3462–3472. doi: 10.1172/JCI37412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degenkolbe E., Konig J., Zimmer J., Walther M., Reissner C., Nickel J., Ploger F., Raspopovic J., Sharpe J., Dathe K., et al. A GDF5 point mutation strikes twice—causing BDA1 and SYNS2. PLoS Genet. 2013;9:e1003846. doi: 10.1371/journal.pgen.1003846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan B.A., Fekete D.M. Manipulating gene expression with replication-competent retroviruses. Methods Cell. Biol. 1996;51:185–218. doi: 10.1016/s0091-679x(08)60629-9. [DOI] [PubMed] [Google Scholar]
- 36.Logan M., Tabin C. Targeted gene misexpression in chick limb buds using avian replication-competent retroviruses. Methods. 1998;14:407–420. doi: 10.1006/meth.1998.0595. [DOI] [PubMed] [Google Scholar]
- 37.Seemann P., Brehm A., Konig J., Reissner C., Stricker S., Kuss P., Haupt J., Renninger S., Nickel J., Sebald W., et al. Mutations in GDF5 reveal a key residue mediating BMP inhibition by NOGGIN. PLoS Genet. 2009;5:e1000747. doi: 10.1371/journal.pgen.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korchynskyi O., ten Dijke P. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 2002;277:4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 39.Hampf M., Gossen M. A protocol for combined Photinus and Renilla luciferase quantification compatible with protein assays. Anal. Biochem. 2006;356:94–99. doi: 10.1016/j.ab.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 40.Korn A.P., Rose D.R. Torsion angle differences as a means of pinpointing local polypeptide chain trajectory changes for identical proteins in different conformational states. Protein Eng. 1994;7:961–967. doi: 10.1093/protein/7.8.961. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.