Abstract
Multicellular tubes consist of polarized cells wrapped around a central lumen and are essential structures underlying many developmental and physiological functions. In Drosophila compound eyes, each ommatidium forms a luminal matrix, the inter-rhabdomeral space, to shape and separate the key phototransduction organelles, the rhabdomeres, for proper visual perception. In an enhancer screen to define mechanisms of retina lumen formation, we identified Actin5C as a key molecule. Our results demonstrate that the disruption of lumen formation upon the reduction of Actin5C is not linked to any discernible defect in microvillus formation, the rhabdomere terminal web (RTW), or the overall morphogenesis and basal extension of the rhabdomere. Second, the failure of proper lumen formation is not the result of previously identified processes of retinal lumen formation: Prominin localization, expansion of the apical membrane, or secretion of the luminal matrix. Rather, the phenotype observed with Actin5C is phenocopied upon the decrease of the individual components of non-muscle myosin II (MyoII) and its upstream activators. In photoreceptor cells MyoII localizes to the base of the rhabdomeres, overlapping with the actin filaments of the RTW. Consistent with the well-established roll of actomyosin-mediated cellular contraction, reduction of MyoII results in reduced distance between apical membranes as measured by a decrease in lumen diameter. Together, our results indicate the actomyosin machinery coordinates with the localization of apical membrane components and the secretion of an extracellular matrix to overcome apical membrane adhesion to initiate and expand the retinal lumen.
Author Summary
Biological tubes are integral units of tissues and organs such as lung, kidney, and the cardiovascular system. The fundamental design of tubes involves a central lumen wrapped by a sheet of cells. To function properly, the tubes require a precise genetic control over their creation, the diametric growth and maintenance of the lumen during development. In the fruit fly, Drosophila melanogaster, the photoreceptor cells of the eye form a tubular structure. The formation of the retinal lumen is critical for separating and positioning the light sensing organelles of each photoreceptor cell to achieve visual sensitivity. In an effort to investigate the mechanisms of Drosophila retinal lumen formation, we identified a contractile machinery that was present at the apical portion of photoreceptor cells. Our data is consistent with the idea that a contractile force contributes to the initial separation of the juxtaposed apical membranes and subsequent enlargement of the luminal space. Our work suggests that building a biological tube requires not only an extrinsic pushing force provided by the growing central lumen, but also a cell intrinsic pulling force powered by contraction of cells lining the lumen. Our findings expand and demonstrate the coordination of several molecular mechanisms to generate a tube.
Introduction
Multicellular tubes are fundamental structures required for the transport of gases, liquids, or cells and are necessary for the generation and function of tissues and organs such as lung, kidney, blood vessels, neural tubes, and mammary gland. The main feature of a tubular network is a luminal space lined by apical membranes of polarized epithelial or endothelial cells. To construct a functional tube, there needs to be mechanisms to first generate a luminal space and then regulate the expansion and determination of the final diametrical size of the lumen. Cells utilize multiple pathways to organize themselves to form an initial tubular network (reviewed in [1]–[3]) and likewise diametric luminal growth appears to be under precise genetic control [4]. To date lumen growth has been characterized as a process of directed and regulated apical secretion of components into a central space and a reorganization of the apical membrane. Secretion likely provides a mechanical expansion force that drives the diametrical growth of the tube lumen [5]–[7]. Secreted components can include solid extracellular matrices of proteoglycans and collagens. Increase in lumen size can also be achieved by increasing the osmotic lumen pressure via ion pumps and channels [8]–[10]. Furthermore, the fusion of secretory vesicles with apical plasma membranes often changes the cells apical domain antigens, which in return drive the expansion of apical membrane permitting an increase in the diameter of the lumen [4], [11], [12].
The Drosophila compound eye provides an ideal model system to study lumen formation. The Drosophila eye consists of approximately 800 individual units known as ommatidia. In each ommatidium, a tubular structure is generated by the concerted efforts of the eight photoreceptors. Over a period ∼60 hours (h), the reorganization of the photoreceptor apical membranes drives a dramatic morphogenesis from a single epithelial sheet to a single tube containing eight cells surrounding a central lumen matrix, termed the inter-rhabdomeral space (IRS) [13]. Furthermore, each photoreceptor projects its corresponding light sensing organelle, the rhabdomere, within the luminal space. Consequentially, the IRS is required to shape the rhabdomeres and optically position each rhabdomere to achieve proper visual sensitivity [14], [15].
Genetic dissection of retinal lumen formation has provided key insights into fundamental questions about lumen formation, such as the mechanism through which adherent juxtaposed membranes separate. To date Drosophila retinal lumen formation is known to be dependent on secretion of an extracellular matrix [16], [17] and a concurrent steric hindrance of Chaoptin (Chp) based adhesion [17]. The major constituent of the IRS matrix is the proteoglycan protein Eyes shut (EYS), which is also known as Spacemaker. Loss of EYS results in a complete failure of the IRS to form [16], [17]. Nonetheless, secretion of EYS is not sufficient for the generation of a continuous lumen. In addition to being secreted, EYS must be localized around the developing apical rhabdomeres through an interaction with the five-pass transmembrane protein Prominin (Prom) [17], [18]. In the absence of Prom, the lumen space is present but not continuous and the residual fusion between rhabdomeres is the result of the adhesion between the rhabdomeric apical membranes mediated by the GPI-anchored membrane protein Chp [17], [19], [20].
Although these previous results demonstrate the interplay between secretion and adhesion, here we reveal an additional mechanism contributing to retinal lumen formation and expansion. Our results implicate that actin and non-muscle myosin II (MyoII) generate a contractile force at the apical domain of photoreceptors to separate the initial juxtaposed membranes. Rhabdomere adhesion is enhanced upon reduction of components of the actomyosin complex or its upstream activators in our sensitized genetic background. Additionally, knockdown of MyoII in a wild-type background led to a narrower lumen space. Temporal profiling of the key molecules for retinal lumen formation indicates the actomyosin complex is the first to localize to the apical surface followed by Prom and EYS. Thus the actomyosin machinery would be providing an initial apical based tension on the Chp based juxtaposed membranes, assisting the initiation and expansion of subsequently deposited extracellular matrix represented by EYS. All together, our genetic analysis has revealed an unappreciated facet of lumen formation and outlined the temporal steps and coordination required to achieve membrane separation.
Results
Reduction of Actin5C enhances rhabdomere adhesion in an eys, prom trans-heterozygote mutant background
The separation of rhabdomeres and formation of the retinal lumen, the IRS, within each ommatidium depends on the fine balance between an adhesion force, provided by Chp [19]–[22], and an expansion force provided by EYS [16], [17] and Prom [17]. When one copy of both eys and prom are removed there is a partial failure to form a continuous open IRS, exhibited by the presence of juxtaposed rhabdomeres (compare Figure 1 A,B). However, the phenotype is not fully penetrant (Figure 1B,E) and manipulating the levels Prom, EYS, or Chp can modulate the appearance of a continuous retinal lumen [17]. Utilizing this sensitized genetic background, we performed a genetic screen to identify other genes required for the formation of the IRS. Defined genomic deletions were introduced into the eys, prom trans-heterozygote (EP-TH) background and scored for the ability to enhance the EP-TH adhesion phenotype and thus Drosophila eyes were screened for the loss of the deep pseudopupil [23]. From our screen, we identified deletion Df(1)ED6829 (www.flybase.org) as a potential candidate. Transmission electron microscopy (TEM) analysis confirmed that rhabdomere fusion was significantly enhanced upon the inclusion of the deletion in the double heterozygous background. The addition of Df(1)ED6829 in the EP-TH resulted in an 41% increase in rhabdomere fusion (Figure 1 C,E).
Figure 1. Reduction of Act5C dosage enhances rhabdomere adhesion.
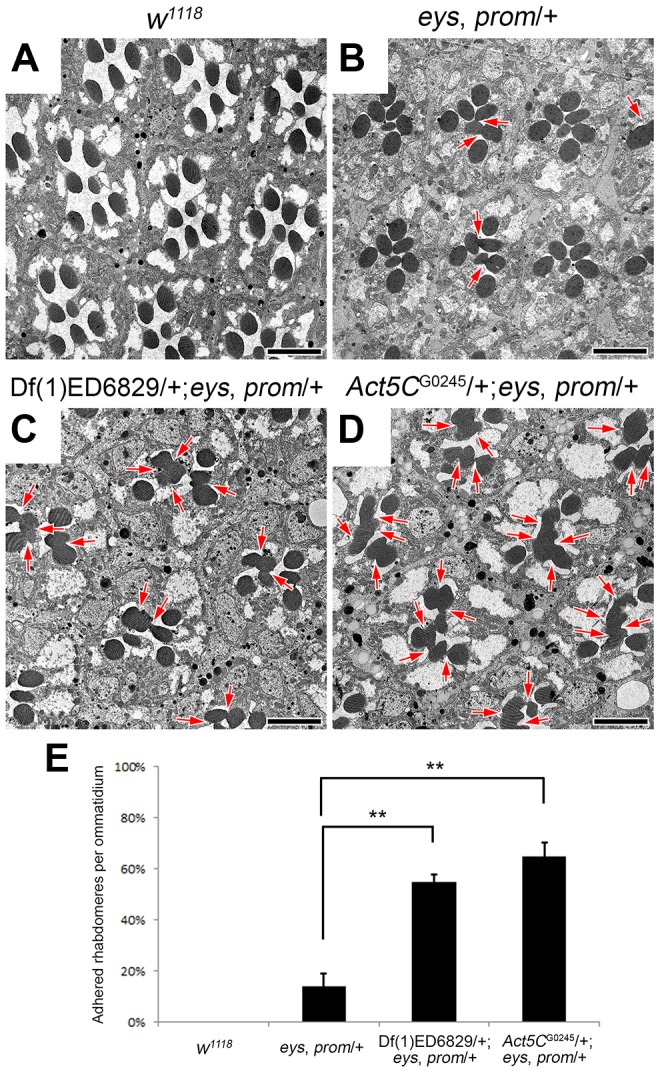
(A–D) Transmission electron micrographs of adult Drosophila ommatidia. (A) w1118, wild type. (B) eys, prom/+. (C) Df(1)ED6829/+; eys, prom/+. The deficiency removes genomic region 5C7-5F3. (D) Act5C G0245/+; eys, prom/+. (E) Quantitative analysis of rhabdomere fusion seen in (A–D). Values represent mean ± SEM. **P<0.01 compared with eys, prom/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
Analysis of overlapping deletions and testing of individual mutants for genes within the deleted interval mapped the responsible locus to Actin5C (Act5C), as alleles of Act5C as well as mild RNAi knockdown of Act5C in the EP-TH background completely phenocopied the phenotype observed with the deficiency (Figure 1D, E and Figure S1). In Drosophila there are six actin genes: Act5C and Act42A are ubiquitously expressed, while Act57B, Act79B, Act87E, and Act88F are muscle-specific [24]–[26]. To test the specificity of our enhancement, we re-examined deletions that individually remove each actin gene. TEM analyses of the corresponding deficiencies for the other five actin genes did not show any enhancement of fused rhabdomeres in the EP-TH background (Figure S2). These data suggested that Act5C was a specific and key component in generating the retinal lumen.
The genetic reduction of Act5C does not alter the dynamics of rhabdomere morphogenesis or structure
Photoreceptors are dependent upon actin for overall structure and integrity. In particular, the microvilli of the rhabdomeres contain an actin cytoskeleton core [27] and the rhabdomeres are supported by the actin-based structure the rhabdomere terminal web (RTW) [28]. The RTW is also responsible for directing delivery of molecules to the rhabdomere [28], [29]. Thus, the enhancement of rhabdomere fusion observed in the triple heterozygote may be an indirect result of disruption of photoreceptor actin based structures. To test this possibility, we examined the ability of the rhabdomere to extend the entire length of the photoreceptor cell body to the cone cell plate [30]. We found the removal of one genetic copy of Act5C in a wild type or EP-TH background does not affect the ability of the rhabdomeres to extend the entire length (Figure S3). Nor did we observe any change in rhabdomere diameter, an indication that microvilli formation and extension is normal (Figure 1). With respect to the formation of the RTW, heterozygous mutation of Act5C does not affect the localization of Moesin (Moe) (Figure S4) and RNAi knockdown of moe in the EP-TH background led to rhabdomere degeneration as observed in a moe mutant [28]. Thus our results do not indicate that the increase in juxtaposed rhabdomeres was the result of a general defect in the actin-based structures of photoreceptors.
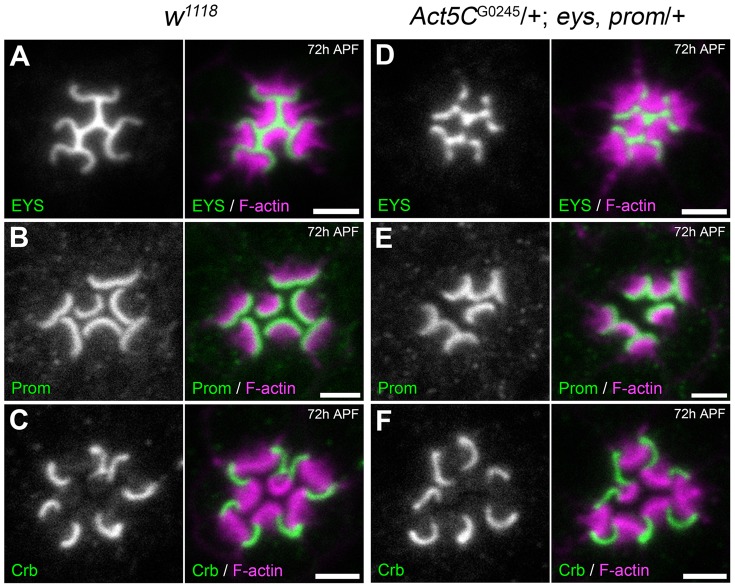
The reduction of Actin5C does not affect spatial localization of EYS, Prominin, or Crumbs
What is the specific primary defect upon the genetic reduction Act5C? Besides structural support, the actin cytoskeleton provides tracks in the cell to allow intracellular transportation of membrane and non-membrane-bound cargos [31], [32]. Furthermore, in mammalian photoreceptors the mammalian Prominin ortholog, Prominin1, directly interacts with actin; actin co-immunoprecipitates with Prominin1 and the binding is attenuated by the Prominin1 R373C mutation [33]. Thus it is conceivable that the increase in rhabdomere fusion upon the genetic reduction of Actin results from either mislocalized or missing apical components or defects in secretion of EYS. We addressed these possibilities by first surveying whether there was any difference in the spatial localization pattern of Prom and EYS in the Act5C/+; EP-TH background. We did not observe any detectable decrease or mislocalized EYS (Figure 2 A,D) or Prom (Figure 2 B,E). In addition to EYS and Prom, the transmembrane protein Crumbs (Crb) has also been implicated in rhabdomere separation and Drosophila salivary gland lumen formation [11], [21], [34], [35]. In photoreceptors, Crb localizes to the stalk membrane, the portion of the apical membrane void of microvilli, and is critical for regulating the length of the stalk membrane. In crb mutant flies neighboring adjacent rhabdomeres remain juxtaposed [21], [34]–[37] implying the shortening of the stalk membrane permitted adjacent rhabdomeres to remain together. In the Act5C/+; EP-TH background we did not observe any visible alteration in Crb localization (Figure 2 C,F). Taken together these results implied that the increase in juxtaposed rhabdomeres, upon the decrease in Act5C genetic dosage, was not due to an obvious mislocalization of known retinal lumen formation proteins.
Figure 2. Localization of Prominin, EYS, and Crumbs in the Act5C/+; EP-TH genetic background.
Confocal immunofluorescence micrographs of wild type (A–C) and Act5C G0245/+; eys, prom/+ (D–F) ommatidium at 72 h APF stained with: (A,D) EYS and F-actin. (B,E) Prominin (Prom) and F-actin. (C,F) Crumbs (Crb) and F-actin. Scale bar, 2 µm.
Loss of non-muscle myosin II results in a discontinuous retinal lumen
To identify the mode of action responsible for the loss of a continuous luminal space in Act5C/+; EP-TH, we next examined the role of the actomyosin network. Actin is well known to interact with non-muscle myosin II (MyoII) to generate a contraction force to induce cellular morphological changes (for reviews see [38]–[40]). Previous studies in Drosophila photoreceptors have demonstrated a role of the actomyosin complex in regulating photoreceptor cell body position, adherens junction formation and apical contraction within the morphogenetic furrow [41]–[43]. Therefore, a plausible explanation was that the reduction of Act5C dosage decreased a contractile force in photoreceptors resulting in inability of the apical membranes to pull away from each other during the initial phase of lumen formation.
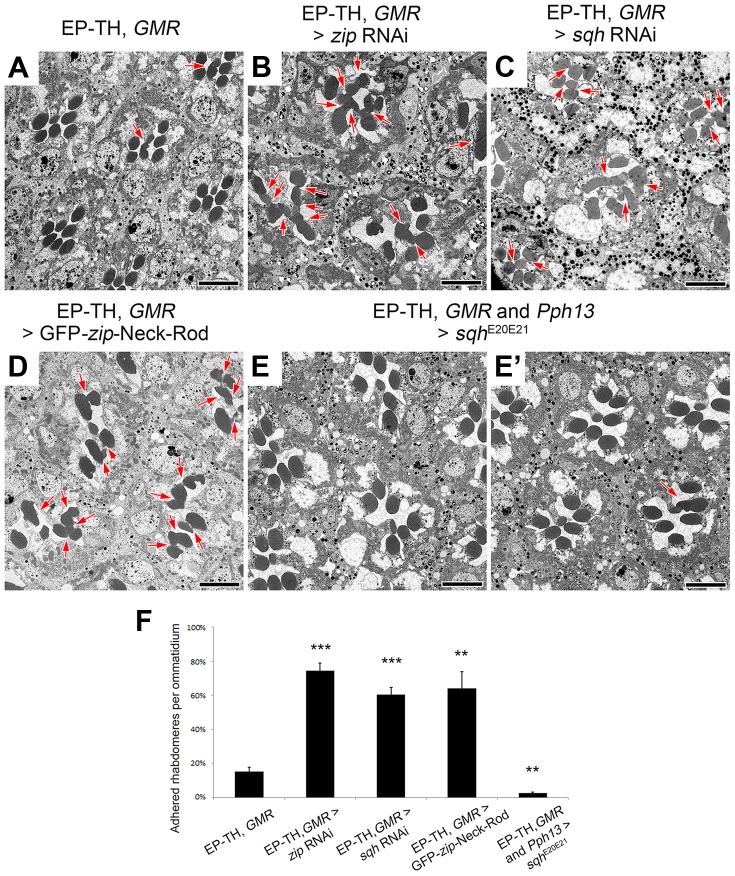
Non-muscle myosin II molecules are hexamers comprised of three pairs of subunits [44]: two heavy chains, encoded by zipper (zip), two regulatory light chains, spaghetti squash (sqh), and two essential light chains [45]–[47]. Unlike the loss of one copy of Act5C, a deletion that removes zip as well as specific zip alleles did not enhance the EP-TH phenotype. Nonetheless, only removing one genetic dosage of zip may not have been sufficient to lower protein levels. In an attempt to further reduce Zip protein levels we employed RNAi against zip. The ability of zip RNAi to decrease Zip protein levels was confirmed by immunofluorescence detection in 48 h after puparium formation (APF) pupal eyes with utilization of zip RNAi flip-out clones (Figure S5 A,B). Upon RNAi knockdown, we observed a significant increase in fused rhabdomeres in the EP-TH background that mimicked the loss of one copy of Act5C (Figure 3 A,B,F). To demonstrate phenotypic specificity, co-expression of UAS-GFP-zip along with UAS-zip-RNAi rescued the rhabdomere adhesion phenotype back to the EP-TH background levels (Figure S5 E,F). To further eliminate the possibility of RNAi off-target effects, we assayed the ability of a dominant-negative (DN) form of Zip (UAS-GFP-zip-Neck-Rod) [48], [49] to enhance rhabdomere fusion. We obtained a significant enhancement with the expression of the dominant-negative form of Zip in the EP-TH background (Figure 3 D,F). Similar to the MyoII heavy chain Zip, reduction of the myosin regulatory light chain, Sqh, also displayed an essential role in retinal lumen formation. RNAi knockdown of sqh, confirmed by immunofluorescence in flip-out clones (Figure S5 C,D), substantially enhanced the presence of juxtaposed rhabdomeres in the EP-TH background (Figure 3 C,F). In contrast, the co-expression of a constitutively active form of Sqh (UAS-sqh E20E21) [50] with the sqh RNAi construct reduced the rhabdomere enhancement back to EP-TH levels (Figure S5 G,H). More importantly, the expression of the constitutively active Sqh alone in the EP-TH background rescued the rhabdomere adhesion. The frequency of observed juxtaposed rhabdomeres was significantly reduced from 15% per ommatidium to 3% (Figure 3 E,F).
Figure 3. Non-muscle myosin II is involved in retinal lumen formation.
(A–E′) Transmission electron micrographs of adult Drosophila ommatidia. (A) eys, prom, GMR-GAL4/+. (B) eys, prom, GMR-GAL4/+; UAS-zip RNAi/+. (C) eys, prom, GMR-GAL4/+; UAS-sqh RNAi/+. (D) eys, prom, GMR-GAL4/+; UAS-GFP-zip-Neck-Rod/+. (E and E′) eys, prom, GMR-GAL4/+; Pph13-Gal4/UAS-sqh E20E21. (F) Quantitative analysis of rhabdomere fusion seen in (A–E′). Values represent mean ± SEM. *** P<0.001, **P<0.01 compared with eys, prom, GMR-Gal4/+. Arrows indicate adhesion between rhabdomeres. Scale bar, 5 µm.
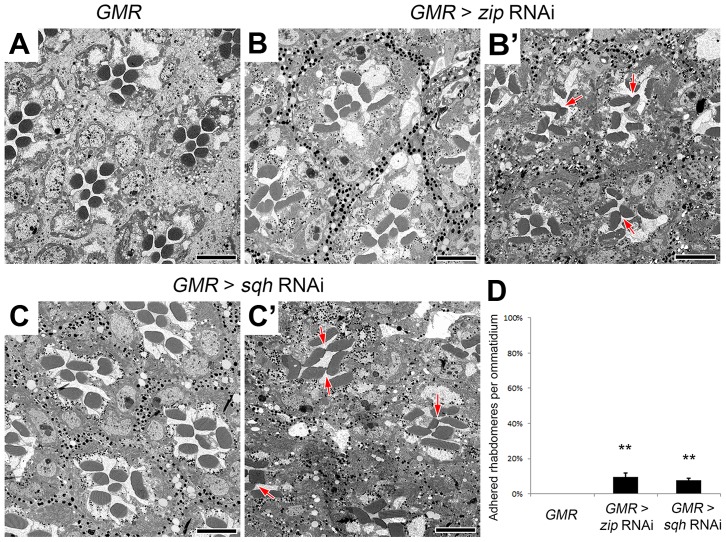
The effect of the reduction of the actomyosin machinery components, Act5C, Zip, and Sqh, on the formation of a continuous IRS was not limited to the sensitized EP-TH background. In an eys or prom single-heterozygous mutant background rhabdomere fusion is never observed (Figure S6 A,C,F,H), but in these single-heterozygous backgrounds the reduction of Act5C, Zip, or Sqh resulted in the appearance of fused rhabdomeres (Figure S6). Notably, in the wild-type background in which neither EYS nor Prom level is altered, the RNAi knockdown of zip, sqh as well as mild Act5C knockdown is sufficient in generating juxtaposed rhabdomeres (Figure 4, Figure S7); strong knockdown of Act5C with the GMR-GAL4 leads to loss of rhabdomere structures (Figure S1 H). These results demonstrated that the actomyosin complex is not only involved in, but is also required and essential for retinal lumen formation.
Figure 4. Non-muscle myosin II is required for retinal lumen formation.
(A–C′) Transmission electron micrographs of adult Drosophila ommatidia. (A) GMR-Gal4/+. (B and B′) GMR-Gal4/+; UAS-zip RNAi/+. (C and C′) GMR-Gal4/+; UAS-sqh RNAi/+. (D) Quantitative analysis of rhabdomere fusion seen in (A–C′). Values represent mean ± SEM. **P<0.01 compared with GMR-Gal4/+. Arrows indicate adhesion between rhabdomeres. Scale bar, 5 µm.
Regulation of the actomyosin machinery during retinal lumen formation
If the actomyosin machinery was a key element in the formation of the retinal lumen we should find common phenotypes among the potential upstream regulators of MyoII. To date there are more than a dozen of kinases reported to phosphorylate and activate MyoII regulatory light chain in invertebrate and vertebrate model organisms (reviewed in [40]). Fortuitously, our assay permitted a screening of potential regulators in Drosophila photoreceptor cells (Table S1). From our limited screen we found three candidates that when knocked-down were capable of enhancing the rhabdomere fusion in the EP-TH background: Rho-kinase (Rok) [51], and the upstream activator of Rok, the small GTPase Rho1 [52], and the upstream transcriptional regulator Snail [53] (Figure 5 A,B,D,E). However, Rok not only activates Sqh through direct phosphorylation, it also indirectly activates Sqh by inhibiting its inhibitor, the Myosin binding subunit (Mbs) of the myosin light chain phosphatase complex [54]. To test the possibility that the regulation of retinal lumen formation involves Mbs-mediated Sqh dephosphorylation, we reasoned that the overexpression of a constitutively active form of Mbs (MbsN300), that lacks the Rok regulatory target site [43] would result in an increase in rhabdomere fusion in the EP-TH background. With the expression of MbsN300, we observed an increase in rhabdomere fusion (Figure 5 C,E), suggesting Mbs is a negative regulator for actomyosin contraction in Drosophila retina. Jointly, these results indicated that in Drosophila photoreceptor cells, Sqh was positively regulated by Rok and Rho1, and negatively regulated by Mbs.
Figure 5. Regulators of Sqh are involved in retinal lumen formation.
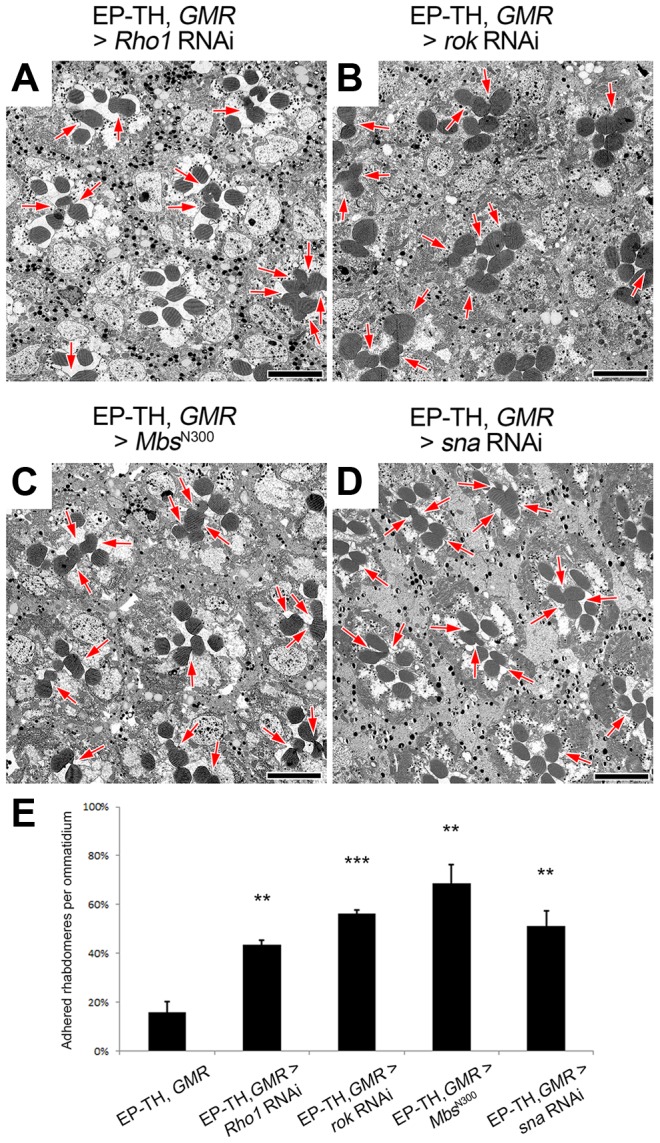
(A–D) Transmission electron micrographs of adult Drosophila ommatidia. (A) eys, prom, GMR-GAL4; UAS-Rho1 RNAi/+. (B) eys, prom, GMR-GAL4/+; UAS-rok RNAi/+. (C) eys, prom, GMR-GAL4/+; UAS-Mbs N300/+. (D) eys, prom, GMR-GAL4/+; UAS-snail RNAi/+. (E) Quantitative analysis of rhabdomere fusion seen in (A–D). Values represent mean ± SEM. *** P<0.001, **P<0.01 compared with eys, prom, GMR-Gal4/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
The actomyosin machinery localizes apically and is required for proper rhabdomere spacing
To investigate how the actomyosin machinery might contribute to retinal lumen formation, we first determined the sub-cellular localization of MyoII. Utilizing a GFP tagged Sqh under the transcriptional control of its native promoter [55], we observed that Sqh-GFP was localized to the apical cytosol of photoreceptor cells just basal to the developing rhabdomeres (Figure 6), which is consistent with Zip antibody staining pattern in Drosophila photoreceptors reported previously [56]. This localization pattern of Sqh was not dependent upon an extracellular matrix, since Sqh-GFP localized normally in the eys null (Figure S8 A,B) and the prom null background (Figure S8 C,D).
Figure 6. Sqh localization during retinal lumen formation.
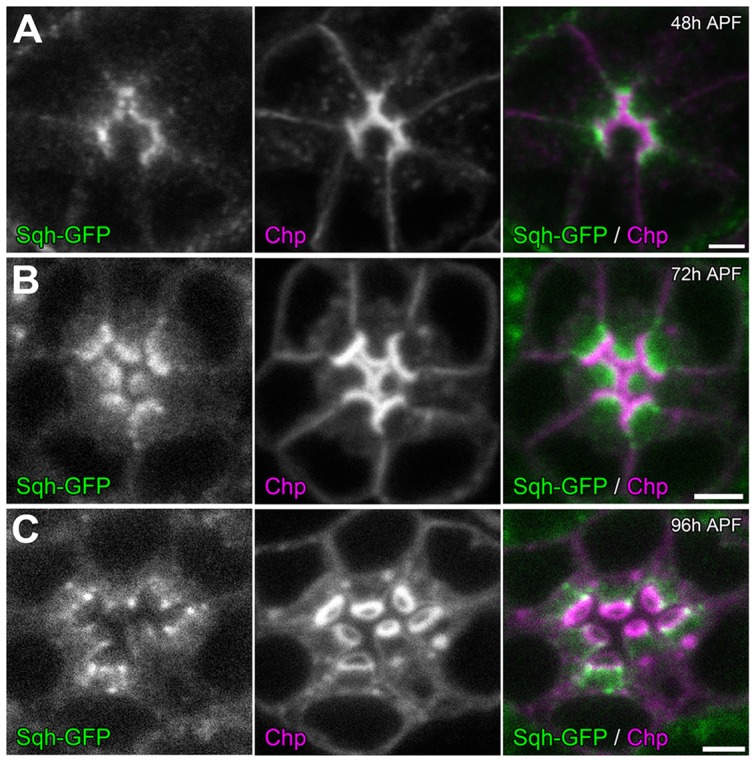
Confocal immunofluorescence micrographs of +/+; sqh-GFP/sqh-GFP (sqh-GFP in green) ommatidium co-stained with Chaoptin (Chp, magenta) at: (A) 48 h APF. (B) 72 h APF. (C) 96 h APF. Chaoptin labels the developing rhabdomeres.
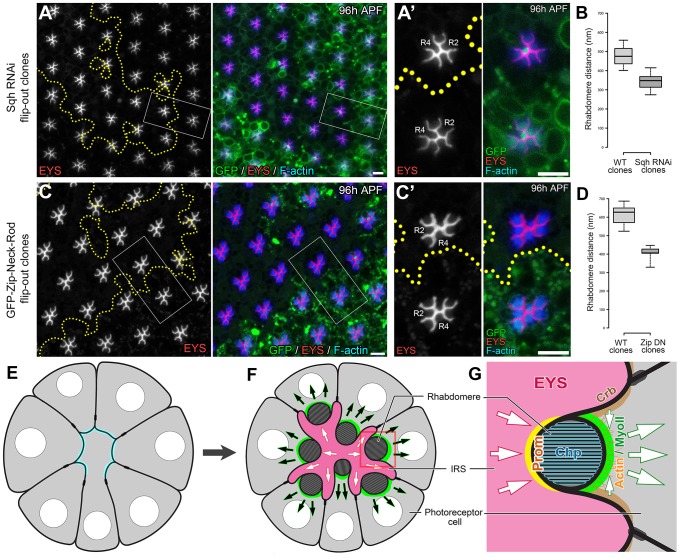
The apical localization pattern of MyoII predicts that the potential contraction may act on the apical membranes. To detect potential defects in retraction of the apical membrane, we investigated whether the distance between apical rhabdomere membranes was altered upon the reduction of MyoII in a wild-type background. We generated sqh RNAi (Figure 7 A) and zip dominant-negative (GFP-Zip-Neck-Rod) (Figure 7 C) flip-out clones and measured the distance between the apical rhabdomeres. In particular we measured the width of the IRS between the apical membranes of rhabdomeres R2 and R4, two diametrically opposing rhabdomeres. The distance between apical rhabdomere membranes, which is equal to the width of the retinal lumen, was visualized by EYS staining. Our assay demonstrated that the lumen width decreased by 28% in sqh RNAi flip-out clones and by 33% in GFP-zip-Neck-Rod flip-out clones compared with their neighboring wild-type clones (Figure 7 A–D). These results are consistent with a hypothesis that the actomyosin machinery generates a contraction force at the apical photoreceptor cell membranes to pull the apical membranes inward to initiate and expand the retinal lumen (Figure 7 E–G). We also tested other aspects of cell morphology, and we found that this apically localized actomyosin machinery is not sufficient to induce a whole cell size change in photoreceptors nor does reduction of MyoII affect the overall extension of the rhabdomeres (Figure S9). However, we also noted changes in rhabdomere width, oblong rhabdomeres, with the knockdown of the actomyosin machinery but this phenotype did not correlate with the enhanced rhabdomere adhesion observed (Figure S10).
Figure 7. Actomyosin contraction is required for increasing the width of the retinal lumen.
(A) RNAi knockdown of Sqh in the flip-out clones. hs-flp/+; GMR>w + STOP>Gal4/+; UAS-sqh RNAi/UAS-mCD8-GFP. (C) Overexpression of a dominant-negative form of Zip in flip-out clones. hs-flp/+; GMR>w + STOP>Gal4/+; UAS-GFP-zip-Neck-Rod/UAS-mCD8-GFP. GFP (green) marks the cells expressing the RNAi or dominant negative constructs, EYS (red) labels the retinal lumen, and F-actin (blue) labels the rhabdomeres. (A, C) Low magnification view. (A′, C′) A magnified view of the highlighted areas. (B, D) Box and whisker plot of the distance between the R2 and the R4 rhabdomeres of the ommatidia shown in (A) (n = 31) and (C) (n = 18), respectively. Mosaic ommatidium containing both wildtype and mutant cells were not quantified for the box plot. Boxes extend from 25th to 75th percentile, with a line at the median. Whiskers extend to the most extreme values (Spear style). (E–G) Model for Drosophila retinal lumen formation. Arrows indicate direction of forces. Scale bar, 5 µm.
Temporal sequence of contraction, secretion, and adhesion
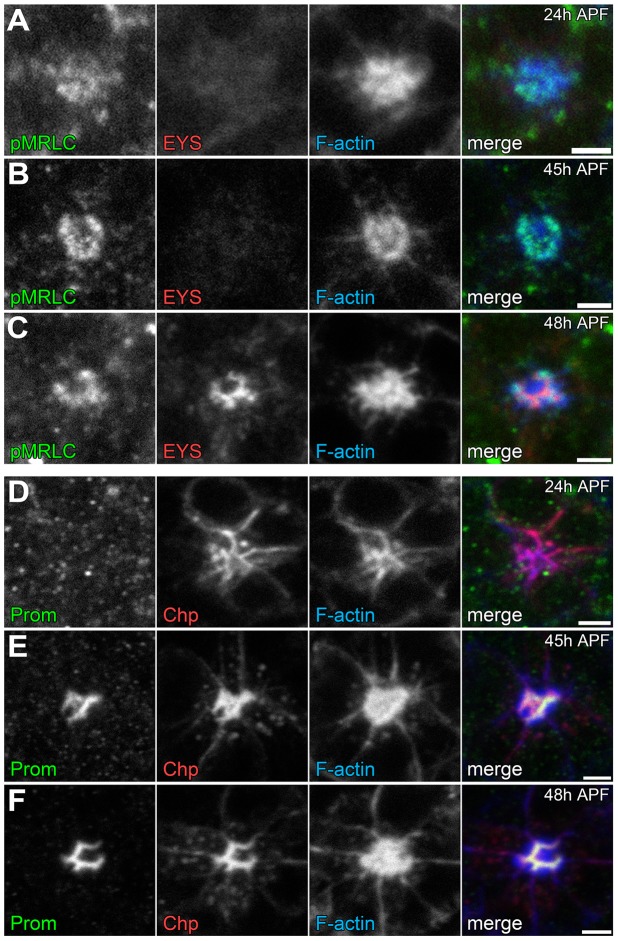
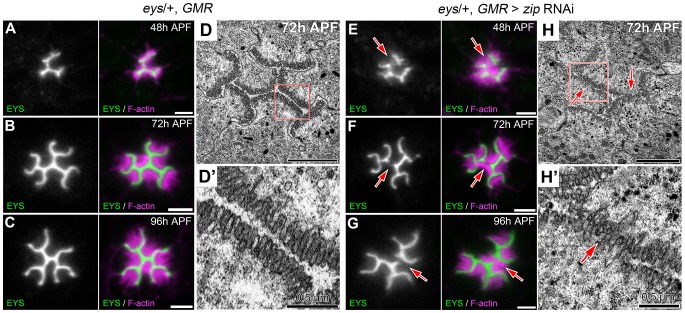
Temporally, we know that the photoreceptor cell apical membranes are juxtaposed to each other as early as 24 h APF [30] and as expected the adhesive membrane protein Chaoptin was detected on the apical surface linking the membranes together (Figure 8 D). Concurrently, we also observed not only an accumulation of F-actin at the apical surface but also the phosphorylated form of Sqh (Figure 8 A). In contrast, at 24 h APF, neither Prom nor EYS was detected on the apical surface (Figure 8 A,D). Subsequently, as the apical membranes initiate their transformation we observed the accumulation of Prom at 45 h APF on the apical surface (Figure 8 E,F) only then followed by the appearance of EYS at 48 h APF (Figure 8 B,C). Consistent with the early localization of MyoII to the apical surface, we can detect MyoII-induced rhabdomere fusion as early as 48 h APF (Figure 9); without MyoII knockdown, the eys heterozygous mutant alone does not lead to rhabdomere fusion (Figures S6 C). TEM analysis at 72 h clearly demonstrated the interlocking of microvilli between rhabdomeres of different photoreceptors (Figure 9 H). These data indicated that the critical developmental role of the actomyosin machinery was occurring prior to 48 h APF.
Figure 8. Temporal profile of the coordination of actomyosin contraction, steric hindrance of adhesion, and secretion of an extracellular matrix.
(A–F) Confocal immunofluorescence micrographs of wild-type ommatidium. (A–C) Ommatidium stained with phospho-Sqh (pMRLC, green), EYS (red), and F-Actin (blue) at: (A) 24 h APF. (B) 45 h APF. (C) 48 h APF. (D–F) Ommatidium stained with Prominin (Prom, green), Chaoptin (Chp, red), and F-Actin (blue) at: (D) 24 h APF. (E) 45 h APF. (F) 48 h APF. Scale bar, 2 µm.
Figure 9. Defects in apical membrane separation are detected early and maintained throughout development.
(A–C, E–G) Confocal immunofluorescence micrographs of (A–C) eys, GMR-Gal4/+ ommatidium and (E–G) eys, GMR-Gal4/+; UAS-zip RNAi/+ ommatidium co-stained with EYS (green) and F-actin (magenta) at (A,E) 48 h APF. (B,F) 72 h APF. (C,G) 96 h APF. (D,H) Transmission electron micrograph of (D,D′) eys, GMR-Gal4/+ ommatidium and (H,H′) eys, GMR-Gal4/+; UAS-zip RNAi/+ ommatidium at 72 h APF, (D′,H′) represents high magnification of boxed region in (D) or (H), respectively. Arrows denote juxtaposed apical rhabdomere membranes. Scale bar, (A–D, E–H) 2 µm, (D′,H′) 0.5 µm.
Discussion
Drosophila retinal lumen formation: a process of chord hollowing initiated by actomyosin machinery
There are several morphological strategies to generate luminal spaces (reviewed in [2], [3]). Chord hollowing is the process in which cells create a de novo lumen between their apical domains, as exemplified by Madin-Darby canine kidney (MDCK) cysts [57]. The process of chord hollowing best describes the mechanism observed in each Drosophila ommatidium. In Drosophila, the encasing of the photoreceptors by the overlying cones cells results in the apical membranes of the eight photoreceptors cells to rotate 90 degrees inward and in the end are now juxtaposed to each other [30]. Subsequently, the depositing of an extracellular matrix between the apical domains generates the retinal lumen, the IRS [16], [17]. As described lumen formation involves a few common design principles [1], [2] and with respect to chord hollowing the critical step being how space is initiated between adherent cells.
Initially, our genetic analysis of mutations for Prom, a glycosylated surface protein, and EYS, a secreted extracellular protein [17], strengthened the notion that there are molecules that provide anti-adhesive properties and the concurrent depositing of an extracellular matrix or osmotic pressure results in the permanent separation of the membranes. In Drosophila photoreceptors, Prom has the potential to act as an anti-adhesive molecule as described for Mucin1 and members of the CD34 family of proteins (reviewed in [2]). Prom is a five-transmembrane protein with two large extracellular loops containing a minimum of four sites that are N-glycosylated [18]. Prom is present on the apical surface before secretion of the extracellular matrix and in combination with EYS provides a barrier to prevent interactions between the adhesive molecule Chp [17], [20], [22]. Nonetheless, the exact mechanism of its anti-adhesive properties remains ambiguous. We cannot separate the role of N-glycosylation from proper trafficking of Prom to the membrane [18]. Furthermore we have not been able to confirm whether Prom's interaction with EYS is essential for membrane separation or whether the anti–adhesive properties of Prom is sufficient to prevent the interaction of Chaoptin between rhabdomeres as long as a force is supplied to keep the membranes apart. With respect to the force, the secretion of EYS fulfills this role. Not only is the separation of the membranes and formation of the retinal lumen dependent on the presence of EYS [16], [17] but alteration of the amount of EYS is sufficient to modulate the diameter of the lumen [17].
Nevertheless, even with these two well-defined parameters, anti-adhesion and secretion, numerous questions remain about membrane separation. For instance, do the photoreceptor cells themselves generate an active force for separating the rhabdomere membranes? Thus our sensitized EP-TH genetic background provided an opportunity to not only address potential questions related to the regulation of Prom and EYS but also reveal additional mechanisms for retinal lumen formation. Here our results demonstrated that the specific reduction in the genetic dosage of Act5C increased the likelihood that the apical membranes remaining juxtaposed. The reduction of Act5C genetic dosage did not affect rhabdomere morphogenesis, the trafficking of components to the apical membrane, or secretion of EYS but rather revealed the critical role of actomyosin machinery in separating the apical membranes; the reduction of the myosin heavy chain, Zip, and the myosin regulatory light chain, Sqh, phenocopied the results obtained with Act5C. In light of the fact that the loss of actomyosin machinery results in rhabdomere fusion in the presence of wild-type levels of Prom and EYS demonstrates that the role of the actomyosin machinery is not an accessory process but a required mechanism for retinal lumen formation.
Thus all together our data advocate a temporal framework for initiation of the retinal luminal space (Figure 7 E–G). Based on our results, the accumulation of the actomyosin complex on the apical surface occurs prior to the secretion of an extracellular matrix. The combination of MyoII and the actin meshwork generates a contractile force that pulls the apical membrane towards the center of the photoreceptor cell. Furthermore, the polarized localization of MyoII, rather than a circumferential cable pattern or a dispersed pattern, suggested that in photoreceptor cells the actomyosin contraction may not be a “purse string” or a “ratchet-like” mechanism which contracts the cell from the periphery to the center, as reported in vertebrate neurulation and in Drosophila mesoderm invagination (see review [38]). We propose that together with the subsequent accumulation of Prom, the actomyosin contraction provides the necessary separation and weakening of inter-apical membrane interactions. Lastly, the secretion of EYS and its interaction with Prom provides an additional separation force and permanent barrier to prevent the adhesive properties of Chp from interacting between rhabdomeric membranes, thus fully establishing and stabilizing the retinal lumen space.
Comparison of actomyosin contraction in lumen formation and tissue morphogenesis
Whether the role of actomyosin contraction is limited to Drosophila retinal lumen formation or chord hollowing remains to be tested. Nonetheless, actomyosin contraction has been implicated in other luminal systems and is likely another core process required for lumen formation. For example, during murine vascular lumen formation, MyoII fails to localize to the apical cell membrane upon pharmacological inhibition of ROCK or its upstream activator vascular endothelial growth factor A, and the vascular tubes formed to a lesser extent [58]. Furthermore, the actomyosin contraction observed in Drosophila photoreceptors may also extend to vertebrate ciliary photoreceptors. Previous work has demonstrated the functional conservation of EYS and Prom among rhabdomeric and ciliary photoreceptors [18]. Interestingly, both Prominin1 and MyoII localize to the basal region of the outer segment where the nascent discs are formed in mammalian photoreceptors [59]–[61]. Therefore, one possibility is that actomyosin contraction is involved in the intra-cellular disk lumen formation. Specifically, actomyosin contraction might be required to pull the plasma ciliary membrane inwards to form and morphologically flatten the nascent discs [62], [63]. Overall, utilization of Drosophila retinal lumen as a model tissue, in particular our EP-TH sensitized genetic background, to perform unbiased screens will further define the mechanisms for activation of the actomyosin machinery and elucidate mechanisms for lumen formation and regulation.
Materials and Methods
Drosophila stocks and clonal analysis
All crosses and staging were performed at 23°C unless otherwise noted. Drosophila stocks used in this study include: prom 1, eys 1 [17], UAS-GFP-zip, UAS-GFP-zip-Neck-Rod (Dr. D. Kiehart), UAS-Mbs N300 (Dr. J. Treisman), sqh-GFP (Dr. A. Martin), GMR>w + STOP>Gal4 (Dr. C. Desplan), and UAS-sqh E20E21 (Dr. M. Birnbaum). The following fly stocks were obtained from the Bloomington Drosophila Stock Center: w1118, GMR-Gal4, UAS-mCD8-GFP, Act5C G0245, Act5C G0009, Act5C G0010, Act5C G0025, Act5C G0177, zip 1, sqh AX3, Df(1)ED6829, Df(1)ED418, Df(2R)ED1484, Df(2R)ED3791, Df(3L)BSC223, Df(3R)ED5613, Df(3R)ED5705, UAS-zip RNAi (TRiP #HMS01618), UAS-sqh RNAi (TRiP #HMS00437), UAS-Rho1 RNAi (TRiP #JF02809), UAS-rok RNAi (TRiP #JF03225), UAS-crb RNAi (TRiP #JF02777), UAS-Act5C RNAi (TRiP #HMS02487), UAS-atg1 RNAi (TRiP #JF02273, and #GL00047), UAS-cta RNAi (TRiP #JF01607, and #HMS02362), UAS-fog RNAi (TRiP #GL00529), UAS-mist RNAi (TRiP #HMS02327), UAS-RhoGEF2 RNAi (TRiP #HMS01118), UAS-sna RNAi (TRiP #HMS01252), UAS-SNF1A RNAi (TRiP #JF01951, #HMS00362, and #GL00004), UAS-sqa RNAi (TRiP #JF02277), UAS-Strn-Mlck RNAi (TRiP #JF02278, #JF02170, and #HMS01665), UAS-T48 RNAi (TRiP #HMS02248), and UAS-twi RNAi (TRiP #JF02003, and #HMS01317). Pph13-Gal4 was generated by inserting the immediate upstream 1.6 kb of genomic DNA extending from the first coding Methionine of the Pph13 locus into pCHS-GAL4. prom-Gal4 was generated by inserting the immediate upstream 3.6 kb of genomic DNA extending from the first coding Methionine of the prominin locus into pCHS-GAL4.
To generate flip-out clones, UAS-zip RNAi, UAS-GFP-zip-Neck-Rod, or UAS-sqh RNAi males were crossed with hs-Flp; GMR>w + STOP>Gal4; UAS-mCD8-GFP females, and the 24 h–48 h larvae were subject to 1 h heatshock at 37°C and then returned to 23°C to generate clones. GMR promoter starts to drive Gal4 expression in photoreceptors of flip-out clones starting from the third instar larval stage. Clones were marked by the presence of mCD8-GFP. To generate MARCM clones, sqh AX3, Frt19A; GMR-Gal4 females were crossed with hs-Flp, tub-Gal80, Frt19A; UAS-mCD8-GFP males, and the third instar larvae were subject to 1 h heatshock at 37°C and then returned to 23°C. Mutant clones were marked by the presence of mCD8-GFP.
Transmission electron microscopy, immunofluorescence staining, and imaging
Drosophila eye samples were prepared for transmission electron microscopy (TEM) as previously described [17]. All crosses were maintained at 23°C and adult heads were fixed within 8 h after eclosion. Standard fixation and staining protocols were used for immunofluorescence staining. Briefly, pupal retinas were staged at 23°C, dissected in PBS, and fixed in PBS containing 4% formaldehyde for 10 min (24 h, 45 h, and 48 h APF pupae, as well as 72 h APF pupae for anti-Prom staining) or 40 min (72 h and 96 h APF pupae). The primary antibodies used were: mouse anti-EYS (mAb 21A6, 1∶50, Developmental Studies Hybridoma Bank) [17]; rabbit anti-Prom (1∶100) [17]; rat anti-Crb (1∶400, Dr. E. Knust) [64]; rabbit anti-Zip (#656, 1∶400, Dr. D. Kiehart) [65]; guinea pig anti-Sqh (GP#21, 1∶400, Dr. D. Kiehart) [66]; mouse anti–phospho–Sqh (Ser19, analogous to D. melanogaster Sqh Ser21) (pMRLC, 1∶100, Cell Signaling Technology); mouse anti-Chp (mAb 24B10, 1∶100, Developmental Studies Hybridoma Bank) [19]; mouse anti-Na+ K+ ATPase alpha subunit (a5, 1∶100, Developmental Studies Hybridoma Bank) [29]; rabbit anti-phospho-Moesin (pMoe) (mAb 41A3, 1∶100, Cell Signaling). Rhodamine (1∶200) or Alexa Fluor 647 (1∶50) conjugated phalloidin (Life Technologies) was used for the detection of F-actin. The FITC or RX conjugated secondary antibodies (1∶200) were obtained from Jackson ImmunoResearch Laboratories. Confocal images were taken on a Leica TCS SP5 and TEM was performed with a JOEL 1010, and all pictures were processed in Adobe Photoshop.
Supporting Information
Reduction of Act5C dosage enhances rhabdomere adhesion. (A–G) Transmission electron micrographs of adult Drosophila ommatidia. (A) eys, prom/+. (B) Df(1)ED418/+; eys, prom/+. The deficiency removes genomic region 5C7-5E4. (C) Act5C G0009/+; eys, prom/+. (D) Act5C G0010/+; eys, prom/+. (E) Act5C G0025/+; eys, prom/+. (F) Act5C G0177/+; eys, prom/+. (G) eys, prom/+; prom-Gal4/UAS-Act5C RNAi. (H) eys, prom, GMR-GAL4/+; UAS-Act5C RNAi/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
Rhabdomere fusion is specific to the reduction in Act5C. (A–F) Transmission electron micrographs of adult Drosophila ommatidia of (A) eys, prom/+ with deficiencies that remove one of the two copies of: (B) Act42A, (C) Act57B, (D) Act79B, (E) Act87E, (F) Act88F. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
Reduction of Act5C dosage does not affect the vertical extension of the rhabdomeres. (A–D) Confocal immunofluorescence micrographs showing the vertical view of adult Drosophila ommatidia. (A) w1118, wild type. (B) Act5C G0245/+. (C) eys, prom/+. (D) Act5C G0245/+; eys, prom/+. Na+ K+ ATPase (NaK, green) labels the basolateral membranes of photoreceptor cells, and F-actin (magenta) labels the rhabdomeres. Scale bar, 10 µm.
(TIF)
The localization of phospho-Moesin is not affected by the reduction of Act5C genetic dosage. (A,B) Confocal immunofluorescence micrographs of 48 h APF Drosophila ommatidium. Phospho-Moesin (pMoe, green) labels the activated form of Moesin, and F-actin (magenta) labels the rhabdomeres. (A) w1118, wild type. (B) Act5C G0245/+. Scale bar, 2 µm.
(TIF)
zip RNAi and sqh RNAi specifically target zipper and spaghetti squash. (A,B) Confocal immunofluorescence micrographs of zip RNAi flip-out clones (hs-flp/+; GMR>w + STOP>Gal4/+; UAS-zip RNAi/UAS-mCD8-GFP) of pupal retinas at 48 h APF visualizing mCD8-GFP (green), Zip (red), and F-Actin (blue). The RNAi expressing cells are marked with GFP and resulted in a reduction of Zip immunoreactivity. (C,D) Confocal immunofluorescence micrographs of sqh RNAi flip-out clones (hs-flp/+; GMR>w + STOP>Gal4/+; UAS-sqh RNAi/UAS-mCD8-GFP) of pupal retinas at 48 h visualizing mCD8-GFP (green), Sqh (red), and F-Actin (blue). The RNAi expressing cells are marked with GFP and resulted in a reduction of Sqh immunoreactivity. Dotted lines delineate wild-type and knockdown cells. (E–H) Transmission electron micrographs of adult Drosophila ommatidia of (E) eys, prom, GMR-GAL4/+; UAS-zip RNAi/UAS-mCD8-GFP. (F) eys, prom, GMR-GAL4/+; UAS-zip RNAi/UAS-GFP-zip. (G) eys, prom, GMR-GAL4/+; UAS-sqh RNAi/UAS-mCD8-GFP. (H) eys, prom, GMR-GAL4/+; UAS-sqh RNAi/UAS-sqh E20E21. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, (A,C) 30 µm, (B,D, E–H) 5 µm.
(TIF)
Reduction of actomyosin components results in rhabdomere fusion in eys or prom single-heterozygote background. (A–J) Transmission electron micrographs of adult Drosophila ommatidia of (A) eys/+. (B) Act5C G0245/+; eys/+. (C) eys, GMR-Gal4/+. (D) eys, GMR-Gal4/+; UAS-zip RNAi/+, (E) eys, GMR-Gal4/+; UAS-sqh RNAi/+. (F) prom/+ (G) Act5C G0245/+; prom/+. (H) prom, GMR-Gal4/+. (I) prom, GMR-Gal4/+; UAS-zip RNAi/+, (J) prom, GMR-Gal4/+; UAS-sqh RNAi/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
RNAi knockdown of Act5C results in rhabdomere fusion in the otherwise wild-type background. (A–D′) Transmission electron micrographs of adult Drosophila ommatidia of (A) w 1118. (B) Act5C G0245/+. (C,C′) +/+; prom-Gal4/UAS-Act5C RNAi. (D,D′) +/+; Pph13-Gal4/UAS-Act5C RNAi. (C′,D′) A magnified view of the highlighted areas in (C) and (D), respectively. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, (A,B,C,D) 5 µm, (C′,D′) 2 µm.
(TIF)
Sqh localization is not dependent on Prominin or EYS. Confocal immunofluorescence micrographs of eys (A,B) and prom (C,D) null mutant ommatidium with sqh-GFP (green) expression, and stained with Chaoptin (Chp, red), and F-Actin (blue) at 48 h APF (A,C) and 72 h APF (B,D). Chaoptin marks the rhabdomeres, and F-actin mainly marks the rhabdomeres but also weakly labels the rhabdomere terminal web. Scale bar, 2 µm.
(TIF)
Reduction of Sqh does not alter the whole cell size. (A–E) Confocal immunofluorescence micrographs of clones of cells lacking Sqh protein in (A–D) horizontal and (E) vertical optical sections. Effects of loss of Sqh by RNAi knockdown (A,B,E) or MARCM (C,D) are imaged at (A,C) 48 h APF, (B,D) 72 h APF, or (E) in adult eyes. (A,B,E) hs-flp/+; GMR>w + STOP>Gal4/+; UAS-sqh RNAi/UAS-mCD8-GFP. (C,D) sqh AX3, Frt19A/hs-flp, tub-Gal80, Frt19A; GMR-Gal4, UAS-mCD8-GFP/+. In all panels, GFP marks the mutant cells. Na+ K+ ATPase (NaK, red in A–D and blue in E) labels the lateral and basal plasma membrane and F-actin (blue in A–D and red in E) labels the developing rhabdomeres. Scale bars, (A–D) 5 µm, (E) 20 µm.
(TIF)
Changes in rhabdomere width do not contribute to the adhesion of rhabdomeres. (A) Transmission electron micrographs of adult eys, prom, GMR-GAL4/+; UAS-sqh RNAi ommatidia. Oblong rhabdomeres are defined as rhabdomeres with their width equal to or greater than twice of their length. Arrows indicate fusion between rhabdomeres, and asterisks label oblong rhabdomeres. (B) Quantitative analysis of rhabdomere fusion in normal ommatidia and in ommatidia with oblong rhabdomere(s). Not statistically significant (NS), P>0.05. Scale bars, 2 µm.
(TIF)
Tested potential actomyosin regulators via RNAi in the EP-TH genetic background.
(DOCX)
Acknowledgments
We thank Drs. M. Birnbaum, C. Desplan, D. Kiehart, E. Knust, A. Martin, J. Treisman, R. Ward, the Bloomington Drosophila Stock Center and Developmental Studies Hybridoma Bank for reagents. We thank the assistance of Dr. J. Powers and the Indiana University Light Microscopy Imaging Center for image generation. We thank Drs. K. Hu, N. Sokol, and J. Kumar for comments on the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded from the American Cancer Society (RSG-10-041-01-DDC) to ACZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Andrew DJ, Ewald AJ (2010) Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev Biol 341: 34–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Datta A, Bryant DM, Mostov KE (2011) Molecular regulation of lumen morphogenesis. Curr Biol 21: R126–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lubarsky B, Krasnow MA (2003) Tube morphogenesis: Making and shaping biological tubes. Cell 112: 19–28. [DOI] [PubMed] [Google Scholar]
- 4. Beitel GJ, Krasnow MA (2000) Genetic control of epithelial tube size in the Drosophila tracheal system. Development 127: 3271–3282. [DOI] [PubMed] [Google Scholar]
- 5. Forster D, Armbruster K, Luschnig S (2010) Sec24-Dependent Secretion Drives Cell-Autonomous Expansion of Tracheal Tubes in Drosophila. Curr Biol 20: 62–68. [DOI] [PubMed] [Google Scholar]
- 6. Grieder NC, Caussinus E, Parker DS, Cadigan K, Affolter M, et al. (2008) gamma COP Is Required for Apical Protein Secretion and Epithelial Morphogenesis in Drosophila melanogaster. Plos One 3: e3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tsarouhas V, Senti KA, Jayaram SA, Tiklova K, Hemphala J, et al. (2007) Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev Cell 13: 214–225. [DOI] [PubMed] [Google Scholar]
- 8. Lowery LA, Sive H (2005) Initial formation of zebrafish brain ventricles occurs independently of circulation and requires the nagie oko and snakehead/atp1a1a.1 gene products. Development 132: 2057–2067. [DOI] [PubMed] [Google Scholar]
- 9. Bagnat M, Cheung ID, Mostov KE, Stainier DYR (2007) Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol 9: 954–U119. [DOI] [PubMed] [Google Scholar]
- 10. Olver RE, Strang LB (1974) Ion Fluxes across Pulmonary Epithelium and Secretion of Lung Liquid in Fetal Lamb. Journal of Physiology-London 241: 327–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Myat MM, Andrew DJ (2002) Epithelial tube morphology is determined by the polarized growth and delivery of apical membrane. Cell 111: 879–891. [DOI] [PubMed] [Google Scholar]
- 12. Omori Y, Malicki J (2006) oko meduzy and related crumbs genes are determinants of apical cell features in the vertebrate embryo. Curr Biol 16: 945–957. [DOI] [PubMed] [Google Scholar]
- 13. Cagan RL, Ready DF (1989) The emergence of order in the Drosophila pupal retina. Dev Biol 136: 346–362. [DOI] [PubMed] [Google Scholar]
- 14. Kirschfeld K (1967) [The projection of the optical environment on the screen of the rhabdomere in the compound eye of the Musca]. Exp Brain Res 3: 248–270. [DOI] [PubMed] [Google Scholar]
- 15.Land MF, Nilsson DE (2002) Animal Eyes: Oxford University Press.
- 16. Husain N, Pellikka M, Hong H, Klimentova T, Choe KM, et al. (2006) The agrin/perlecan-related protein eyes shut is essential for epithelial lumen formation in the Drosophila retina. Dev Cell 11: 483–493. [DOI] [PubMed] [Google Scholar]
- 17. Zelhof AC, Hardy RW, Becker A, Zuker CS (2006) Transforming the architecture of compound eyes. Nature 443: 696–699. [DOI] [PubMed] [Google Scholar]
- 18. Nie J, Mahato S, Mustill W, Tipping C, Bhattacharya SS, et al. (2012) Cross species analysis of Prominin reveals a conserved cellular role in invertebrate and vertebrate photoreceptor cells. Dev Biol 371: 312–320. [DOI] [PubMed] [Google Scholar]
- 19. Reinke R, Krantz DE, Yen D, Zipursky SL (1988) Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell 52: 291–301. [DOI] [PubMed] [Google Scholar]
- 20. Van Vactor D Jr, Krantz DE, Reinke R, Zipursky SL (1988) Analysis of mutants in chaoptin, a photoreceptor cell-specific glycoprotein in Drosophila, reveals its role in cellular morphogenesis. Cell 52: 281–290. [DOI] [PubMed] [Google Scholar]
- 21. Gurudev N, Yuan M, Knust E (2014) chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells. Biol Open 3: 332–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krantz DE, Zipursky SL (1990) Drosophila chaoptin, a member of the leucine-rich repeat family, is a photoreceptor cell-specific adhesion molecule. EMBO J 9: 1969–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Francesc N, Kirschfe K (1971) Phenomena of Pseudopupil in Compound Eye of Drosophila. Kybernetik 9: 159–182. [DOI] [PubMed] [Google Scholar]
- 24. Fyrberg EA, Bond BJ, Hershey ND, Mixter KS, Davidson N (1981) The actin genes of Drosophila: protein coding regions are highly conserved but intron positions are not. Cell 24: 107–116. [DOI] [PubMed] [Google Scholar]
- 25. Fyrberg EA, Mahaffey JW, Bond BJ, Davidson N (1983) Transcripts of the six Drosophila actin genes accumulate in a stage- and tissue-specific manner. Cell 33: 115–123. [DOI] [PubMed] [Google Scholar]
- 26. Wagner CR, Mahowald AP, Miller KG (2002) One of the two cytoplasmic actin isoforms in Drosophila is essential. Proceedings of the National Academy of Sciences of the United States of America 99: 8037–8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arikawa K, Hicks JL, Williams DS (1990) Identification of actin filaments in the rhabdomeral microvilli of Drosophila photoreceptors. J Cell Biol 110: 1993–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Karagiosis SA, Ready DF (2004) Moesin contributes an essential structural role in Drosophila photoreceptor morphogenesis. Development 131: 725–732. [DOI] [PubMed] [Google Scholar]
- 29. Li BX, Satoh AK, Ready DF (2007) Myosin V, Rab11, and dRip11 direct apical secretion and cellular morphogenesis in developing Drosophila photoreceptors. J Cell Biol 177: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Longley RL Jr, Ready DF (1995) Integrins and the development of three-dimensional structure in the Drosophila compound eye. Dev Biol 171: 415–433. [DOI] [PubMed] [Google Scholar]
- 31. Tuxworth RI, Titus MA (2000) Unconventional myosins: anchors in the membrane traffic relay. Traffic 1: 11–18. [DOI] [PubMed] [Google Scholar]
- 32. Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480. [DOI] [PubMed] [Google Scholar]
- 33. Yang Z, Chen Y, Lillo C, Chien J, Yu Z, et al. (2008) Mutant prominin 1 found in patients with macular degeneration disrupts photoreceptor disk morphogenesis in mice. J Clin Invest 118: 2908–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Izaddoost S, Nam SC, Bhat MA, Bellen HJ, Choi KW (2002) Drosophila Crumbs is a positional cue in photoreceptor adherens junctions and rhabdomeres. Nature 416: 178–183. [DOI] [PubMed] [Google Scholar]
- 35. Pellikka M, Tanentzapf G, Pinto M, Smith C, McGlade CJ, et al. (2002) Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 416: 143–149. [DOI] [PubMed] [Google Scholar]
- 36. Chartier FJ, Hardy EJ, Laprise P (2012) Crumbs limits oxidase-dependent signaling to maintain epithelial integrity and prevent photoreceptor cell death. J Cell Biol 198: 991–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Johnson K, Grawe F, Grzeschik N, Knust E (2002) Drosophila crumbs is required to inhibit light-induced photoreceptor degeneration. Curr Biol 12: 1675–1680. [DOI] [PubMed] [Google Scholar]
- 38. Martin AC (2010) Pulsation and stabilization: contractile forces that underlie morphogenesis. Dev Biol 341: 114–125. [DOI] [PubMed] [Google Scholar]
- 39. Sawyer JM, Harrell JR, Shemer G, Sullivan-Brown J, Roh-Johnson M, et al. (2010) Apical constriction: a cell shape change that can drive morphogenesis. Dev Biol 341: 5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol 10: 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Corrigall D, Walther RF, Rodriguez L, Fichelson P, Pichaud F (2007) Hedgehog signaling is a principal inducer of Myosin-II-driven cell ingression in Drosophila epithelia. Dev Cell 13: 730–742. [DOI] [PubMed] [Google Scholar]
- 42. Escudero LM, Bischoff M, Freeman M (2007) Myosin II regulates complex cellular arrangement and epithelial architecture in Drosophila. Dev Cell 13: 717–729. [DOI] [PubMed] [Google Scholar]
- 43. Lee A, Treisman JE (2004) Excessive Myosin activity in mbs mutants causes photoreceptor movement out of the Drosophila eye disc epithelium. Mol Biol Cell 15: 3285–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Korn ED, Hammer JA 3rd (1988) Myosins of nonmuscle cells. Annual review of biophysics and biophysical chemistry 17: 23–45. [DOI] [PubMed] [Google Scholar]
- 45. Edwards KA, Chang XJ, Kiehart DP (1995) Essential light chain of Drosophila nonmuscle myosin II. J Muscle Res Cell Motil 16: 491–498. [DOI] [PubMed] [Google Scholar]
- 46. Karess RE, Chang XJ, Edwards KA, Kulkarni S, Aguilera I, et al. (1991) The regulatory light chain of nonmuscle myosin is encoded by spaghetti-squash, a gene required for cytokinesis in Drosophila. Cell 65: 1177–1189. [DOI] [PubMed] [Google Scholar]
- 47. Kiehart DP, Lutz MS, Chan D, Ketchum AS, Laymon RA, et al. (1989) Identification of the gene for fly non-muscle myosin heavy chain: Drosophila myosin heavy chains are encoded by a gene family. EMBO J 8: 913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Franke JD, Montague RA, Kiehart DP (2005) Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol 15: 2208–2221. [DOI] [PubMed] [Google Scholar]
- 49. Franke JD, Montague RA, Kiehart DP (2010) Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Dev Biol 345: 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Winter CG, Wang B, Ballew A, Royou A, Karess R, et al. (2001) Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell 105: 81–91. [DOI] [PubMed] [Google Scholar]
- 51. Amano M, Ito M, Kimura K, Fukata Y, Chihara K, et al. (1996) Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J Biol Chem 271: 20246–20249. [DOI] [PubMed] [Google Scholar]
- 52. Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, et al. (1996) Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. EMBO J 15: 2208–2216. [PMC free article] [PubMed] [Google Scholar]
- 53. Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M (1992) dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev 6: 1518–1530. [DOI] [PubMed] [Google Scholar]
- 54. Kimura K, Ito M, Amano M, Chihara K, Fukata Y, et al. (1996) Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273: 245–248. [DOI] [PubMed] [Google Scholar]
- 55. Royou A, Field C, Sisson JC, Sullivan W, Karess R (2004) Reassessing the role and dynamics of nonmuscle myosin II during furrow formation in early Drosophila embryos. Molecular Biology of the Cell 15: 838–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Baumann O (2004) Spatial pattern of nonmuscle myosin-II distribution during the development of the Drosophila compound eye and implications for retinal morphogenesis. Dev Biol 269: 519–533. [DOI] [PubMed] [Google Scholar]
- 57. Martin-Belmonte F, Yu W, Rodriguez-Fraticelli AE, Ewald AJ, Werb Z, et al. (2008) Cell-polarity dynamics controls the mechanism of lumen formation in epithelial morphogenesis. Curr Biol 18: 507–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Strilic B, Kucera T, Eglinger J, Hughes MR, McNagny KM, et al. (2009) The molecular basis of vascular lumen formation in the developing mouse aorta. Dev Cell 17: 505–515. [DOI] [PubMed] [Google Scholar]
- 59. Chaitin MH, Coelho N (1992) Immunogold localization of myosin in the photoreceptor cilium. Invest Ophthalmol Vis Sci 33: 3103–3108. [PubMed] [Google Scholar]
- 60. Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, et al. (2000) A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum Mol Genet 9: 27–34. [DOI] [PubMed] [Google Scholar]
- 61. Williams DS, Hallett MA, Arikawa K (1992) Association of myosin with the connecting cilium of rod photoreceptors. J Cell Sci 103 Pt 1: 183–190. [DOI] [PubMed] [Google Scholar]
- 62. Steinberg RH, Fisher SK, Anderson DH (1980) Disc morphogenesis in vertebrate photoreceptors. J Comp Neurol 190: 501–508. [DOI] [PubMed] [Google Scholar]
- 63. Williams DS (1991) Actin filaments and photoreceptor membrane turnover. Bioessays 13: 171–178. [DOI] [PubMed] [Google Scholar]
- 64. Richard M, Grawe F, Knust E (2006) DPATJ plays a role in retinal morphogenesis and protects against light-dependent degeneration of photoreceptor cells in the Drosophila eye. Dev Dyn 235: 895–907. [DOI] [PubMed] [Google Scholar]
- 65. Kiehart DP, Feghali R (1986) Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol 103: 1517–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Franke JD, Boury AL, Gerald NJ, Kiehart DP (2006) Native nonmuscle myosin II stability and light chain binding in Drosophila melanogaster. Cell Motil Cytoskeleton 63: 604–622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Reduction of Act5C dosage enhances rhabdomere adhesion. (A–G) Transmission electron micrographs of adult Drosophila ommatidia. (A) eys, prom/+. (B) Df(1)ED418/+; eys, prom/+. The deficiency removes genomic region 5C7-5E4. (C) Act5C G0009/+; eys, prom/+. (D) Act5C G0010/+; eys, prom/+. (E) Act5C G0025/+; eys, prom/+. (F) Act5C G0177/+; eys, prom/+. (G) eys, prom/+; prom-Gal4/UAS-Act5C RNAi. (H) eys, prom, GMR-GAL4/+; UAS-Act5C RNAi/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
Rhabdomere fusion is specific to the reduction in Act5C. (A–F) Transmission electron micrographs of adult Drosophila ommatidia of (A) eys, prom/+ with deficiencies that remove one of the two copies of: (B) Act42A, (C) Act57B, (D) Act79B, (E) Act87E, (F) Act88F. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
Reduction of Act5C dosage does not affect the vertical extension of the rhabdomeres. (A–D) Confocal immunofluorescence micrographs showing the vertical view of adult Drosophila ommatidia. (A) w1118, wild type. (B) Act5C G0245/+. (C) eys, prom/+. (D) Act5C G0245/+; eys, prom/+. Na+ K+ ATPase (NaK, green) labels the basolateral membranes of photoreceptor cells, and F-actin (magenta) labels the rhabdomeres. Scale bar, 10 µm.
(TIF)
The localization of phospho-Moesin is not affected by the reduction of Act5C genetic dosage. (A,B) Confocal immunofluorescence micrographs of 48 h APF Drosophila ommatidium. Phospho-Moesin (pMoe, green) labels the activated form of Moesin, and F-actin (magenta) labels the rhabdomeres. (A) w1118, wild type. (B) Act5C G0245/+. Scale bar, 2 µm.
(TIF)
zip RNAi and sqh RNAi specifically target zipper and spaghetti squash. (A,B) Confocal immunofluorescence micrographs of zip RNAi flip-out clones (hs-flp/+; GMR>w + STOP>Gal4/+; UAS-zip RNAi/UAS-mCD8-GFP) of pupal retinas at 48 h APF visualizing mCD8-GFP (green), Zip (red), and F-Actin (blue). The RNAi expressing cells are marked with GFP and resulted in a reduction of Zip immunoreactivity. (C,D) Confocal immunofluorescence micrographs of sqh RNAi flip-out clones (hs-flp/+; GMR>w + STOP>Gal4/+; UAS-sqh RNAi/UAS-mCD8-GFP) of pupal retinas at 48 h visualizing mCD8-GFP (green), Sqh (red), and F-Actin (blue). The RNAi expressing cells are marked with GFP and resulted in a reduction of Sqh immunoreactivity. Dotted lines delineate wild-type and knockdown cells. (E–H) Transmission electron micrographs of adult Drosophila ommatidia of (E) eys, prom, GMR-GAL4/+; UAS-zip RNAi/UAS-mCD8-GFP. (F) eys, prom, GMR-GAL4/+; UAS-zip RNAi/UAS-GFP-zip. (G) eys, prom, GMR-GAL4/+; UAS-sqh RNAi/UAS-mCD8-GFP. (H) eys, prom, GMR-GAL4/+; UAS-sqh RNAi/UAS-sqh E20E21. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, (A,C) 30 µm, (B,D, E–H) 5 µm.
(TIF)
Reduction of actomyosin components results in rhabdomere fusion in eys or prom single-heterozygote background. (A–J) Transmission electron micrographs of adult Drosophila ommatidia of (A) eys/+. (B) Act5C G0245/+; eys/+. (C) eys, GMR-Gal4/+. (D) eys, GMR-Gal4/+; UAS-zip RNAi/+, (E) eys, GMR-Gal4/+; UAS-sqh RNAi/+. (F) prom/+ (G) Act5C G0245/+; prom/+. (H) prom, GMR-Gal4/+. (I) prom, GMR-Gal4/+; UAS-zip RNAi/+, (J) prom, GMR-Gal4/+; UAS-sqh RNAi/+. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, 5 µm.
(TIF)
RNAi knockdown of Act5C results in rhabdomere fusion in the otherwise wild-type background. (A–D′) Transmission electron micrographs of adult Drosophila ommatidia of (A) w 1118. (B) Act5C G0245/+. (C,C′) +/+; prom-Gal4/UAS-Act5C RNAi. (D,D′) +/+; Pph13-Gal4/UAS-Act5C RNAi. (C′,D′) A magnified view of the highlighted areas in (C) and (D), respectively. Arrows indicate the incomplete separation between rhabdomeres. Scale bar, (A,B,C,D) 5 µm, (C′,D′) 2 µm.
(TIF)
Sqh localization is not dependent on Prominin or EYS. Confocal immunofluorescence micrographs of eys (A,B) and prom (C,D) null mutant ommatidium with sqh-GFP (green) expression, and stained with Chaoptin (Chp, red), and F-Actin (blue) at 48 h APF (A,C) and 72 h APF (B,D). Chaoptin marks the rhabdomeres, and F-actin mainly marks the rhabdomeres but also weakly labels the rhabdomere terminal web. Scale bar, 2 µm.
(TIF)
Reduction of Sqh does not alter the whole cell size. (A–E) Confocal immunofluorescence micrographs of clones of cells lacking Sqh protein in (A–D) horizontal and (E) vertical optical sections. Effects of loss of Sqh by RNAi knockdown (A,B,E) or MARCM (C,D) are imaged at (A,C) 48 h APF, (B,D) 72 h APF, or (E) in adult eyes. (A,B,E) hs-flp/+; GMR>w + STOP>Gal4/+; UAS-sqh RNAi/UAS-mCD8-GFP. (C,D) sqh AX3, Frt19A/hs-flp, tub-Gal80, Frt19A; GMR-Gal4, UAS-mCD8-GFP/+. In all panels, GFP marks the mutant cells. Na+ K+ ATPase (NaK, red in A–D and blue in E) labels the lateral and basal plasma membrane and F-actin (blue in A–D and red in E) labels the developing rhabdomeres. Scale bars, (A–D) 5 µm, (E) 20 µm.
(TIF)
Changes in rhabdomere width do not contribute to the adhesion of rhabdomeres. (A) Transmission electron micrographs of adult eys, prom, GMR-GAL4/+; UAS-sqh RNAi ommatidia. Oblong rhabdomeres are defined as rhabdomeres with their width equal to or greater than twice of their length. Arrows indicate fusion between rhabdomeres, and asterisks label oblong rhabdomeres. (B) Quantitative analysis of rhabdomere fusion in normal ommatidia and in ommatidia with oblong rhabdomere(s). Not statistically significant (NS), P>0.05. Scale bars, 2 µm.
(TIF)
Tested potential actomyosin regulators via RNAi in the EP-TH genetic background.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.