Abstract
An innovative high-throughput medium development method based on media blending was successfully used to improve the performance of a Chinese hamster ovary fed-batch medium in shaking 96-deepwell plates. Starting from a proprietary chemically-defined medium, 16 formulations testing 43 of 47 components at 3 different levels were designed. Media blending was performed following a custom-made mixture design of experiments considering binary blends, resulting in 376 different blends that were tested during both cell expansion and fed-batch production phases in one single experiment. Three approaches were chosen to provide the best output of the large amount of data obtained. A simple ranking of conditions was first used as a quick approach to select new formulations with promising features. Then, prediction of the best mixes was done to maximize both growth and titer using the Design Expert software. Finally, a multivariate analysis enabled identification of individual potential critical components for further optimization. Applying this high-throughput method on a fed-batch, rather than on a simple batch, process opens new perspectives for medium and feed development that enables identification of an optimized process in a short time frame.
Keywords: medium optimization, media blending, high-throughput, CHO cells, monoclonal antibody, fed-batch, (micro-scale) cell culture
Introduction
Process development for protein therapeutics is increasingly dependent on high-throughput (HT) technologies to accelerate the screening of many conditions and the optimization of cell culture process outputs. Automated HT experimentation provides opportunities to explore a large design space by using full factorial experimental design and decrease costs by reducing raw materials, culture media, labor and time.1-3 Among the numerous HT systems available, microwell plates, first used for analytical applications, have become an important tool for microbial and mammalian cell culture applications during the last ten years. Intense efforts were made to understand suspension culture conditions within these devices, by characterizing oxygen mass transfer rates and mixing conditions in particular, to confirm their efficiency in supporting cell culture needs. Their integration into standard lab automation liquid handling platforms that enable simultaneous loading, sampling and feeding of cells, and the incorporation of fluorescence patch sensors into wells to perform pH, DO and OD measurements, have made them an efficient scale-down tool for bioprocess development studies.4-9
Medium optimization is an important step in process development as medium components at suboptimal concentrations might be limiting for cell growth or productivity, and therefore might directly affect process performance.10 On the other hand, medium components might also have an effect on secreted proteins, more particularly on their glycosylation, which is essential for their bioactivity and stability in vivo.11,12 The traditional strategy used for medium development, relying on the variation of one factor at a time (OFAT) while keeping the others constant, is laborious and time-consuming and does not account for synergistic interactions of components. Therefore, new technologies such as design of experiments (DoE) and statistical analyses, which enable the testing of several components at a time and identification of their interactions, have been implemented.13-15 Several strategies for medium optimization have been described.16,17 For example, optimization can be based on spent medium analysis,18 on metabolite flux analyses or on metabolomics,19-21 allowing rebalance of components in subsequent experiments. On the other hand, in the high-throughput approach where statistical DoE is linked to automation and small cell culture devices enable testing of several hundreds of media formulations, tests are usually performed by monitoring critical process outputs, e.g., cell growth, protein titers.3,22-24 When working with complex biological systems such as recombinant mammalian cell cultures, mixture designs to evaluate combinations of different defined formulations can be an important tool for media optimization.16,22,25 This approach is particularly interesting when testing numerous components because it avoids component solubility issues that might occur using factorial designs. Through evaluation of the performance of the various new mixtures obtained by media blending, optimal concentration ranges of the various medium components can be identified. Jordan et al.26 recently described a novel high-throughput methodology based on an extended media blending strategy that was used to reshuffle 20 amino acids in one round of experiments. Several significantly improved viable cell densities and titers of a Chinese hamster ovary (CHO) cell batch culture producing a monoclonal antibody (mAb) resulted from 192 mixtures prepared by media blending from 10 formulations.
In this study, we describe a new blending design for the medium development of a fed-batch cell culture process, enabling the optimization of all medium components of a proprietary medium in one single experiment. The ability to simultaneously test all media components presents the advantage that no potentially critical factors are missed. Blends were tested on both the cell expansion and the fed-batch production phases, ensuring that the best production media identified can also be used as expansion media. The resulting huge amount of data was analyzed by three approaches, a scoring and ranking of media blends based on their potential to improve cell growth and productivity, and two statistical approaches using Design Expert and multivariate analysis (MVA) (Fig. 1). The robustness of the methodology was verified by repeating the entire blending experiment under similar conditions. This study represents the first of a global three-step strategy that will enable us to develop and optimize a fed-batch process in 18 weeks (Fig. 2). As shown here, in the first step basal medium is optimized while keeping the feed constant. In the next step, media candidates identified during the first step are tested in a second round of experiments in combination with a panel of feed blends generated from a certain number of feed formulations. The third step consists of optimizing key media components (previously identified by MVA) and the feeding strategy.
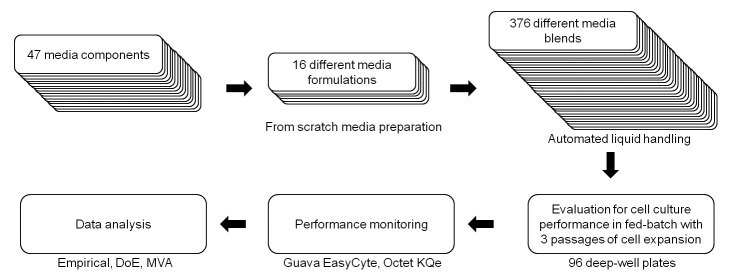
Figure 1. High-throughput media blending method for the medium development of a fed-batch cell culture process. Sixteen media formulations were designed from 43 of 47 components. Media blending following a custom-made mixture DoE resulted in 376 different blends which were evaluated in 96-DWP for cell culture performance in fed-batch. Data were first analyzed empirically, and then by statistical methodologies.
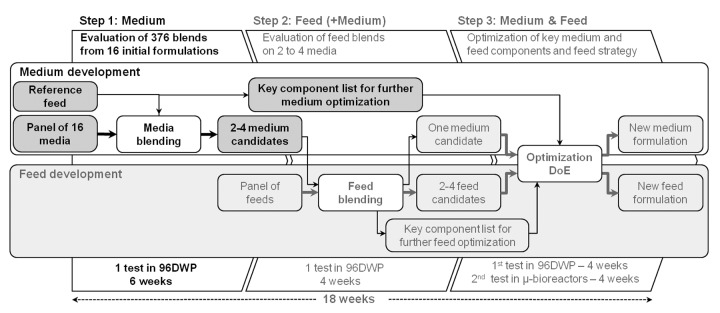
Figure 2. Improved development strategy using high-throughput cell culture methods based on media and feed blending. Medium and feed development and optimization of a fed-batch process using a 3 step strategy.
Results
Media formulation design and preparation
A first-generation proprietary medium formulation designed for an industrial fed-batch process was further improved by media blending. The goal of this exercise was to optimize the concentrations of 43 of 47 components. Of the four remaining components, glucose and NaHCO3 were kept constant, while NaCl and NaOH were used for osmolality and pH adjustments, respectively. Sixteen media formulations were designed with the 43 components (Table 1). For each component, three levels were chosen (low = 0, intermediate = 1 and high = 2). The components and their testing range are given in Table 2. To choose the component concentration for each level, a preliminary cell culture experiment with concentrated proprietary medium (0.25 to 3x) was performed, and cell growth and titer were measured. Based on these experimental data, as well as on component concentrations in proprietary media formulations at 1x and scientific knowledge from literature, concentrations corresponding to the three levels were selected. Level 1 was close to the concentrations found in the first-generation proprietary medium (Ctrl1) for most of the components, and F2 with all components at level 1 was considered as a second control (Ctrl2). These controls were performed to assess experiment and plate-to-plate variability. Except for the first three formulations with all components at the same level (respectively 0, 1 and 2), each formulation was randomly designed regarding each component level. Eighty random designs of the 16 formulations were generated and assessed for correlations between components. The design that best minimized these correlations was chosen in order to maximize the design space of the experiment.
Table 1. Media formulation design.
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F15 | F16 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 |
| C2 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 2 | 0 | 1 | 2 |
| C3 | 0 | 1 | 2 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 1 | 0 | 2 | 2 | 0 | 0 |
| C4 | 0 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 2 |
| C5 | 0 | 1 | 2 | 2 | 2 | 2 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | 1 |
| C6 | 0 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 2 | 0 | 1 | 0 | 2 | 0 | 1 | 2 |
| C7 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 2 | 2 | 1 | 2 | 0 | 1 | 1 | 1 | 0 |
| C8 | 0 | 1 | 2 | 0 | 2 | 1 | 1 | 2 | 2 | 1 | 0 | 2 | 0 | 0 | 0 | 2 |
| C9 | 0 | 1 | 2 | 2 | 0 | 2 | 2 | 1 | 0 | 1 | 1 | 2 | 0 | 2 | 0 | 2 |
| C10 | 0 | 1 | 2 | 0 | 2 | 2 | 1 | 0 | 1 | 0 | 2 | 2 | 0 | 2 | 2 | 1 |
| C11 | 0 | 1 | 2 | 2 | 1 | 1 | 2 | 0 | 2 | 0 | 2 | 2 | 1 | 2 | 0 | 0 |
| C12 | 0 | 1 | 2 | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| C13 | 0 | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 1 | 2 | 2 | 0 | 1 | 0 | 1 | 2 |
| C14 | 0 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 2 | 1 | 2 |
| C15 | 0 | 1 | 2 | 0 | 2 | 0 | 1 | 1 | 1 | 2 | 0 | 2 | 2 | 1 | 1 | 0 |
| C16 | 0 | 1 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 2 | 1 | 0 | 2 | 2 |
| C17 | 0 | 1 | 2 | 2 | 2 | 0 | 0 | 2 | 1 | 2 | 0 | 2 | 1 | 1 | 0 | 2 |
| C18 | 0 | 1 | 2 | 0 | 1 | 2 | 1 | 1 | 1 | 2 | 0 | 0 | 0 | 2 | 1 | 2 |
| C19 | 0 | 1 | 2 | 0 | 0 | 1 | 2 | 2 | 2 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| C20 | 0 | 1 | 2 | 1 | 0 | 1 | 2 | 2 | 2 | 0 | 1 | 1 | 0 | 2 | 2 | 0 |
| C21 | 0 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 0 | 0 | 2 | 0 | 0 | 1 |
| C22 | 0 | 1 | 2 | 2 | 1 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 1 | 0 | 2 | 1 |
| C23 | 0 | 1 | 2 | 2 | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 0 |
| C24 | 0 | 1 | 2 | 2 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 2 | 0 | 2 |
| C25 | 0 | 1 | 2 | 0 | 2 | 0 | 2 | 1 | 2 | 2 | 2 | 0 | 2 | 0 | 2 | 2 |
| C26 | 0 | 1 | 2 | 2 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 2 | 1 | 0 | 1 | 0 |
| C27 | 0 | 1 | 2 | 2 | 1 | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 0 |
| C28 | 0 | 1 | 2 | 1 | 0 | 2 | 1 | 0 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 1 |
| C29 | 0 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 0 | 1 | 0 | 2 | 2 | 1 | 0 | 2 |
| C30 | 0 | 1 | 2 | 2 | 2 | 0 | 0 | 2 | 2 | 0 | 2 | 2 | 2 | 0 | 0 | 1 |
| C31 | 0 | 1 | 2 | 2 | 0 | 2 | 1 | 2 | 2 | 0 | 2 | 2 | 0 | 2 | 2 | 0 |
| C32 | 0 | 1 | 2 | 2 | 2 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 0 |
| C33 | 0 | 1 | 2 | 1 | 0 | 2 | 1 | 1 | 1 | 2 | 0 | 2 | 0 | 1 | 1 | 1 |
| C34 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 0 | 0 | 1 | 1 |
| C35 | 0 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | 0 | 0 | 2 | 2 | 0 | 2 | 1 | 1 |
| C36 | 0 | 1 | 2 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 0 | 2 | 0 |
| C37 | 0 | 1 | 2 | 2 | 2 | 2 | 2 | 0 | 2 | 0 | 1 | 1 | 0 | 1 | 1 | 2 |
| C38 | 0 | 1 | 2 | 2 | 0 | 1 | 2 | 2 | 1 | 2 | 0 | 1 | 0 | 2 | 1 | 2 |
| C39 | 0 | 1 | 2 | 1 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 0 | 2 |
| C40 | 0 | 1 | 2 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 2 | 1 | 2 | 0 | 2 | 1 |
| C41 | 0 | 1 | 2 | 2 | 2 | 2 | 0 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 0 | 0 |
| C42 | 0 | 1 | 2 | 2 | 0 | 1 | 0 | 2 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 1 |
| C43 | 0 | 1 | 2 | 0 | 2 | 1 | 0 | 0 | 2 | 1 | 2 | 2 | 2 | 0 | 2 | 0 |
This matrix shows 16 media formulations (F1-F16) designed with 43 components (C1-C43). Values 0 (low), 1(mid) and 2 (high) represent relative concentrations of each component. Except for F1, F2 and F3 with all components at the same level, each formulation was randomly designed regarding each component level
Table 2. Tested component concentration ranges.
| Components | Level 0 (mM) | Level 2 (mM) |
|---|---|---|
| NaH2PO4 | 1.7 | 10 |
| L-Leucine | 2 | 9 |
| L-Lysine | 1 | 6 |
| Glycine | 0 | 3 |
| L-Methionine | 0.4 | 2 |
| L-Glutamic acid | 1 | 4 |
| L-Phenylalanine | 0.5 | 3 |
| L-Proline | 0.7 | 6 |
| L-Threonine | 0.7 | 6 |
| L-Tryptophan | 0.5 | 2 |
| L-Valine | 1 | 7 |
| Magnesium Sulfate | 0.1 | 1.5 |
| Calcium Chloride | 0.1 | 1.05 |
| myo-Inositol | 0.07 | 0.7 |
| Sodium pyruvate | 0.8 | 4 |
| D-Biotin | 0.0008 | 0.01 |
| Choline Chloride | 0.1 | 1 |
| L-Aspargine | 3 | 9 |
| Folic acid | 0.006 | 0.04 |
| Niacinamide (B3) | 0.03 | 0.15 |
| D-Pantothenic acid x 1/2Ca | 0.015 | 0.15 |
| L-Serine | 1 | 8 |
| Potassium Chloride | 1 | 10 |
| Pyridoxine | 0.005 | 0.05 |
| L-Aspartic acid | 0.8 | 2.4 |
| Riboflavin | 0.0003 | 0.003 |
| Thiamine | 0.008 | 0.04 |
| Ferric ammonium citrate (mg/L) | 1 mg/L | 10 mg/L |
| Vitamin B12 | 0.0003 | 0.004 |
| Hypoxanthine | 0.008 | 0.04 |
| Thymidine | 0.0015 | 0.006 |
| Putrescine | 0.006 | 0.03 |
| Ethanolamine | 0.1 | 0.5 |
| Zinc Sulfate | 0.004 | 0.02 |
| Cupric sulfate | 0.00004 | 0.0008 |
| Pluronic (g/L) | 0.5 g/L | 2.0 g/L |
| L-Tyrosine | 0.7 | 3 |
| Sodium Selenite | 0.00001 | 0.00006 |
| L-Alanine | 0 | 3 |
| L-Arginine | 1 | 3 |
| L-Cysteine | 1 | 3 |
| L-Histidine | 0.4 | 3 |
| L-Isoleucine | 1 | 6 |
43 among 47 medium components were tested at three levels, level 0 (low), level 1 (intermediate) and level 2 (high). Level 1 is not shown but represents an intermediate concentration between level 0 and level 2 for each component. Glucose (6g/L) and NaHCO3 (2g/L) were kept constant, while NaCl and NaOH were used for osmolality and pH adjustments.
Process performance in deepwell plates
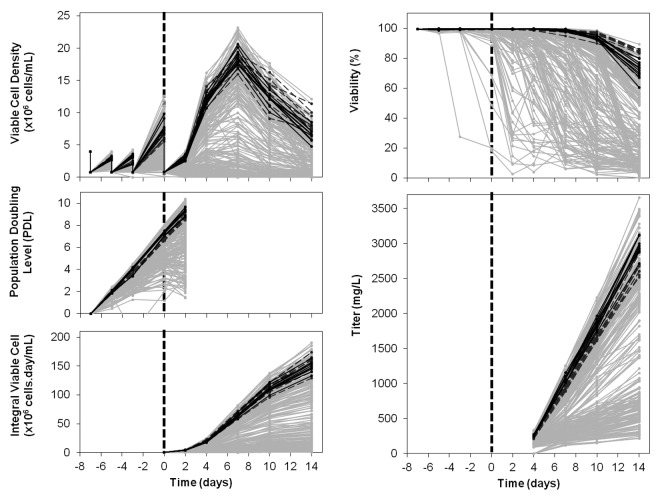
The design and mixing of the 16 formulations resulted in 376 different media blends that were added into 96-deepwell plates (DWP) containing CHO cells producing a mAb, using a robotic platform. An example of media blending mixtures in some of the 396 wells (376 media blends and 20 controls) is shown in Table 3. Mixtures were tested on both cell expansion and fed-batch production phases. Figure 3 shows the process performance obtained with the 376 different media blends in terms of population doubling level (PDL), cell growth, viability, integral of viable cells (IVC), and titer. All outputs showed broad ranges of results, depending on tested conditions. During the expansion phase, some media blends were able to increase cell growth rate by 20% as shown by PDL data, and to reduce doubling time to 20 h from 24 h in controls (data not shown). This was confirmed during the production phase, with up to 20% IVC improvements, from 156 (for controls) to 190 x 106 cells.day/mL (for the best condition). Several conditions did not allow growth or induced important cell aggregation. Large variations in growth profiles were observed, from no growth to 23.7 x 106 cells/mL (controls at 18.0 x 106 cells/mL). If ~50 conditions improved cell growth, only 10 showed equal or increased cell viability at harvest. This might be due to new nutrient limitations resulting from improved growth, and to the same feed regimen applied to all conditions. Regarding the titer, an improvement of up to 40% was observed, with a maximum titer around 3.7g/L. Ctrl 2 (F2 with all components at level 1) showed a significant and robust titer improvement (around 13%) compared with Ctrl 1.
Table 3. Examples of blending mixtures.
| Run | Well | Plate | Ctrl 1 | F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | F15 | F16 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | A02 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3 | A03 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 16 | B06 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0.7 | 0 | 0 | 0 | 0 | 0 |
| 94 | B04 | 2 | 0 | 0 | 0 | 0 | 0 | 0.3 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 95 | B05 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 130 | E10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 | 0.5 | 0 | 0 |
| 231 | H01 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 |
| 239 | H09 | 3 | 0 | 0 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.3 |
| 249 | A09 | 4 | 0 | 0 | 0 | 0 | 0.3 | 0 | 0.7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 311 | H01 | 4 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0.5 | 0 | 0 | 0 | 0 | 0 | 0 |
| 398 | H08 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.7 | 0 | 0.3 | 0 | 0 | 0 | 0 |
| 399 | H09 | 5 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
12 blending mixture recipes from 16 formulations are shown as examples. One control (Ctrl 1: proprietary medium) is shown in run 3 and another control (Ctrl 2: F2 formulation: mid-point of each component) in Run 95
Figure 3. Viable cell density, viability, IVC, PDL and titer. Data from day -7 to day 14 (viable cell density and viability), from day -7 to day 2 (PDL) and from day 0 to day 14 (IVC and titer) for 376 media blends and 20 controls. Dashed black lines: Ctrl 1; full black lines: Ctrl 2; gray lines: all other tested conditions.
Data analysis process
To get the best output of the large data set obtained from the media blending experiment, three options were chosen, as shown in Figure 4. A first approach was to analyze data of individual outputs (viable cell density, titer, IVC, PDL, viability) for each condition on a spreadsheet, and to score and rank the various tested conditions. An example of this approach is given in Table 4, which shows process performance data obtained for some of the blending experiment conditions (described in Table 3), and a calculated improvement score for each output and a global score and rank for each condition. This approach represents a quick way to select the most promising conditions.
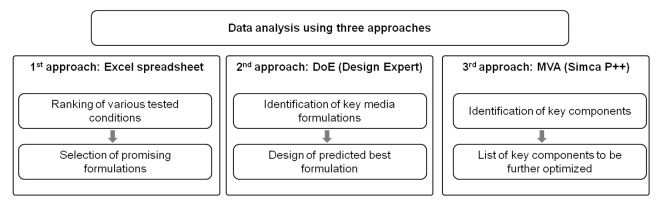
Figure 4. Data analysis process. Three strategies were applied: 1st level of analysis in Excel: determination of an improvement score for each output vs control, a global score for each mixture and a rank to select best conditions; 2nd level of analysis by Design Expert, modeling each output to predict best formulation; 3rd level of analysis: multivariate analysis using SIMCA-P++ to identify key components.
Table 4. Examples of process performance data and of scores and ranks.
| Run | Well | Plate | IVC | Viability | Titer | PDL | Improvement score for each output (%) | Global score | Rank | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (x106 cells.day/mL) | (%) | (mg/L) | IVC | Viability | Titer | PDL | |||||||
| 2 | A02 | 1 | 157 | 64 | 2442 | 8.8 | 1.0 | - | - | - | 1.7 | 127 | |
| 3 | A03 | 1 | Ctrl 1 | 162 | 83 | 2601 | 8.7 | 3.9 | 1.3 | - | - | 12.3 | 95 |
| 16 | B06 | 1 | 180 | 89 | 2470 | 9.7 | 15.5 | 8.7 | - | 8.9 | 81.1 | 6 | |
| 94 | B04 | 2 | 12 | 31 | 583 | 4.6 | - | - | - | - | 0 | 320* | |
| 95 | B05 | 2 | Ctrl 2 | 145 | 68 | 3126 | 9.5 | - | - | 16.8 | 6.6 | 47.6 | 27 |
| 130 | E10 | 2 | 185 | 63 | 3468 | 10 | 18.7 | - | 29.6 | 12.8 | 117 | 1 | |
| 231 | H01 | 3 | 167 | 67 | 3657 | 9.5 | 7.2 | - | 36.6 | 7.2 | 100.4 | 3 | |
| 239 | H09 | 3 | 175 | 84 | 2869 | 10.0 | 12.4 | 2.0 | 7.2 | 12.4 | 69.9 | 11 | |
| 249 | A09 | 4 | 30 | 17 | 778 | 7.0 | - | - | - | - | 0 | 261* | |
| 311 | H01 | 4 | 186 | 60 | 3278 | 10.3 | 19.1 | - | 22.5 | 16.6 | 111.9 | 2 | |
| 398 | H08 | 5 | 149 | 68 | 2619 | 9.2 | - | - | - | 3.7 | 7.8 | 110 | |
| 399 | H09 | 5 | 105 | 60 | 2686 | 8.8 | - | - | 0.3 | - | 0.7 | 131 | |
The left part of the table shows data for IVC, viability and titer on day 14 and PDL on day 2 for cells incubated with some of the 376 blending mixtures (see Table 3 for blending mixture recipes). The right part of the table shows individual improvement scores for IVC, viability, titer and PDL vs. control (expressed in %), the global score for each blending mixture (corresponding to the addition of individual scores normalized vs. maximum titer score), and the rank based on the global score. *For those conditions having a global score of zero, ranking was based on the amount of titer.
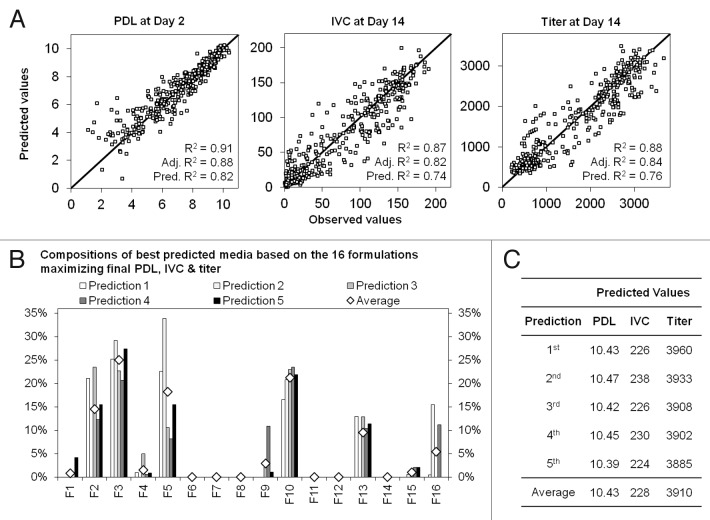
The two other approaches used statistical tools enabling a more in-depth data analysis. A prediction of the best mixtures maximizing both cell growth and titer was obtained using the Design Expert software. Each output was modeled regarding the different formulation mixtures. Main effect and the 1st level of interactions were assessed by ANOVA. A good correlation between experimental values and predictions from the model was obtained for PDL, IVC and titer with R2 around 0.9 and predicted-R2 between 0.7 and 0.8 (Fig. 5A), enabling best mixture predictions. Figure 5B shows the composition of the 5 best predicted media, plus a 6th medium representing the average of the 5 best predicted media, based on 16 formulations and maximizing final PDL, IVC and titer. Figure 5C shows predicted values for PDL, IVC and titer for the 5 best predictions and for the average of these 5 best predictions.
Figure 5.Data processing with Design Expert. A. Correlation between experimental and predicted values for models designed for PDL at day 2, IVC (x 106 cells.day/mL) and titer (mg/L) at day 14. B. Composition of the 5 best predicted media based on 16 formulations maximizing final PDL, IVC and titer and composition of a sixth medium representing the average of the 5 best predicted media. C. Predicted values for PDL, IVC and titer for the 5 best predictions and for the 6th medium (average of 5 predictions).
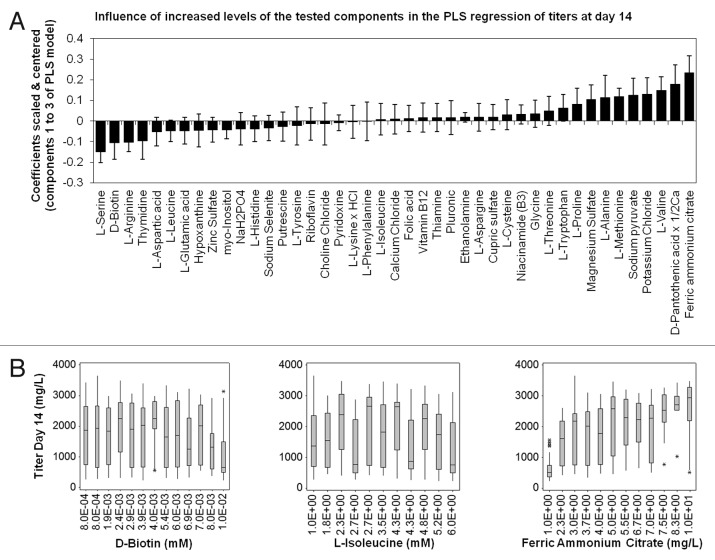
The third level of analysis, based on MVA, used SIMCA-P++ software. A model was created for the titer using partial least squares (PLS) regressions. Three components were used to create a relatively good model with respect to the low number of tested conditions compared with the number of evaluated factors. The R2 (an indication of how well the model fits experimental data) and the Q2 (an estimation of the model predictive ability) for the 3rd component were 0.514 and 0.438, respectively. Figure 6A shows the influence of individual components on titer on day 14 determined by PLS regressions. Ferric ammonium citrate, panthotenic acid and valine appeared among components having a strong positive effect on titer, while serine, biotin and arginine were among those having a negative effect. Scatter plots of 3 components with either a highly positive, a highly negative or a neutral effect on titer are shown in Figure 6B. Ferric ammonium citrate showed poor performance at low concentration (titer less than 1g/L at 1mg/L), but then titer increased to stabilize around 3g/L at 5mg/L. Best results were obtained between 7.5 and 10 mg/L. The titer was not substantially affected by the biotin concentration between 0.8 and 4 μM, but decreased at higher concentrations. On the other hand, isoleucine had no clear effect on titer between 1 and 6 mM. Potentially, there might be additional components with a significant positive or negative effect.
Figure 6.MVA with SIMCA-P++. A. Influence of individual components on the titer on day 14. B. Box-plots showing titer distribution on day 14 vs. tested component concentration for a component with a highly negative effect (Biotin), no effect (Isoleucine) and a highly positive effect (Ferric ammonium citrate) on titer on day 14. * represent outliers.
The PLS analysis alone will not be able to reliably attribute the effects to all the components, because of existing correlations between certain of them. To assess this, a correlation matrix was drawn (data not shown). A high correlation was observed for two pairs of factors, methionine vs asparagine and putrescine vs glutamic acid. Candidate components for further optimization were selected based on two criteria, their tendency to influence the model and their correlation with other important factors in the test. In total, 13 factors can be proposed based on these criteria. Factors influencing the model are ferric ammonium citrate, pantothenic acid, valine, methionine, arginine, biotin and serine, while factors correlating with an important factor in the MVA are aspartic acid, asparagine, cupric sulfate, cysteine, Vitamin B12 and sodium selenite (correlates with a factor that correlates with an important factor in the MVA). Further optimization of these factors using, for example, a DoE approach should lead to the identification of media formulations with potentially improved performance.
Robustness of the media blending method
To evaluate the robustness of the method, the experiment with media blends was repeated under similar conditions. Figure 7A shows the final titer distribution on each plate of the 1st and the 2nd blending experiment. Globally, titer distribution was comparable for all plates of both experiments; only plate 4 of the 1st experiment showed lower titers, which might be linked to a technical problem such as wrong dilution or wrong pipetting. Nevertheless, as shown in Figure 7B, when considering the 20 best conditions obtained for titer and IVC from the 1st experiment, 18 were in close correlation with data obtained in the 2nd experiment, and the R2 was around 0.7 for both IVC and titer. This demonstrates the robustness of the blending method for the best performing conditions. More variations were observed for conditions inducing lower performance, linked to cell clumping, metabolic factors or physical parameters. The next step after identification of the best performing media formulations will be to test them at larger scale to confirm predictability.
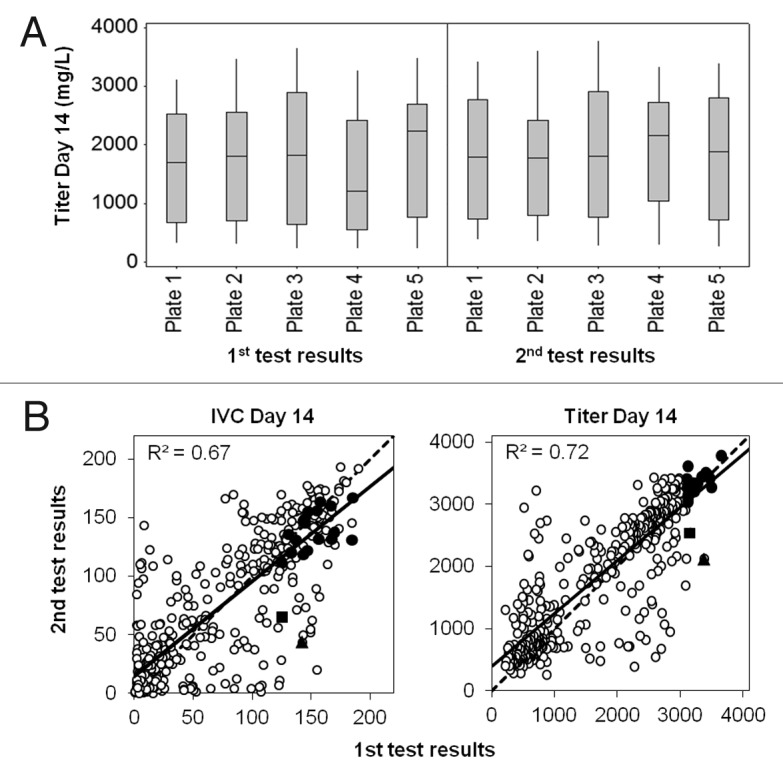
Figure 7.Robustness of the blending experiment method. A. Box-plots showing titer distribution on day 14 for the 5 plates of the first and the 2nd blending experiment. B. Correlation between the first and the second media blending experiment regarding IVC (x 106 cells.day/mL) and titer (mg/L) obtained on day 14. Open circles represent the 376 tested conditions and black symbols represent the 20 best conditions for titer in the first experiment (circles represent the 18 conditions that were highly reproducible whereas the square and the triangle represent the two conditions lacking reproducibility).
Discussion
The biotechnology industry is strongly motivated to develop high performing processes in a minimal time frame to meet increasing market demands and reduce manufacturing costs. Many efforts have focused on media optimization, as a well-balanced media composition is essential for maximal viable cell density and productivity, two major elements of a fed-batch process.16,17,24,27 Usually, medium and feed development of a fed-batch process are performed sequentially because of the large number of experiments required for a simultaneous optimization. For example, Zhang et al.15 sequentially developed a medium and a feed for a fed-batch process for CHO cells expressing recombinant antibody by using a Plackett-Burman design to screen active factors for cell growth and antibody production, followed by a central composite design to optimize their concentration, and by a feeding design based on stoichiometric ratios of different nutrients improving productivity. Nevertheless, the outcome of a successive optimization strategy might not always be ideal because basal medium and feed medium might have interrelated impacts on cell culture performance. Indeed, an improved basal medium can alter the metabolism and growth of cells, which then may require a modified feed. Therefore, sequential optimization of some elements have to be repeated, or sequential medium and feed optimization have to be followed by a final round of integrated optimization of feed and process settings, as proposed by Jiang et al.28 Here, we describe an innovative high-throughput methodology based on media blending for medium development of a fed-batch cell culture process that already integrates a reference feed in the basal medium optimization process. Starting from a proprietary medium composed of 47 components, 16 formulations were designed by varying 43 of those over 3 different levels. The only factors excluded from our blending design were glucose, which was part of the feed and was not limiting during the first 3 d of culture, NaOH, which was needed for pH adjustment, NaHCO3 as the principal pH buffer, and NaCl for osmolality adjustment. Media blending, based on a custom-made mixture DoE considering binary blends, resulted in 376 blends that were tested in 96-DWP on process performance of a CHO fed-batch culture. Testing the different blends already in the expansion phase ensures that the best identified production media can also be used as expansion media. Indeed, some blends were already able to increase cell growth by 20% during the cell expansion phase, and this increase was confirmed during the production phase. Regarding the titer, an important improvement of up to 40% was observed with certain blends. Globally, best conditions generally improved IVC, titer and specific productivity, but some conditions showed no IVC increase but an increase in titer and specific productivity.
The strategy used for data analyses included 3 levels. The first level was an empirical analysis of the effect of each media blend on the cell culture process performance. By scoring and ranking all blending conditions regarding their potential to improve process outputs, best conditions became readily available and could be further tested at larger scale for confirmation. This method represents a simple and quick way for medium optimization without the use of complex statistical methodologies. The two other levels of analyses used statistical tools, enabling a more in-depth evaluation. By using mathematical models, the first tool, Design Expert software, enabled prediction of best mixtures maximizing final PDL, IVC and titer. A great advantage of this method is that several criteria could be analyzed together to determine synergistic responses between criteria. Moreover, it allowed ranking of the initial 16 formulations, e.g., to identify the formulations that were systematically present in poor performing mixes and could be replaced by others to be tested in a new DoE. The modeling tool is also useful to simulate untested new mixtures with different blending ratios and not limited to two formulations, which will enable the prediction of the best possible blend.
The ultimate level of analysis based on MVA used SIMCA-P++ software. The model created for the titer using partial least square regression enabled identification of ferric ammonium citrate, panthothenic acid, valine, methionine, arginine, biotin and serine as factors with the most influence on the titer, while other factors such as aspartic acid, asparagine, cupric sulfate, cysteine, Vitamine B12 and sodium selenite were selected because they correlated with an important factor in the MVA. It should be noted that we actually cannot distinguish between a true effect or a “false” effect due to such a correlation. The benefit of this third method is that it enables the identification of key media components that can be further evaluated, and should lead to the identification of new media formulations with potentially improved performance.
In conclusion, our high-throughput media blending approach is a robust and rapid method for medium optimization of a fed-batch process. Data analysis by simple ranking based on critical process outputs, e.g., cell growth, viability, titer, provides an easy and quick tool to determine best performing media formulations among almost 400 different blends, which can be rapidly confirmed at larger scales. On the other hand, statistical tools enable a more in-depth analysis, allowing prediction of best performing media formulations and identification of critical media components for further optimization. Compared with traditional medium development strategies, this method greatly reduces costs and development time and enhances the possibility of achieving an improved and more consistent performance within one experiment. The whole media blending process, including data analysis, was performed within 6 weeks. This methodology can also be applied to feed development and opens new perspectives for fed-batch process development and optimization, and for other activities such as cell line screening and cell line stability studies.
Materials and Methods
Media formulation preparation
The 16 formulations were prepared by weighing individual components. For each formulation: glucose concentration was fixed to 33.3 mM to prevent any limitation during the first 3 d of culture; NaOH concentration was adjusted to obtain a pH of 7.0 before NaHCO3 addition; NaHCO3 concentration was fixed to 23.8 mM to maintain buffering capacity; NaCl concentration was adjusted to reach an osmolality around 315 mOsm/kg.
Automated media blending
High-throughput media blending was performed on a liquid handling workstation (Biomek FX, Beckman-Coulter, Inc. Fullerton, CA, USA). A total of 376 mixtures were directly blended into five square-shaped 96-DWP (Greiner Bio-One # 780271). On each plate, 16 wells were kept available for reagents necessary for titer determination. The 16 formulations were blended following a custom-made mixture DoE with binary blends. The candidate points of the design were vertices (1 formulation at 100%), center of edges (2 formulations at 50%), and third of edges (2 formulations, one at 33% and the other at 67%). In addition to the 376 mixtures, 20 controls were performed to assess experimental and plate-to-plate variability. Controls were either proprietary medium (Ctrl 1: this reference medium is not part of the media blending design and its exact composition is undisclosed) or F2 formulation (Ctrl 2: intermediate level for each component).
Cell cultures in 96-DWP
Experiments were performed using a CHO-S cell line producing a mAb. The cells were first expanded in shake tubes or shake bottles in proprietary medium. Seven days before starting the fed-batch process (day -7), the cells were centrifuged, re-suspended in F1 formulation (lowest level of each component; Table 1) at a concentration of 5 x 106 cells/mL and then seeded at 0.75 x 106 cells/mL into 5 square-shaped 96-DWP previously filled with the 376 different blends and 20 controls (final volume per well of 450 μL). All small volume liquid handlings (below 500µL) were performed with the robotic platform. The plates were then incubated with vented lids to minimize evaporation6 in a shaker incubator at 37°C, 5% CO2, 90% humidity and 320 rpm agitation (ISF1-X, Kuhner AG). Three passages were performed under the same conditions for each of the different media mixtures, on day -5, -3 and 0 (day of the start of the fed-batch). At each passage, the cells were diluted to 0.75 x 106 cells/mL and re-incubated in a new set of five 96-DWP containing media blends and controls. Samples were taken for growth and viability assessment (Guava Easy-Cyte, Merck Millipore). The 3 passages before the fed-batch inoculation were performed in media with MSX while the fed-batch was performed without. After the expansion phase, the fed-batch process was started with cells seeded at 0.75 x 106 cells/mL into various media blends, and feeds were added on day 2, 4, 7 and 10. The feeding system consisted in a glucose solution at 400 g/L, a chemically-defined main feed containing over 30 components and a highly concentrated alkaline amino acid solution. Prior to each feeding and at the end of the culture (day 14), samples (below 40 μL) were taken for growth and viability assessment and titer determination.
PDL, IVC, titer and specific productivity (PCD) determination
PDL was calculated during expansion phase (day -7 to day 0) and during the first two days of fed-batch culture, according to the following formula:
PDL = [1 / log10 (2)] x log10(TCD t / VCD t-1) + PDL t-1 with TCD t = total cell density at time t and VCD t-1 = viable cell density at time t-1.
IVC (106 cells.day/mL) was calculated during the fed-batch culture, according to the following formula: IVC t = IVC t-1 + (VCD t + VCD t-1) / 2 x Δt, where IVC t = IVC at time t, IVC t-1 = IVC at the previous cell counting (IVC 0 = initial IVC = cell density of fed-batch inoculation), VCD t = viable cell density at time t, VCD t-1 = viable cell density at time t-1 and Δt = difference between time t and t-1.
Titer quantification of the mAb produced during the fed-batch process was performed with the Octet KQe (ForteBio) using Protein A sensors. Each sample was diluted 20 x into a dilution buffer (PBS pH = 7.4, BSA 0.1g/L, Tween 20 at 1%). Regeneration buffer was glycin 2 M and neutralization buffer was the dilution buffer.
Specific productivity (PCD in pg/cells.day) was calculated according to the following formula: PCD t = Titer t / IVC t.
Data analysis
Spreadsheet analysis
Data were compiled in a spreadsheet and different media blending conditions were scored and ranked according to their potential to improve process performance. First, an improvement score was determined for each output (IVC, viability, titer, PDL), defined as the percentage of improvement vs. control. A global score was then calculated for each condition by adding individual scores previously normalized against maximum titer score (normalization was performed against maximum titer because titer showed a higher percentage of improvement than other outputs). Ranking global scores of all blending conditions enabled determination of best formulations. For those conditions having a global score of zero, ranking was based on the amount of titer.
Analysis by Design Expert
Design Expert software (V8.1, StatEase) was used to analyze each output (PDL, IVC, and titer mainly), generating reduced quadratic mixture models for each one and at each time point. The analysis of variance (ANOVA) of each model indicated the main factors (key formulations) with a significant influence on PDL, IVC and titer. A square root transformation was applied to titer and IVC and a power transformation to PDL to improve their models. Models were then used to predict best mixtures from the 16 formulations to maximize both growth and production. An “average best” mixture was designed.
Analysis by SIMCA-P++
The analysis was based on MVA and used SIMCA-P++ software (V12, U-Metrics). PLS regressions were performed for each output to study the effects of each component in order to identify key components. To detect a possible correlation between components of the MVA, a correlation matrix was drawn and correlation factors were determined. A correlation was considered as strong if the correlation factor was above 0.8, medium if it was between 0.6 and 0.8, and negligible if it was below 0.6.
Acknowledgments
We acknowledge our colleagues from BPS Downstream and Analytical for support.
Glossary
Abbreviations:
- ANOVA
analysis of variance
- CHO
Chinese hamster ovary
- Ctrl
control
- DO
dissolved oxygen
- DoE
design of experiments
- DWP
deepwell plate
- HT
high-throughput
- IVC
integral viable cell density
- MVA
multivariate analysis
- OD
optical density
- OFAT
one factor at a time
- PCD
picogram per cells.day
- PDL
population doubling level
- PLS
partial least squares
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Amanullah A, Otero JM, Mikola M, Hsu A, Zhang J, Aunins J, et al. Novel micro-bioreactor high throughput technology for cell culture process development: Reproducibility and scalability assessment of fed-batch CHO cultures. Biotechnol Bioeng. 2010;106:57–67. doi: 10.1002/bit.22664. [DOI] [PubMed] [Google Scholar]
- 2.Bareither R, Pollard D. A review of advanced small-scale parallel bioreactor technology for accelerated process development: current state and future need. Biotechnol Prog. 2011;27:2–14. doi: 10.1002/btpr.522. [DOI] [PubMed] [Google Scholar]
- 3.Barrett TA, Wu A, Zhang H, Levy MS, Lye GJ. Microwell engineering characterization for mammalian cell culture process development. Biotechnol Bioeng. 2010;105:260–75. doi: 10.1002/bit.22531. [DOI] [PubMed] [Google Scholar]
- 4.Baboo JZ, Galman JL, Lye GJ, Ward JM, Hailes HC, Micheletti M. An automated microscale platform for evaluation and optimization of oxidative bioconversion processes. Biotechnol Prog. 2012;28:392–405. doi: 10.1002/btpr.1500. [DOI] [PubMed] [Google Scholar]
- 5.Chen A, Chitta R, Chang D, Amanullah A. Twenty-four well plate miniature bioreactor system as a scale-down model for cell culture process development. Biotechnol Bioeng. 2009;102:148–60. doi: 10.1002/bit.22031. [DOI] [PubMed] [Google Scholar]
- 6.Duetz WA. Microtiter plates as mini-bioreactors: miniaturization of fermentation methods. Trends Microbiol. 2007;15:469–75. doi: 10.1016/j.tim.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 7.Funke M, Diederichs S, Kensy F, Müller C, Büchs J. The baffled microtiter plate: increased oxygen transfer and improved online monitoring in small scale fermentations. Biotechnol Bioeng. 2009;103:1118–28. doi: 10.1002/bit.22341. [DOI] [PubMed] [Google Scholar]
- 8.Micheletti M, Lye GJ. Microscale bioprocess optimisation. Curr Opin Biotechnol. 2006;17:611–8. doi: 10.1016/j.copbio.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 9.Wen Y, Zang R, Zhang X, Yang S-T. A 24-microwell plate with improved mixing and scalable performance for high throughput cell cultures. Process Biochem. 2012;47:612–8. doi: 10.1016/j.procbio.2011.12.023. [DOI] [Google Scholar]
- 10.Kim DY, Lee JC, Chang HN, Oh DJ. Effects of supplementation of various medium components on Chinese hamster ovary cell cultures producing recombinant antibody. Cytotechnol. 2005;47:37–49. doi: 10.1007/s10616-005-3775-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gawlitzek M, Estacio M, Fürch T, Kiss R. Identification of cell culture conditions to control N-glycosylation site-occupancy of recombinant glycoproteins expressed in CHO cells. Biotechnol Bioeng. 2009;103:1164–75. doi: 10.1002/bit.22348. [DOI] [PubMed] [Google Scholar]
- 12.Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology. 2009;19:936–49. doi: 10.1093/glycob/cwp079. [DOI] [PubMed] [Google Scholar]
- 13.Lee GM, Kim EJ, Kim NS, Yoon SK, Ahn YH, Song JY. Development of a serum-free medium for the production of erythropoietin by suspension culture of recombinant Chinese hamster ovary cells using a statistical design. J Biotechnol. 1999;69:85–93. doi: 10.1016/S0168-1656(99)00004-8. [DOI] [PubMed] [Google Scholar]
- 14.Sandadi S, Ensari S, Kearns B. Application of fractional factorial designs to screen active factors for antibody production by chinese hamster ovary cells. Biotechnol Prog. 2006;22:595–600. doi: 10.1021/bp050300q. [DOI] [PubMed] [Google Scholar]
- 15.Zhang H, Wang H, Liu M, Zhang T, Zhang J, Wang X, Xiang W. Rational development of a serum-free medium and fed-batch process for a GS-CHO cell line expressing recombinant antibody. Cytotechnol: published on line 21 August 2012. [DOI] [PMC free article] [PubMed]
- 16.Jerums M, Yang X. Optimization of cell culture media. BioProcess International. 2005;3:38–44. [Google Scholar]
- 17.Zhang M, Lawshé A, Koskie K, Johnson T, Caple MV, Ross JS. Rapid development and optimization of cell culture media. BioPharm International. 2008;21:60–8. [Google Scholar]
- 18.Xie L, Wang DI. Stoichiometric analysis of animal cell growth and its application in medium design. Biotechnol Bioeng. 1994;43:1164–74. doi: 10.1002/bit.260431122. [DOI] [PubMed] [Google Scholar]
- 19.Dietmair S, Hodson MP, Quek LE, Timmins NE, Chrysanthopoulos P, Jacob SS, et al. Metabolite profiling of CHO cells with different growth characteristics. Biotechnol Bioeng. 2012;109:1404–14. doi: 10.1002/bit.24496. [DOI] [PubMed] [Google Scholar]
- 20.Selvarasu S, Ho YS, Chong WPK, Wong NSC, Yusufi FNK, Lee YY, et al. Combined in silico modeling and metabolomics analysis to characterize fed-batch CHO cell culture. Biotechnol Bioeng. 2012;109:1415–29. doi: 10.1002/bit.24445. [DOI] [PubMed] [Google Scholar]
- 21.Xing Z, Kenty B, Koyrakh I, Borys M, Pan SH, Li ZJ. Optimizing amino acid composition of CHO cell culture medium for a fusion protein production. Process Biochem. 2011;46:1423–9. doi: 10.1016/j.procbio.2011.03.014. [DOI] [Google Scholar]
- 22.Didier C, Etcheverrigary M, Kratje R, Goicoechea HC. Crossed mixture design and multiple response analysis for developing complex culture media used in recombinant protein production. Chemom Intell Lab Syst. 2007;86:1–9. doi: 10.1016/j.chemolab.2006.07.007. [DOI] [Google Scholar]
- 23.Girard P, Jordan M, Tsao M, Wurm FM. Small-scale bioreactor system for process development and optimization. Biochem Eng J. 2001;7:117–9. doi: 10.1016/S1369-703X(00)00110-8. [DOI] [PubMed] [Google Scholar]
- 24.Hodge G. Media development for mammalian cell culture. BioPharm International. 2005;18:1–4. [Google Scholar]
- 25.Rispoli F, Shah V. A new efficient mixture screening design for optimization of media. Biotechnol Prog. 2009;25:980–5. doi: 10.1002/btpr.225. [DOI] [PubMed] [Google Scholar]
- 26.Jordan M, Voisard D, Berthoud A, Tercier L, Kleuser B, Baer G, et al. Cell culture medium improvement by rigourous shuffling of components using media blending. Cytotechnol. 2013;65:31–40. doi: 10.1007/s10616-012-9462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Vijayasankaran N, Shen A. (Y), Kiss R, Amanullah A. Cell culture processes for monoclonal antibody production. MAbs. 2010;2:455–77. doi: 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z, Droms K, Geng Z, Casnocha S, Xiao Z, Gorfien S, et al. Fed-batch cell culture process optimization – a rationally integrated approach. BioProcess International. 2012;10:40–5. [Google Scholar]