Abstract
Introduction
Previous studies showed liver dysfunction after severe burn, and that this is associated with activation of endoplasmic reticulum (ER) stress. Autophagy is a catabolic process to maintain cellular organelle balance; ER stress is associated with autophagy signaling cascades. We thus sought to determine whether autophagy signals were associated with damage in the liver after burn, and further whether burn associated ER stress activates autophagy signals in hepatocytes.
Methods
C57BL6 male mice received a 25% total body surface area full thickness (TBSA) scald burn, and liver was harvested at 24 hours after burn. HepG2 cells were stimulated with an ER stress inducer thapsigargin (TG) for 24 hours to mimic ER stress in vitro. TUNEL staining was performed on liver histology sections. Autophagy was assessed by immunoblotting. Statistical analysis was by Student’s t-test and significance was accepted at p<0.05.
Results
TUNEL positive stained hepatocytes increased in burned animals with a significant elevation of caspase 3 activity (p<0.05). A marked increase in hepatic ATG3, ATG5 and LC3B was detected as well (p<0.05). Expression of Beclin-1, LC3A and LC3B increased in HepG2 cells in response to TG, similar to the response seen in vivo. Cytosolic ATP dropped significantly, and AMPK and mTOR were phosphorylated as well in response to TG (p<0.05).
Conclusion
ER stress, which occurs in hepatocytes after severe injury, is associated with autophagy and liver damage following severe burn. In response to ER stress, activated autophagy is associated with AMPK/mTOR pathway.
Keywords: thermal injury, mouse liver, endoplasmic reticulum (ER), autophagy, apoptosis
INTRODUCTION
Acute and prolonged hepatic dysfunction has been described repeatedly following severe burn in both pre-clinical models and in patients (1, 2). Previous studies showed that intracellular calcium elevation and endoplasmic reticulum (ER) stress play a role in mediating these changes (3). Through the unfolded protein response (UPR) pathway, ER stress is activated to maintain cell integrity in response to the loss of ER stored calcium and unfolded protein accumulation (4). However, prolonged ER stress is beyond the capacity of cells to compensate, and directs cells toward apoptosis. We observed that severe burn caused hepatic ER stress associated with apoptosis and decreased hepatic function for up to 4 weeks after injury (5). However, the mechanism by which injury associated ER stress subsequently mediates liver damage and dysfunction following severe burn is not defined.
Autophagy, including macrophagy, microphagy and chaperone-mediated autophagy, is a common and fundamental catabolic process in cellular organelle homeostasis. In response to starvation or other pathophysiological events, cellular organelles and cytosolic particles such as mitochondria and glycogen are sequestered into an autophagosome, which subsequently fuses with an endosome and/or lysosome, to form an autolysosome. The consequent activation of ubiquitin systems results in proteolysis and recycling of macromolecules (6). Though the mechanism of autophagy formation is still being investigated, a group of ATG (autophagy-related) proteins have been identified in eukaryotic cells which are expressed with the elongating phagophore and autophagosome formation (7). About 18 out of 30 ATG proteins including Atg1–10, Atg12–14, Atg16–18, Atg29, and Atg31 interact each other and accumulate consequently for final autophagosome formation (8).
Signal regulation of autophagy in the process of initiation, elongation, and maturation has been studied years. (9) The mammalian target of rapamycin (mTOR) is a critical regulator of autophagy induction. Protein kinase B (Akt) and the mitogen-activated protein kinase (MAPK) signals activate mTOR through the interaction of tuberous sclerosis complex (TSC) 1/2, v Rheb and the mammalian target of rapamycin complex 1/2 (mTORC1/2) (10). Activated mTOR blocks the interaction between ATG13 and the phosphorylation of serine-threonine kinase ATG1 thus suppressing autophagy (11); AMP activated protein kinase (AMPK) is a key energy sensor which regulates cellular metabolism to maintain energy homeostasis. AMPK stimulates autophagy initiation by deactivating mTOR (12). Under starvation, AMPK promotes autophagy by directly activating and phosphorylating mammalian autophagy-initiating kinase Ulk1, a homologue of yeast ATG1 (13). AMPK is also required for initiation of ATG1 homologs in mammals such as UNC-51-like kinase 1 (ULK1) mediated autophagy by its binding capacity of ULK1 (14).
Recent studies indicate that autophagy is induced by ER stress (9, 15). Ciechomska et al reported recently that cyclosporine A (CsA) induced ER stress via autophagy in malignant glioma cells (16). Hoyer-Hansen et al reported that intracellular calcium activates AMPK/mTOR pathways through calcium calmodulin dependent kinase (CamKK) (17). We thus posited that perhaps ER stress after severe burn activates autophagy signals in hepatocytes leading to hepatocyte apoptosis and finally hepatic dysfunction. In a pre-clinical model, we found that severe burn increased apoptosis in the liver with activation of ER stress and initiation of the autophagy cascade. Furthermore we specified that the ER stress similar to that induced by severe burn activated autophagy signals associated with phosphorylated AMPK/mTOR in vitro.
MATERIALS AND METHODS
Burned Mice
Adult male C57BL/6 mice (6 to 8 weeks old) were purchased from Jackson Laboratory (Bar Harbor, ME) and allowed to acclimate for one week prior to the experiment. Mice were housed in a temperature-controlled cubicle with a 12-hour light/dark cycle with laboratory chow and water ad libitum. All animal procedures were performed in adherence to the National Institutes of Health Guide for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee of the University of Texas Southwestern Medical Center at Dallas, TX.
A well-established method with minor modifications was applied to induce a fullthickness scald burn (18). Eight mice were anesthetized with 1.2% avertin (250mg/kg body weight i.p.). The dorsal and lateral surfaces were shaved and 1 ml of 0.9% saline was injected subcutaneously along the spinal column to protect the spinal cord during the burn. Mice were placed to a mold with an opening providing a 12.5% total body surface area burn. Mice were then immersed in 97°C water for 10 seconds on the dorsal and 2 seconds on the ventral side. Lactated Ringers solution (1 ml) was then administered intraperitoneally for resuscitation and 0.05 mg/kg body weight of buprenorphine was administered subcutaneously for analgesia. Sham animals (n=8) underwent anesthesia and shaving, but were immersed in water at room temperature (25°C). Mice were housed in separate cages for the duration of the experiment and pair fed, and sacrificed at 24 hours after burn. Liver tissue was excised immediately and either stored in 10% neutral buffered formalin (NBF) or snap-frozen in liquid nitrogen and subsequently at −80°C.
Terminal deoxyuridine nick end labeling (TUNEL) Immunofluorescent staining
Fixed liver tissues were paraffin embedded, and sectioned. Tissue sections (5µm) were deparaffinised and rehydrated followed by TUNEL immunofluorescent staining following the manufacturer’s recommended procedure (Promega, Madison, WI). Briefly, Sections were treated with 100µl of the 20µg/ml proteinase K solution for 10 minutes and then fixed in 4% methanol-free formaldehyde solution. Sections were incubated with 50ul of Terminal Deoxynucleotidyl Transferase (rTdT) reaction buffer for 1 hour at 37°C. Sections were finally treated with 10µl of ProLong Gold anti-fade reagent with DAPI nuclear staining (Invitrogen, Carlsbad, CA) for nuclear counterstaining. The images were captured for each slide under a Zeiss Axio Observer Epifluorescence Microscope equipped with filter sets for DAPI (445nm) and fluorescein 12-dUTP (520nm) with Hamamatsu Orca 1024BT camera monochrome digital camera. TUNEL positive cells were artificially stained green in the nucleus.
In Vitro Hepatocytes
Human hepatocellular liver carcinoma cells (HepG2) were cultured in DMEM low glucose medium containing 10% fetal bovine serum (FBS), 2mM L-Glutamine. 0.25% trypsin-EDTA was applied for passage when cell confluence was over 80%. Cells and medium were purchased from ATCC Cell Biology (Manassas, VA). The day before experiment, cells were transferred into 6-well plates and the culture medium was replaced without FBS. Based on a previous study (19), cells were treated with 100nM of TG, an ER stress inducer, for 24 hours. Cell lysate was collected and protein was extracted for further biological and molecular analysis. The experiment was conducted four times independently.
Cytosolic caspase 3 activity was measured by a fluorometric method. Briefly 20µg of extracted protein sample incubated with 5mM of Z-DEVD-R110 in reaction buffer (5mM PIPES, pH 7.4, 1mM EDTA, 0.05% Triton, 5mM DTT) for 30 minutes at room temperature while avoiding light, the absorbance were then detected by Fluorescence Reader Flurostar (BMG LAB TECH, Durham, NC) with excitation wavelength at 485nm and emission wavelength at 520nm. Caspase 3 substrate Z-DEVD-R110 was purchased from American Peptide Company Inc. (Sunnyvale, CA).
Cytosolic ATP levels were measured in vitro following the product instructions provided by the manufacturer (BioVision, Milpitas, CA).
Immunoblotting
Approximately 30 mg of frozen tissue was homogenized in T-PER Tissue Protein Extraction Reagent plus Halt Protease Inhibitor Cocktail (Thermo Scientific, Rockford, CA). The homogenate was centrifuged at 20,000 xg for 30 minutes at 4°C and the pellet discarded. Protein concentration was measured with a BioSpektrometer kinetic spectrometer (Eppendorf, Hauppauge, NY) using the Lowry protein assay method. Thirty micrograms of each protein sample was subsequently analyzed by SDS-PAGE and Western blotting. Band intensities were quantified with the GeneSnap/GeneTools software (Syngene, Frederick, MD). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin were utilized as loading controls. All antibodies including ATG3, ATG5, LC3A, LC3B, Beclin-1, ULK1, mTOR, AMPK and GAPDH antibodies were purchased from Cell Signaling Tech, Inc. (Danvers, MA). SuperSignal West Pico Chemiluminescent Substrate was purchased from Thermo Scientific Inc, (Rockford, IL).
Statistical Analysis was performed using a Student`s t-test to compare differences between groups; all data were normally distributed. Data are expressed as the Mean ± SEM. Significance was accepted at p<0.05.
RESULTS
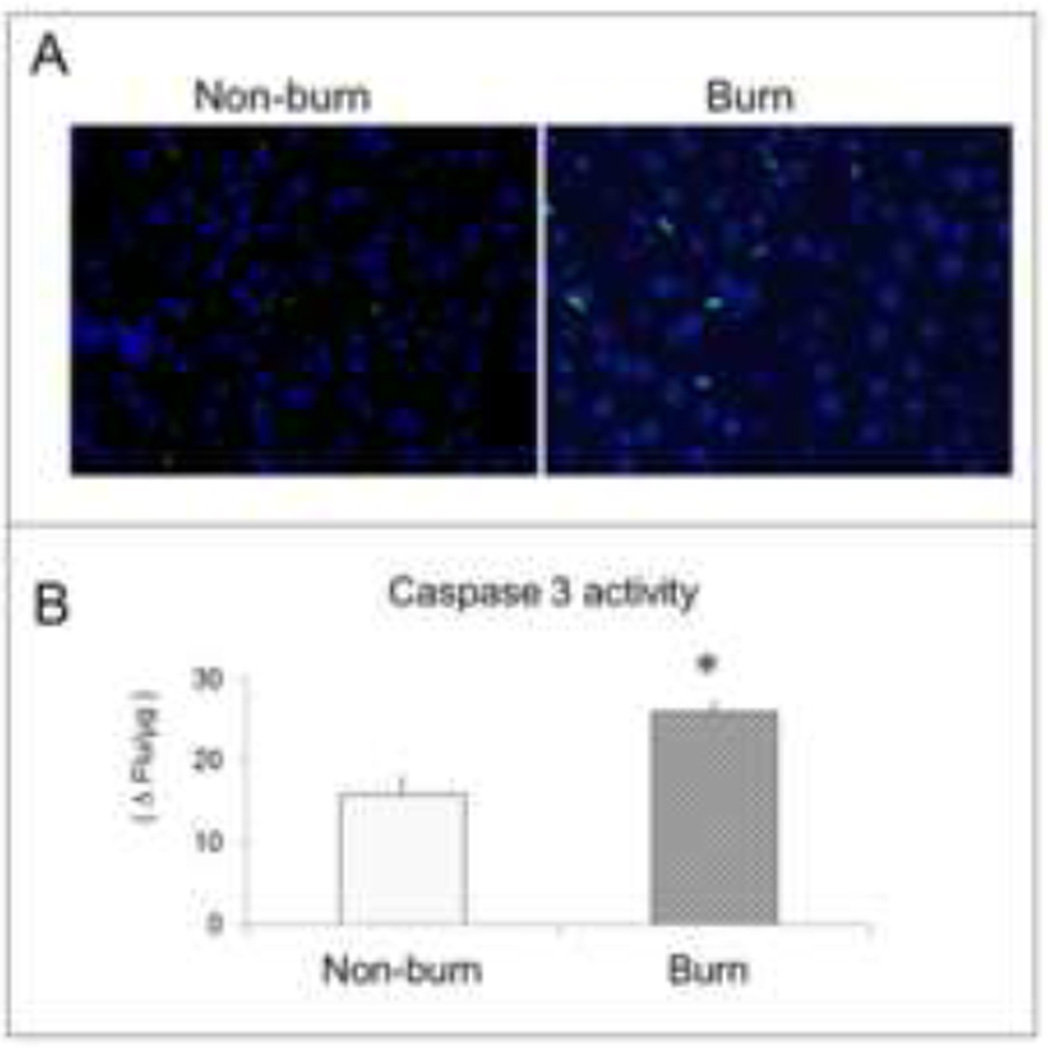
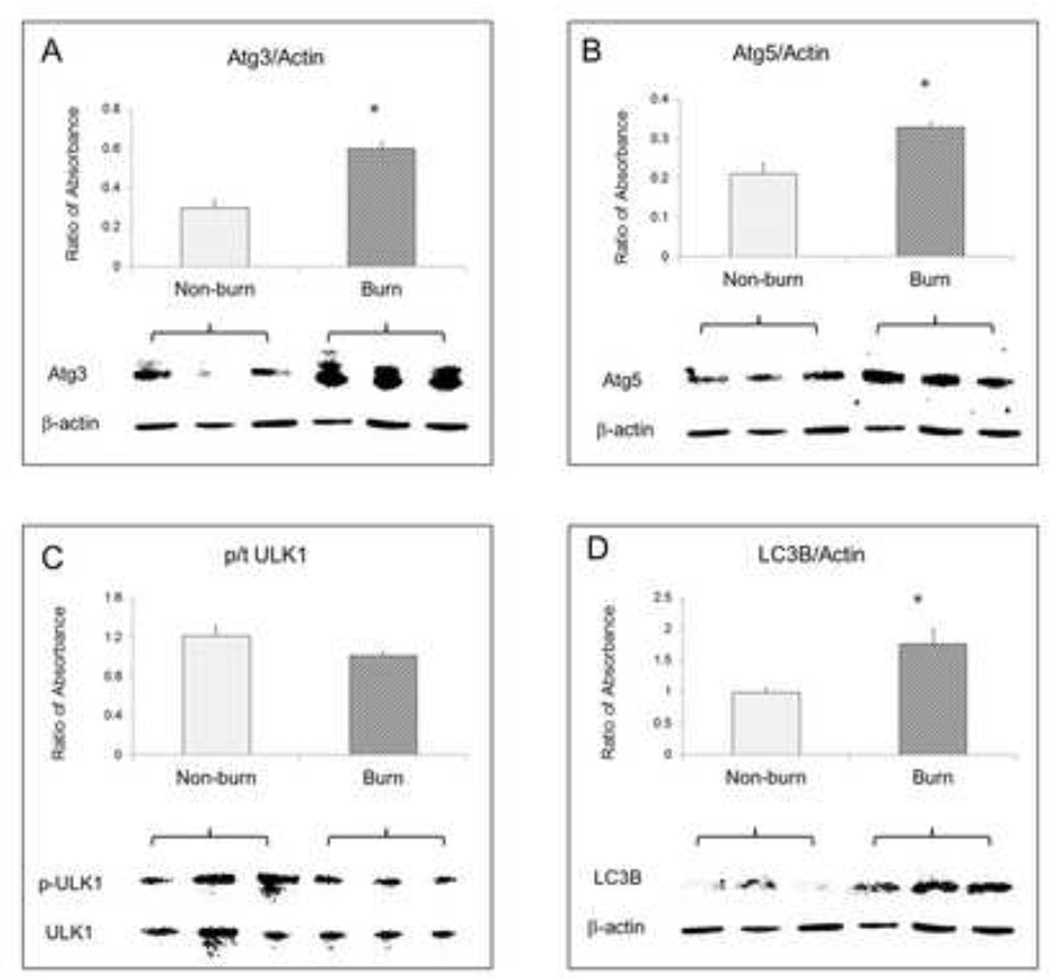
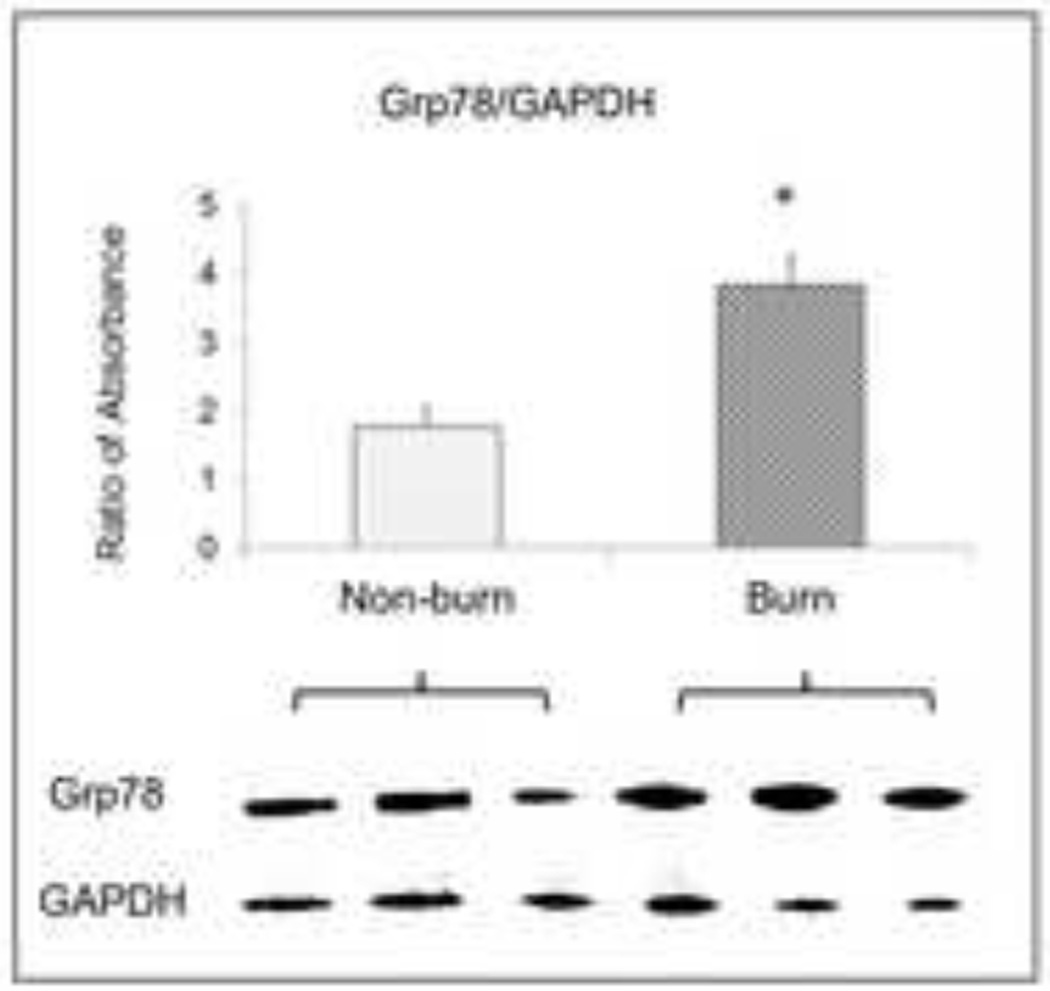
TUNEL positive cells increased in the burned animals [Fig 1A]. Cytosolic caspase-3 activity was also significantly higher in liver tissue at 24 hours after burn [Fig 1B] (p<0.05). After normalisation with GAPDH, absorbance ratios for ATG3, ATG5 and LC3B were significantly higher in burned mice compared to sham (p<0.05) indicating autophagy signaling in the liver after severe burn [Fig 2]. The absorbance ratio of GRP78 was significantly higher in liver from burn animals indicating ER stress as well which was similar to our previous studies [Fig 3].
Figure 1.
(A) TUNEL immunofluorescent staining showed positive apoptotic cells stained with green nuclei in liver tissue from non-burn (left) and burn animals (right); (B) liver tissue caspase 3 level was measured with 5 µM of Z-DEVD-R110 reaction and quantified as the change in fluorescence per minute per microgram protein. Data are mean ± SEM. * p<0.05, burned vs. non-burn.
Figure 2.
Western blot data and statistical analysis results showed expression of autophagy signals including (A) ATG3, (B) ATG5, (C) ULK1, and (D) LC3B in mouse liver 24 hours after burn, β-actin as loading control. Data are mean ± SEM. * p<0.05, burned vs. non-burn.
Figure 3.
Western blot data and statistical analysis results showed expression of ER stress protein GRP78 in liver homogenate from burn animals. Data are mean ± SEM. * p<0.05, burned vs. non-burn.
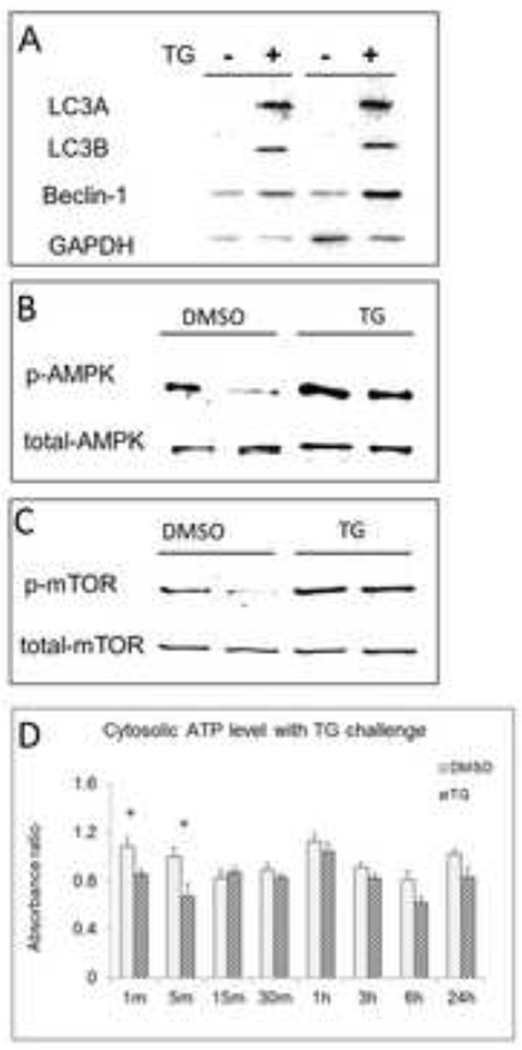
In vitro, we found expression of the autophagy marker Light Chain 3 (LC3) isoforms LC3A and LC3B significantly increased in HepG2 after TG challenge. Beclin-1 also increased [Fig 4A]. These findings indicate an increase in autophagy related signals in association with ER stress. Cytosolic ATP levels dropped significantly immediately after TG challenge (p<0.05), and recovered to normal levels within 15 minutes [Fig 4B]; Western blot data showed that the ratio of phosphorylated AMPK to total AMPK increased in HepG2 cells with TG challenge, and phosphorylated mTOR increased in response to TG induced ER stress as well [Fig 4C].
Figure 4.
In vitro, western blot data showing the expression of (A) autophagy makers LC3A and LC3B, Beclin-1, (B) phosphorylation of AMPK, and (C) mTOR in TG treated HepG2 cells. (D) Cytosolic ATP level alteration in HepG2 cell with TG challenge. Data are mean ± SEM. * p<0.05, TG treated group vs. vehicle treated group.
DISCUSSION
In the current study, we observed that expression of autophagy signals including ATG3, ATG5 and LC3B significantly increased in mouse liver 24 hours after burn. Furthermore, utilisation of in vitro model that was similar to burn induced ER stress in its induction of autophagy, we found that ER stress inducer TG decreased cytosolic ATP level and activated AMPK /mTOR signals in HepG2 cells.
Severe burn induces ER stress in the cells of many organ systems, suggesting that ER stress is a major cellular response to injury, and this process is associated with organ damage and functional impairment (20). ER stress was reported to be associated with injured liver after hemorrhage trauma (21). We previously observed that liver impairment associated with hepatic ER stress in response to severe burn (5, 20). Like previous studies, we found that hepatic ER stress occurred after burn with GRP78 expression augment, and also liver damage with increased cell death in the current study.
Further investigating the mechanism of autophagy activation in response to ER stress, we found that cellular energy consumption with decreased ATP level at the earlier time in TG treated HepG2 cell. This cellular change was similar to metabolic response after burn. Cellular stress after burn mostly includes oxidative stress and ER stress, leading to the consequence of metabolic alteration with energy consumption and redistribution to maintain homeostasis (22). Metabolic processes that use adenosine triphosphate (ATP) as an energy source that is converted to downstream mediators. Under conditions of energy consumption and after severe burn, adenylate cyclase converts ATP to AMP, thus diminishing levels of ATP and increasing AMP; this appears to be maximal at 72 hours after injury (23). The resulting decreased ratio of ATP to AMP is associated with AMPK activation (22) and subsequent phosphorylation of mTOR to initiate autophagy signaling pathways. In vitro study, we observed that ER stress inducer TG stimulated phosphorylation of AMPK/mTOR, indicating hepatic autophagy activation is associated with AMPK/ mTOR pathway in response to ER stress.
Autophagy has been considered as cytoprotective when compared to apoptosis (24). Xiao, R’s group confirmed that autophagy has a protective effect on myocardiocytes in burned rats (25). Cell death occurs when autophagy is inhibited, and Beclin-1 has been currently considered as one of key link connecting these discriminate events (26). The intracellular relationship of ER stress induced autophagy and apoptosis due in our in vitro data. Beclin-1 significantly increased in the HepG2 response to ER stress. However, to our surprise, we did not see the beclin-1 changes in vivo either in total amount or in its activated form. The current data suggests complicated mechanisms not fully explained for the role of autophagy in hepatic damage in response to severe burn. Other linkage pathways such as P53 induced damagerelated autophagy modulator (DRAM) should be investigated in the future. (27)
Autophagy signaling is not normal in critically ill patients (28), therefore regulation is a potential therapeutic target to improve for disease treatment. The synthesized peptide Tatbeclin-1 induces the autophagy process; mice treated with Tat-beclin-1 were resistant to several infectious diseases (29). Other potential mediators related to AMPK/mTOR pathway regulation may also be considered, and may be effective at improving hepatic function after burn through the mechanism of autophagy. Such compounds could include rapamycin (30) or pharmacological inhibition of autophagy with chloroquine or 3-Methyladenine (3-MA) which protects cells from zVAD-induced death (31). Further investigation of above potential candidates has been considered by using our burn animal model.
In conclusion, severe burn induces the cascade of autophagy in the liver associated with ER stress, and that contributes to hepatic damage after injury. Moreover, ER stress activated hepatic autophagy signaling associated with AMPK/ mTOR pathway. The current findings lighten a new direction to preserve liver homeostasis after burn, and further develop a protective strategy for burn and trauma patients with liver impairment.
ACKNOWLEDGEMENT
We wish to thank Kevin Despain, Ming-Mei Liu, and David Maass from Department of Surgery Core Laboratory for their technical support.
This work was supported by funds from the Golden Charity Guild Charles R Baxter, MD Chair and Checklist and Decision Support in Nutritional Care for Burned Patients. United States Army Medical Research Administration, TATRC, Department of Defense W81XWH-12-2-0074-01.
Footnotes
Presented at the 2013 Annual Meeting of the American Burn Association, Palm Springs, CA
DECLARATION:
The authors have no relevant disclosures.
REFERENCES
- 1.Ding HQ, Zhou BJ, Liu L, Cheng S. Oxidative stress and metallothionein expression in the liver of rats with severe thermal injury. Burns : journal of the International Society for Burn Injuries. 2002;28:215–221. doi: 10.1016/s0305-4179(02)00018-9. [DOI] [PubMed] [Google Scholar]
- 2.Jeschke MG, Gauglitz GG, Kulp GA, Finnerty CC, Williams FN, et al. Long-term persistance of the pathophysiologic response to severe burn injury. PloS one. 2011;6:e21245. doi: 10.1371/journal.pone.0021245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jeschke MG, Gauglitz GG, Song J, Kulp GA, Finnerty CC, et al. Calcium and Er Stress Mediate Hepatic Apoptosis after Burn Injury. J Cell Mol Med. 2009 doi: 10.1111/j.1582-4934.2008.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- 5.Song J, Finnerty CC, Herndon DN, Boehning D, Jeschke MG. Severe burn-induced endoplasmic reticulum stress and hepatic damage in mice. Mol Med. 2009;15:316–320. doi: 10.2119/molmed.2009.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annual review of pathology. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N. Autophagy: process and function. Genes & development. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki K, Ohsumi Y. Molecular machinery of autophagosome formation in yeast, Saccharomyces cerevisiae. FEBS letters. 2007;581:2156–2161. doi: 10.1016/j.febslet.2007.01.096. [DOI] [PubMed] [Google Scholar]
- 9.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annual review of genetics. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, et al. Regulation of mammalian autophagy in physiology and pathophysiology. Physiological reviews. 2010;90:1383–1435. doi: 10.1152/physrev.00030.2009. [DOI] [PubMed] [Google Scholar]
- 11.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. The Journal of biological chemistry. 1998;273:3963–3966. doi: 10.1074/jbc.273.7.3963. [DOI] [PubMed] [Google Scholar]
- 12.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature cell biology. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee JW, Park S, Takahashi Y, Wang HG. The association of AMPK with ULK1 regulates autophagy. PloS one. 2010;5:e15394. doi: 10.1371/journal.pone.0015394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoyer-Hansen M, Jaattela M. Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell death and differentiation. 2007;14:1576–1582. doi: 10.1038/sj.cdd.4402200. [DOI] [PubMed] [Google Scholar]
- 16.Ciechomska IA, Gabrusiewicz K, Szczepankiewicz AA, Kaminska B. Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine Ainduced cell death. Oncogene. 2013;32:1518–1529. doi: 10.1038/onc.2012.174. [DOI] [PubMed] [Google Scholar]
- 17.Hoyer-Hansen M, Jaattela M. AMP-activated protein kinase: a universal regulator of autophagy? Autophagy. 2007;3:381–383. doi: 10.4161/auto.4240. [DOI] [PubMed] [Google Scholar]
- 18.Toliver-Kinsky TE, Cui W, Murphey ED, Lin C, Sherwood ER. Enhancement of dendritic cell production by fms-like tyrosine kinase-3 ligand increases the resistance of mice to a burn wound infection. J Immunol. 2005;174:404–410. doi: 10.4049/jimmunol.174.1.404. [DOI] [PubMed] [Google Scholar]
- 19.Song J, Zhang XJ, Boehning D, Brooks NC, Herndon DN, et al. Measurement of Hepatic Protein Fractional Synthetic Rate with Stable Isotope Labeling Technique in Thapsigargin Stressed HepG2 Cells. International journal of biological sciences. 2012;8:265–271. doi: 10.7150/ijbs.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeschke MG, Finnerty CC, Herndon DN, Song J, Boehning D, et al. Severe Injury Is Associated With Insulin Resistance, Endoplasmic Reticulum Stress Response, and Unfolded Protein Response. Annals of surgery. 2012;255:370–378. doi: 10.1097/SLA.0b013e31823e76e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jian B, Hsieh CH, Chen J, Choudhry M, Bland K, et al. Activation of endoplasmic reticulum stress response following trauma-hemorrhage. Biochimica et biophysica acta. 2008;1782:621–626. doi: 10.1016/j.bbadis.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horton JW. Free radicals and lipid peroxidation mediated injury in burn trauma: the role of antioxidant therapy. Toxicology. 2003;189:75–88. doi: 10.1016/s0300-483x(03)00154-9. [DOI] [PubMed] [Google Scholar]
- 23.Gore DC, Rinehart A, Asimakis G. Temporal changes in cellular energy following burn injury. Burns : journal of the International Society for Burn Injuries. 2005;31:998–1002. doi: 10.1016/j.burns.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Boya P, Gonzalez-Polo RA, Casares N, Perfettini JL, Dessen P, et al. Inhibition of macroautophagy triggers apoptosis. Molecular and cellular biology. 2005;25:1025–1040. doi: 10.1128/MCB.25.3.1025-1040.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiao R, Teng M, Zhang Q, Shi XH, Huang YS. Myocardial autophagy after severe burn in rats. PloS one. 2012;7:e39488. doi: 10.1371/journal.pone.0039488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang R, Zeh HJ, Lotze MT, Tang D. The Beclin 1 network regulates autophagy and apoptosis. Cell death and differentiation. 2011;18:571–580. doi: 10.1038/cdd.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nature reviews. Molecular cell biology. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 28.Vanhorebeek I, Gunst J, Derde S, Derese I, Boussemaere M, et al. Insufficient activation of autophagy allows cellular damage to accumulate in critically ill patients. The Journal of clinical endocrinology and metabolism. 2011;96:E633–E645. doi: 10.1210/jc.2010-2563. [DOI] [PubMed] [Google Scholar]
- 29.Shoji-Kawata S, Sumpter R, Leveno M, Campbell GR, Zou Z, et al. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013;494:201–206. doi: 10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell death and differentiation. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
- 31.Wu YT, Tan HL, Huang Q, Kim YS, Pan N, et al. Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy. 2008;4:457–466. doi: 10.4161/auto.5662. [DOI] [PubMed] [Google Scholar]