Abstract
Background
The rice PLASTOCHRON (PLA) genes PLA1 and PLA2 regulate leaf maturation and the temporal pattern of leaf initiation. Although the function of PLA genes in the leaf initiation process has been analyzed, little is known about how they affect leaf growth. Previously, we suggested that PLA1 and PLA2 function downstream of the gibberellin (GA) signal transduction pathway. In the present study, we examined the phenotype of a double mutant of pla and slender rice 1 (slr1), which is a constitutive GA response mutant. By analyzing these double mutants, we discuss the relationship between PLA-related and GA-dependent pathways and the possible function of PLA genes in leaf growth.
Findings
Single slr1 and pla mutants exhibited elongated and dwarf phenotypes in the vegetative stage, respectively. The stature and leaf size of the pla1/slr1 and pla2/slr1 double mutants were intermediate between those of the pla and slr1 single mutants. However, the effects of slr1 on leaf elongation were markedly suppressed in the pla1 and pla2 mutant backgrounds. On the other hand, the change in cell length in the double mutants was almost the same as that in the single mutants. An expression analysis of genes involved in GA biosynthesis and catabolism indicated that feedback regulation functioned normally in the pla/slr1 double mutants.
Conclusions
Our genetic results confirm that PLA genes regulate leaf growth downstream of the GA pathway. Our findings also suggest that PLA1 and PLA2 are partly required for GA-dependent leaf elongation, mainly by affecting cellular proliferation.
Keywords: Rice, PLASTOCHRON 1, PLASTOCHRON 2, Gibberellin, SLENDER RICE 1
Findings
Rice plastochron (pla) mutants show a short plastochron and small precocious leaves. PLA1 and PLA2 encode a cytochrome P450, CYP78A11, and an RNA-binding protein, respectively. They are expressed in leaf primordia and regulate the leaf initiation rate and leaf maturation (Miyoshi et al.[2004]; Kawakatsu et al.[2006]). Thus, PLA1 and PLA2 play important roles in leaf development. Previously, we showed that PLA1 and PLA2 function downstream of the gibberellin (GA) signal transduction pathway (Mimura et al.[2012]), and that pla1 and pla2 plants exhibited reduced sensitivity to GA treatment. In addition, GA treatment induced PLA1 and PLA2 expression. In accordance with these results, the expression levels of PLA genes were increased in slender rice 1 (slr1), which is a constitutively active GA signaling mutant, and decreased in slr1-D, which shows reduced sensitivity to GA. However, genetic evidence for the interaction between PLA genes and GA signaling genes is lacking. In the present study, we constructed pla1/slr1 and pla2/slr1 double mutants to investigate the genetic relationships between PLA genes and the GA signaling pathway.
Phenotypes of pla1 and slr1 double mutants
slr1 is a constitutive GA response mutant that is caused by a loss-of-function of DELLA, which is a key factor in the repression of GA responses (Ikeda et al.[2001]). slr1 mutants showed elongated leaves and internodes. In contrast, pla mutants showed dwarfism and small leaves. PLA1 encodes the cytochrome P450 family protein CYP78A11, which is a member of the CYP78A subfamily (Miyoshi et al.[2004]). Many reports have shown that CYP78A family genes regulate organ growth (e.g., seed or fruit size) in several plant species (Anastasiou et al.[2007]; Fang et al.[2012]; Chakrabarti et al.[2013]; Sotelo-Silveira et al.[2013]). It has also been suggested that CYP78A family members are involved in producing an as yet unidentified substance that functions as a mobile growth regulator (Anastasiou et al.[2007]; Adamski et al.[2009]; Eriksson et al.[2010]).
To determine the genetic interaction between SLR1 and PLA1, we generated pla1/slr1 double mutants by crossing SLR1 heterozygous plants with PLA1 heterozygous plants. At the 3-week-old seedling stage, the pla1/slr1 double mutants showed intermediate phenotypes (Figure 1A–C, Table 1). However, the effects of the slr1 mutation on plant height and leaf size in the pla1 background were weaker than those in wild type. The slr1 plants were 53% taller than the wild-type plants, whereas the height of the pla1/slr1 double mutant was 23% that of the pla1 single mutant (Figure 1B). In terms of leaf length, the effect of the slr1 mutation was much more obvious in the wild-type background than in the pla1 mutant background. The third leaf sheath of slr1 was 123% longer than that of wild type, whereas that of the pla1/slr1 double mutant was only 53% that of the pla1 single mutant (Figure 1C). These results suggest that PLA1 activity is partly necessary for leaf elongation in slr1 mutant plants.
Figure 1.
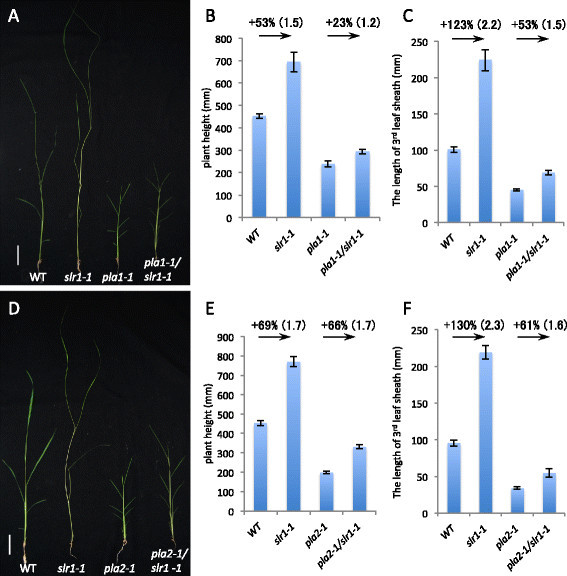
Phenotypes of the pla1/slr1 and pla2/slr1 double mutants. (A–C) Phenotypes of wild-type, slr1-1, pla1-1, and pla1-1/slr1-1 plants. (D–F) Phenotypes of wild-type, slr1-1, pla2-1, and pla2-1/slr1-1 plants. (A, D) Seedlings at 3 weeks after germination (DAG). (B, E) Plant height at 3 weeks after germination. (C, F) Length of the third leaf sheath. The values represent means ± SE (n > 5). Fold-increase are shown in parenthesis after% increase. The scale bars indicate 5 cm.
Table 1.
Seedling phenotypes of the pla1-1/slr1-1 and pla2-1/slr1-1 double mutants at 3 weeks after germination
| Genotype | Plant height (mm) | Leaf number | 1st Leaf (mm) | 2nd LB (mm) | 2nd LS (mm) | 3rd LB (mm) | 3rd LS (mm) | 4th LB (mm) | 4th LS (mm) | n |
|---|---|---|---|---|---|---|---|---|---|---|
| WT |
453 ± 11.3 |
5.6 ± 0.16 |
19.3 ± 0.6 |
21.0 ± 1.4 |
45.7 ± 1.7 |
79.0 ± 3.9 |
100.7 ± 3.5 |
153.7 ± 5.5 |
166.6 ± 5.4 |
10 |
|
slr1-1 |
693 ± 43.3 |
4.8 ± 0.17 |
27.8 ± 2.5 |
25 ± 2.6 |
109.5 ± 8.5 |
186.0 ± 17.5 |
224.3 ± 14.4 |
381.2 ± 29.8 |
321.8 ± 23.1 |
6 |
|
pla1-1 |
238 ± 13.0 |
7.8 ± 0.16 |
14.9 ± 0.7 |
12.6 ± 1.0 |
27.6 ± 1.5 |
29.1 ± 1.9 |
45.0 ± 1.4 |
35.1 ± 0.8 |
62.6 ± 1.9 |
8 |
|
pla1-1/slr1-1 |
294 ± 10.8 |
7.8 ± 0.31 |
15.0 ± 1.2 |
10.9 ± 1.5 |
36.3 ± 3.7 |
26.9 ± 4.1 |
69.0 ± 3.2 |
47.8 ± 3.3 |
92.3 ± 4.4 |
8 |
| WT |
454 ± 12.3 |
5.9 ± 0.17 |
20.4 ± 0.8 |
20.1 ± 1.0 |
44.1 ± 1.8 |
77.3 ± 4.2 |
95.5 ± 4.1 |
157.3 ± 4.8 |
142.5 ± 6.7 |
11 |
|
slr1-1 |
771 ± 25.0 |
5.0 ± 0.21 |
30.5 ± 1.7 |
25.2 ± 1.5 |
109.7 ± 6.3 |
162.5 ± 11.9 |
219.2 ± 9.4 |
396.7 ± 14.2 |
344.2 ± 18.2 |
10 |
|
pla2-1 |
199 ± 6.7 |
9.3 ± 0.15 |
14.5 ± 0.9 |
4.6 ± 0.5 |
23.0 ± 1.6 |
12.6 ± 1.0 |
34.0 ± 1.7 |
27.6 ± 2.2 |
46.4 ± 1.5 |
10 |
| pla2-1/slr1-1 | 332 ± 8.9 | 8.4 ± 0.51 | 16.8 ± 1.6 | 4.8 ± 0.9 | 34.4 ± 4.2 | 16.8 ± 1.3 | 54.8 ± 5.7 | 40.0 ± 4.8 | 75.4 ± 5.5 | 5 |
LB: leaf blade, LS: leaf sheath. The values indicate the means ± SE. n indicate the number of seedlings examined in this study.
Cell size is one of the factors determining leaf size. To clarify how cell size contributes to leaf elongation in slr1 and pla1/slr1 double mutants, we compared the lengths of epidermal cells on the adaxial side of the third leaf sheath in each mutant (Figure 2A). Our results indicate that the effects of the slr1 mutation on cell size were comparable between the wild-type and pla1 backgrounds. Cell length was increased by 17% in slr1 single mutant plants and by 14% in pla1/slr double mutant plants compared to the corresponding genotypes. These results indicate that PLA1 contributes mainly to cell proliferation in GA-dependent leaf elongation.
Figure 2.
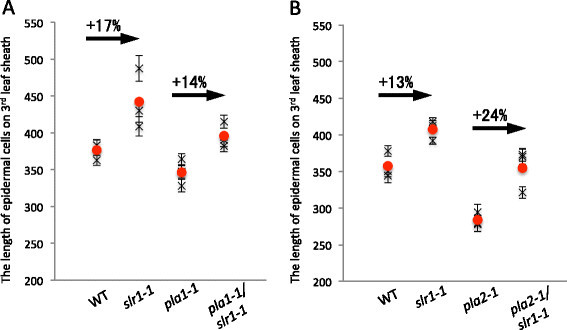
Length of epidermal cells on the adaxial side of the third leaf sheath. (A) Epidermal cell length in wild-type, slr1-1, pla1-1, and pla1-1/slr1-1 plants. (B) Epidermal cell length in wild-type, slr1-1, pla2-1, and pla2-1/slr1-1 plants. Crosses indicate average values of the cells in one sample (average ± SE; n > 100). Closed circles indicate average values of three independent samples.
Phenotypes of pla2 and slr1 double mutants
PLA2 encodes an RNA-binding protein; however, its target RNAs have yet to be elucidated (Kawakatsu et al.[2006]). Similar to the pla1/slr1 double mutants, we examined the phenotype of pla2/slr1 double mutants. The stature and leaf size of the double mutants were intermediate between those of the pla2 and slr1 mutants (Figure 1D–F, Table 1). With regard to plant height, 3-week-old slr1 and pla2/slr1 seedlings were 69% and 66% taller than wild-type and pla2 seedlings, respectively (Figure 1E). With regard to the length of the third leaf sheath, those of the slr1 and pla2/slr1 plants were 130% and 61% longer than in the corresponding genotypes, respectively (Figure 1F). These results suggest that PLA2 is also at least partially involved in GA-dependent leaf elongation.
Next, we measured the length of epidermal cells on the adaxial side of the third leaf sheath. The cells of the pla2/slr1 double mutant were elongated by 24% compared to the pla2 single mutant; whereas those of slr1 were 13% longer than in wild type (Figure 2B). These results indicate that normal GA-dependent cell elongation occurred in the pla2/slr1 double mutants. Accordingly, the suppression of the slr1 phenotype in the pla2/slr1 double mutant may have been due to a reduction in cell number.
Expression of genes involved in GA biosynthesis and catabolism in pla/slr1 double mutants
The content of bioactive GA is maintained through feedback regulation (Dai et al.[2007]; Olszewski et al. [2002]; Yamaguchi [2008]). To investigate whether feedback regulation for GA homeostasis occurred normally in our pla/slr1 double mutant plants, we examined the expression levels of two GA biosynthetic genes GA3 oxidase2 (GA3ox2) and GA20 oxidase2 (GA20ox2), and two GA catabolism genes GA2 oxidase1 (GA2ox1) and GA2ox4, in these mutants by real-time PCR (Figure 3, Additional file 1: Table S1). In pla1 and pla2 mutant plants, the expression levels of these GA biosynthetic and GA catabolism genes were comparable to those in wild-type controls. Thus, PLA1 and PLA2 do not affect the expression of genes involved in GA metabolism. The expression of GA3ox2 was slightly decreased in the slr1 mutant plants, as reported previously (Dai et al.[2007]). The expression of GA20ox2 was not decreased in slr1 mutant, indicating that the expression of GA20ox2 may not be under GA feedback regulation in slr1 mutant. In contrast to GA3ox2 gene, the expression levels of both the GA catabolism genes in slr1 mutant were increased compared to wild type. Similar to the levels seen in the slr1 mutant, GA3ox2 expression was downregulated and GA2ox1 and GA2ox4 expression was upregulated in the pla1/slr1 and pla2/slr1 double mutant plants. These results indicate that the feedback mechanism and GA response were normal in the pla/slr1 double mutants, at least at the transcriptional level.
Figure 3.
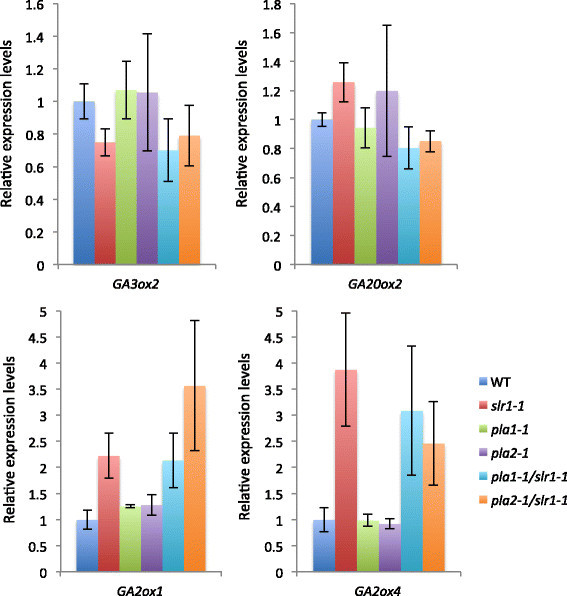
Relative expression levels of GA biosynthetic ( GA3ox2, GA20ox2 ) and GA catabolism ( GA2ox1 , GA2ox4 ) genes in wild-type, slr1-1 , pla1-1 , pla2-1 , pla1-1/slr1-1 , and pla2-1/slr1-1 plants. The expression levels in the mutants are represented relative to that in wild type (assigned a value of 1). The values indicate the means of three biological samples ± SE. Actin1 was used as an internal control. GA3ox2, GA20ox2, GA2ox4 and Actin1 were quantified using TaqMan probes. GA2ox1 was quantified by SYBR green. The primers and probes for each gene are listed in Additional file 1: Table S1.
GA is involved in various developmental processes, including seed germination, stem elongation, flowering, and pollen maturation (Olszewski et al. [2002]; Yamaguchi [2008]). Microarray studies have identified several genes involved in the GA pathway (Yazaki et al.[2003]; Yang et al.[2004]; Jan and Komatsu [2006]). However, the genetic regulation downstream of the GA pathway in leaf development is poorly understood. Previous studies suggested that PLA gene products function downstream of GA. In this study, we demonstrated genetic interactions between PLA genes and SLR1, a central regulator of GA signaling, supporting our previous results.
Our analysis suggests that the intermediate phenotypes of the pla/slr1 double mutants were probably due to a reduction in cell number. There are two explanations for why the absence of PLA functions partly suppressed the slr1 phenotype. First, PLA genes are involved in cellular proliferation in the GA-dependent pathway. Recent studies have indicated that GA promotes not only cell expansion but also cellular proliferation through the regulation of cell cycle inhibitor genes (Achard et al.[2009]). In addition, GA controls the transition from cell proliferation to expansion in maize leaves (Nelissen et al.[2012]). Thus, GA can influence cell number during leaf development, and it is possible that PLA functions are required for cell proliferation rather than cell elongation downstream of the GA pathway. Second, defects in PLA genes affect the duration and/or timing of cellular proliferation, resulting in a decrease in the total cell number in the leaves of pla/slr1 mutants. Previous studies suggested that PLA1 and PLA2 genes regulate the rate of leaf maturation (Kawakatsu et al.[2006]) and that the small leaves in pla mutants were due to precocious leaf maturation. Thus, it is possible that the duration of cell proliferation in developing leaves is insufficient in pla/slr1 double mutants, resulting in suppression of the slr1 phenotype.
Our results indicate that PLA genes partly regulate leaf size by affecting cell proliferation via the GA-dependent pathway. However, it remains unclear how PLA genes regulate cellular proliferation in the GA signaling pathway. Further study is required to clarify the molecular mechanisms underlying the regulatory roles of PLA gene expression in GA-dependent leaf development.
Abbreviations
GA: Gibberellin
PLA: PLASTOCHRON
SLR1: SLENDER RICE1
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
MM performed the experiments and drafted the manuscripts. JI conceived the study and drafted manuscripts. Both authors read and approved the final manuscript.
Additional file
Supplementary Material
List of primers used in real-time PCR assays.
Contributor Information
Manaki Mimura, Email: 7588361782@mail.ecc.u-tokyo.ac.jp.
Jun-Ichi Itoh, Email: ajunito@mail.ecc.u-tokyo.ac.jp.
Acknowledgements
We would like to thank Dr. H. Kitano (Nagoya University) for kind gift of slr1-1 seeds. We also thank R. Soga and K. Ichikawa (The Institute for Sustainable Agro-ecosystem Services, University of Tokyo) for their assistance in cultivating rice plants at the Experimental Farm of the University of Tokyo. This work was partially funded by JSPS KAKENHI 24380005 (J.I.) and Grant-in-Aid for JSPS Fellows 25–8135 (M.M.)
References
- Anastasiou E, Kenz S, Gerstung M, MacLean D, Timmer J, Fleck C, Lenhard M. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signaling. Dev Cell. 2007;13:843–856. doi: 10.1016/j.devcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol. 2009;19:1188–1193. doi: 10.1016/j.cub.2009.05.059. [DOI] [PubMed] [Google Scholar]
- Adamski NM, Anastasiou E, Eriksson S, O'Neill CM, Lenhard M. Local maternal control of seed size by KLUH/CYP78A5-dependent growth signaling. Proc Natl Acad Sci U S A. 2009;106:20115–20120. doi: 10.1073/pnas.0907024106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti M, Zhang N, Sauvage C, Muños S, Blanca J, Cañizares J, Diez MJ, Schneider R, Mazourek M, McClead J, Causse M, van der Knaap E. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc Natl Acad Sci U S A. 2013;110:17125–17130. doi: 10.1073/pnas.1307313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Zhao Y, Ma Q, Hu Y, Hedden P, Zhang Q, Zhou DX. The rice YABBY1 gene is involved in the feedback regulation of gibberellin metabolism. Plant Physiol. 2007;144:121–133. doi: 10.1104/pp.107.096586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Stransfeld L, Adamski NM, Breuninger H, Lenhard M. KLUH/CYP78A5-dependent growth signaling coordinates floral organ growth in Arabidopsis. Curr Biol. 2010;20:527–532. doi: 10.1016/j.cub.2010.01.039. [DOI] [PubMed] [Google Scholar]
- Fang W, Wang Z, Cui R, Li J, Li Y. Maternal control of seed size by EOD3/CYP78A6 in Arabidopsis thaliana. Plant J. 2012;70:929–939. doi: 10.1111/j.1365-313X.2012.04907.x. [DOI] [PubMed] [Google Scholar]
- Ikeda A, Ueguchi-Tanaka M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J. Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell. 2001;13:999–1010. doi: 10.1105/tpc.13.5.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A, Komatsu S. Functional characterization of gibberellin-regulated genes in rice using microarray system. Genomics Proteomics Bioinformatics. 2006;4:137–144. doi: 10.1016/S1672-0229(06)60026-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakatsu T, Itoh J, Miyoshi K, Kurata N, Alvarez N, Veit B, Nagato Y. PLASTOCHRON2 regulates leaf initiation and maturation in rice. Plant Cell. 2006;18:612–625. doi: 10.1105/tpc.105.037622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura M, Nagato Y, Itoh J. Rice PLASTOCHRON genes regulate leaf maturation downstream of the gibberellin signal transduction pathway. Planta. 2012;235:1081–1089. doi: 10.1007/s00425-012-1639-5. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Ahn BO, Kawakatsu T, Ito Y, Itoh J, Nagato Y, Kurata N. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci U S A. 2004;101:875–880. doi: 10.1073/pnas.2636936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelissen H, Rymen B, Jikumaru Y, Demuynck K, Van Lijsebettens M, Kamiya Y, Inzé D, Beemster GT. A local maximum in gibberellin levels regulates maize leaf growth by spatial control of cell division. Curr Biol. 2012;22:1183–1187. doi: 10.1016/j.cub.2012.04.065. [DOI] [PubMed] [Google Scholar]
- Olszewski N, Sun TP, Gubler F. Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell. 2002;14:S61–S80. doi: 10.1105/tpc.010476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo-Silveira M, Cucinotta M, Chauvin AL, Chávez Montes RA, Colombo L, Marsch-Martínez N, de Folter S. Cytochrome P450 CYP78A9 is involved in Arabidopsis reproductive development. Plant Physiol. 2013;162:779–799. doi: 10.1104/pp.113.218214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- Yang GX, Jan A, Shen SH, Yazaki J, Ishikawa M, Shimatani Z, Kishimoto N, Kikuchi S, Matsumoto H, Komatsu S. Microarray analysis of brassinosteroids- and gibberellin-regulated gene expression in rice seedlings. Mol Genet Genomics. 2004;271:468–378. doi: 10.1007/s00438-004-0998-4. [DOI] [PubMed] [Google Scholar]
- Yazaki J, Kishimoto N, Nagata Y, Ishikawa M, Fujii F, Hashimoto A, Shimbo K, Shimatani Z, Kojima K, Suzuki K, Yamamoto M, Honda S, Endo A, Yoshida Y, Sato Y, Takeuchi K, Toyoshima K, Miyamoto C, Wu J, Sasaki T, Sakata K, Yamamoto K, Iba K, Oda T, Otomo Y, Murakami K, Matsubara K, Kawai J, Carninci P, Hayashizaki Y, Kikuchi S. Genomics approach to abscisic acid- and gibberellin-responsive genes in rice. DNA Res. 2003;10:249–261. doi: 10.1093/dnares/10.6.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in real-time PCR assays.


