SUMMARY
Diet greatly influences gene expression and physiology. In mammals, elucidating the effects and mechanisms of individual nutrients is challenging due to the complexity of both the animal and its diet. Here we used an interspecies systems biology approach with Caenorhabditis elegans and two if its bacterial diets, Escherichia coli and Comamonas aquatica, to identify metabolites that affect the animal’s gene expression and physiology. We identify vitamin B12 as the major dilutable metabolite provided by Comamonas aq. that regulates gene expression, accelerates development and reduces fertility, but does not affect lifespan. We find that vitamin B12 has a dual role in the animal: it affects development and fertility via the methionine/S-Adenosylmethionine (SAM) cycle and breaks down the short-chain fatty acid propionic acid preventing its toxic buildup. Our interspecies systems biology approach provides a paradigm for understanding complex interactions between diet and physiology.
INTRODUCTION
Our diet provides building blocks for development and reproduction, as well as energy to sustain daily cellular and organismal activities. Complex diets consist of macronutrients such as carbohydrates, fats and proteins, but also provide micronutrients such as vitamins that function as cofactors in metabolic reactions. In mammals, nutrients are provided not only by diet but are also synthesized by the gut microbiota (Hooper et al., 2002). A major challenge is to unravel the contributions of individual metabolites to cellular and organismal physiology, and to dissect the metabolic and genetic underpinnings of physiological responses to changing diets.
The nematode C. elegans is an emerging model to study the effects of diet on life history traits such as developmental rate, fertility and aging (Coolon et al., 2009; Gracida and Eckmann, 2013; MacNeil et al., 2013). C. elegans is a relatively simple model organism composed of fewer than 1000 somatic cells. It lives in temperate climates around the globe and subsists on diets of various bacterial species growing on rotting vegetation. These bacteria also inhabit the C. elegans intestine to serve as its microbiota (Felix and Duveau, 2013). In the laboratory, C. elegans are grown monoxenically on E. coli OP50, but many other bacterial strains and species have been fed to worms as well (Avery and Shtonda, 2003; Coolon et al., 2009; MacNeil et al., 2013; Soukas et al., 2009). Bacteria supply C. elegans with metabolites that can greatly affect its life history traits. For instance, bacterially derived nitric oxide and folate extend and limit the lifespan of the animal, respectively (Gusarov et al., 2013; Virk et al., 2012). The effects of these metabolites were identified by a hypothesis-driven approach (nitric oxide) or serendipitously by a mutation in the bacteria (folate). Since both C. elegans and its bacterial diet are genetically tractable, we reasoned that this predator-prey combination could be used for the unbiased identification of nutrients that drive transcriptional and physiological responses in the animal.
We previously found that, relative to E. coli OP50, a diet of Comamonas DA1877 accelerates C. elegans development and decreases fertility and lifespan (MacNeil et al., 2013). These physiological effects are accompanied by dramatic changes in gene expression (MacNeil et al., 2013). For instance, the acyl-CoA dehydrogenase-encoding gene acdh-1 is repressed several hundred fold on the Comamonas DA1877 diet relative to E. coli OP50. We created transgenic animals harboring the acdh-1 promoter driving expression of the green fluorescent protein (GFP) to generate a transgenic “dietary sensor” strain with which the transcriptional response to the Comamonas DA1877 diet can be readily monitored in living animals (MacNeil et al., 2013). Remarkably, the effects of Comamonas DA1877 on C. elegans gene expression and development persist even when these bacteria are mixed in small amounts with the E. coli OP50 diet, indicating that Comamonas DA1877 generates one or more dilutable compounds to which C. elegans responds. Using the dietary sensor strain, we identified a C. elegans network consisting of metabolic and regulatory genes that, when perturbed, interferes with the transcriptional response to Comamonas DA1877 (Watson et al., 2013).
Here, we used an interspecies systems biology approach to identify bacterial metabolites that affect C. elegans gene expression and life history traits. We performed genetic screens in E. coli and Comamonas to identify bacterial genes that, when mutated, result in aberrant repression or activation of Pacdh-1::GFP in C. elegans. We performed a secondary metabolite screen by supplementing 25 candidate metabolites to Pacdh-1::GFP animals. Eight compounds activated the dietary sensor, including branched chain amino acids, threonine and propionic acid. Two compounds repressed the sensor – methyl-cobalamin (Me-Cbl) and adenosyl-cobalamin (Ado-Cbl), the two biologically active forms of vitamin B12. We demonstrate that vitamin B12 is generated by Comamonas DA1877 but not by E. coli, and that it drives many of the gene expression and physiological changes in C. elegans induced by the Comamonas diet. Interestingly all eight activating metabolites are closely connected in the C. elegans metabolic network to the two enzymes that require vitamin B12 as a cofactor. We find that vitamin B12 fulfills two important physiological roles in C. elegans: it regulates development through the synthesis of the major methyl donor SAM, and alleviates toxic buildup of the short-chain fatty acid propionic acid. Our interspecies systems biology approach provides a powerful paradigm for gaining insight into the complex interactions between diet, metabolic regulation and physiology.
RESULTS
Genetic Screens in Bacterial Diets
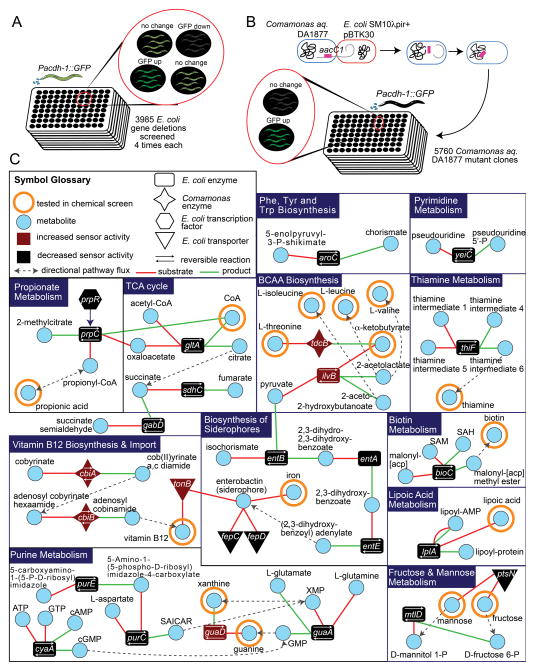
We previously determined that Comamonas DA1877 bacteria must produce a dilutable compound to which C. elegans responds with altered gene expression programs, accelerated development, reduced fertility and reduced lifespan (MacNeil et al., 2013). We reasoned that we could discover bacterial metabolites that affect C. elegans by identifying bacterial genes involved in the generation, processing or transport of these small molecules. We used the C. elegans Pacdh-1::GFP dietary sensor as a reporter for diet-induced gene expression changes: GFP expression is high when the animals are fed E. coli OP50, but barely detectable when they are fed Comamonas DA1877. We performed genetic screens in each of these bacteria to identify mutant strains that, when fed to C. elegans alter GFP expression.
First, we fed dietary sensor animals the Keio E. coli BW25113 collection, which contains deletion mutants for 3,985 of the 4,290 protein-coding genes (Baba et al., 2006). We visually examined whether GFP expression was decreased or increased relative to animals fed the wild type strain (Figure 1A). In total, 70 mutant E. coli strains decreased GFP expression, and seven caused an increase compared to the parent strain (Table S1). Second, we performed a transposon-based mutagenesis screen of Comamonas DA1877 bacteria (Figure 1B). Using a non-replicating transposon, we generated 5,760 Comamonas DA1877 mutants each with a single transposon insertion. Five mutant Comamonas DA1877 strains failed to repress GFP expression when fed to the sensor strain.
Figure 1. Bacterial Screens.
(A) Diagram of the Keio E. coli deletion collection screen.
(B) Diagram of Comamonas DA1877 transposon mutagenesis screen.
(C) Network of metabolites implicated by bacterial screens. SAICAR = 5-Amino-4-imidazole-N-succinocarboxamide ribonucleotide; [acp] = acyl carrier protein.
See also Figure S1, Tables S1 and S2.
To facilitate the mapping of transposon insertions and identification of the disrupted genes, we sequenced the Comamonas DA1877 genome and annotated protein-coding sequences and RNA genes (Figure S1A). Since there was no systematic report on the taxonomic identity of Comamonas DA1877 before this study, we first used the single 16S rRNA gene to identify this strain at the species level as Comamonas aquatica, and we will henceforth refer to Comamonas DA1877 as Comamonas aq. DA1877.
We reasoned that bacterial mutants identified affect C. elegans gene expression due to either the buildup or reduction in particular metabolites. A total of 77 E. coli and 5 Comamonas aq. DA1877 genes (Table S2) were identified in the bacterial screens. We focused on bacterial genes encoding metabolic enzymes, transporters, and transcription factors known to regulate metabolic operons. Mapping these genes onto bacterial metabolic networks revealed that perturbation of several different pathways affected GFP expression in C. elegans (Figure 1C). For instance, mutations in enzymes from E. coli purine metabolism, propionic acid metabolism, the TCA cycle, and the biosynthesis of siderophores (iron scavengers) decreased dietary sensor activity. Conversely, E. coli and Comamonas mutations in branched chain amino acid biosynthesis and vitamin B12 biosynthesis/import resulted in increased C. elegans dietary sensor activity.
A Metabolite Screen in C. elegans Identifies Vitamin B12 as the Candidate Dilutable Comamonas Molecule
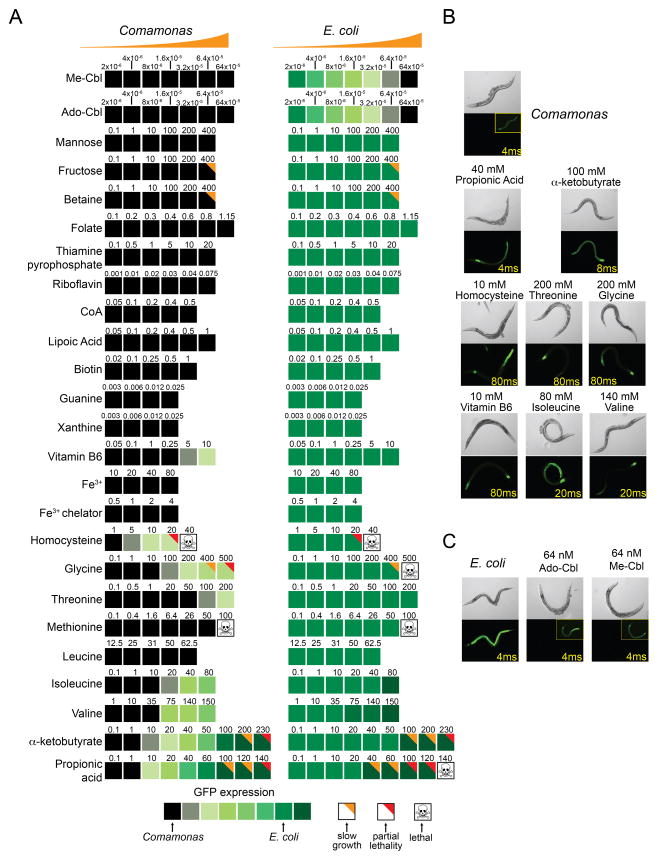
We selected a subset of metabolites implicated by the bacterial screens for use in a secondary screen in which their effect on the C. elegans dietary sensor was tested by direct supplementation to either diet. We focused on 18 metabolites implicated by bacterial genes encoding metabolic enzymes, transporters, and transcription factors known to regulate metabolic operons. We included 7 additional metabolites that were implicated from our earlier C. elegans genetic screens (Watson et al., 2013). We supplemented the metabolites at various concentrations to the Pacdh-1::GFP sensor strain fed either bacterial diet (Figure 2A). Six metabolites caused mild, dose-dependent increases in GFP expression when supplemented to the Comamonas aq. DA1877 diet, including the amino acids valine, isoleucine, threonine and glycine, as well as homocysteine and vitamin B6. Two metabolites, α-ketobutyrate and propionic acid, more dramatically increased GFP expression when supplemented to Comamonas aq. DA1877 and also further increased GFP expression on the E. coli OP50 diet(Figure 2B, Table S3).
Figure 2. Metabolite Screen.
(A) Summary of the observed effects of supplementing metabolites to Pacdh-1::GFP animals fed either Comamonas aq. DA1877 (left column) or E. coli OP50 (right column). Shades of green represent relative GFP expression. Numbers indicate metabolite concentrations (in mM).
(B) Pacdh-1::GFP animals fed Comamonas aq. DA1877 bacteria alone or supplemented with the indicated compounds that activate GFP expression. Exposure time is indicated in yellow. Exposure time for inset images is 400 ms.
(C) Pacdh-1::GFP animals fed E. coli OP50 bacteria supplemented with the indicated compounds that repress GFP expression. Exposure time is indicated in yellow. Exposure time for inset images is 400 ms.
See also Table S3.
Only two metabolites mimicked the strong repressive effect of the Comamonas aq. diet on the Pacdh-1::GFP sensor when supplemented to E. coli OP50: the two biologically active forms of vitamin B12, Ado-Cbl Me-Cbl (Figure 2C). Vitamin B12 is an attractive candidate to be the dilutable molecule produced by Comamonas aq. DA1877 (MacNeil et al., 2013) for several reasons. First, two of five Comamonas aq. DA1877 genes identified in the transposon screen encode vitamin B12 biosynthetic enzymes (cbiA and cbiB, Table S2 and Figure S1B). Second, vitamin B12 was also implicated by the E. coli deletion collection screen, even though they do not synthesize this cofactor (see below). A deletion in tonB, which encodes a protein required for vitamin B12 import (Kadner, 1990), resulted in even greater levels of GFP expression when fed to the sensor strain (Figure S1C). Third, Ado- and Me-Cbl were the only repressors identified in the chemical screen, and both robustly repress Pacdh-1::GFP in low (nM) doses. This fits with the observation that mixing small amounts of the Comamonas aq. DA1877 diet into the E. coli OP50 diet is sufficient to exert gene expression and physiological changes (MacNeil et al., 2013). Finally, mutations in the two C. elegans enzymes that use vitamin B12 interfere with the transcriptional response to the Comamonas aq. DA1877 diet (Watson et al., 2013). Vitamin B12 is an essential nutrient for most animals, but is only synthesized by some species of bacteria (Bender, 2003). It is used as a cofactor by the same two enzymes in all vitamin B12-dependent animals: methylmalonyl-CoA mutase (MUT; MMCM-1 in C. elegans), which is involved in propionyl-CoA breakdown, and methionine synthase (MS; METR-1 in C. elegans), which is involved in the methionine/SAM cycle. Methylmalonyl-CoA mutase uses Ado-Cbl whereas methionine synthase uses Me-Cbl (Figure S1D). We previously found that both of these vitamin B12-dependent enzymes, as well as others in their respective pathways, are required for Comamonas DA1877-induced gene expression changes (Watson et al., 2013). Taken together, these results suggest that vitamin B12 may be the dilutable molecule provided by the Comamonas aq. DA1877 diet that repressesPacdh-1::GFP.
Vitamin B12-Producing Bacteria Repress the Dietary Sensor
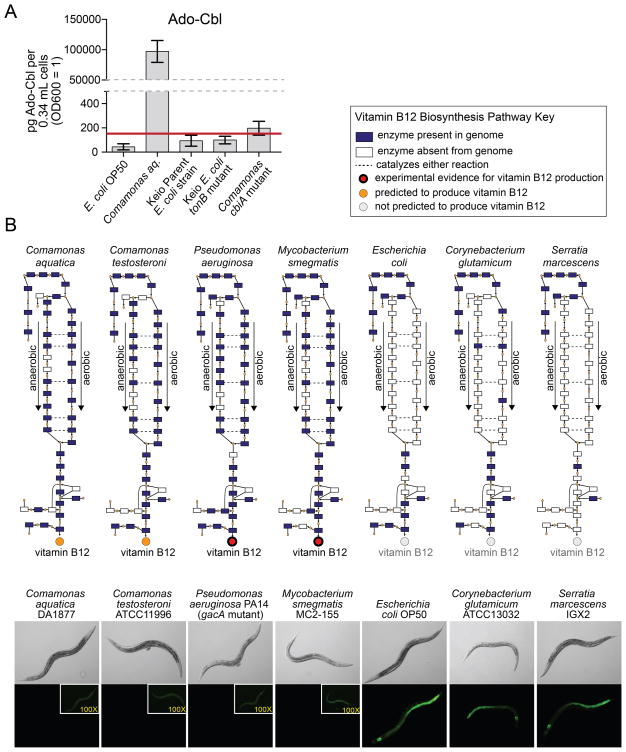
Our results predict that vitamin B12 levels are dramatically higher in the Comamonas aq. DA1877 than the E. coli OP50 diet. To directly compare the amounts of vitamin B12 we measured Ado-Cbl by mass spectrometry. Ado-Cbl levels are much higher in Comamonas aq. DA1877 than E. coli OP50 (Figure 3A), but are reduced to background in the Comamonas aq. cbiA mutant, indicating that this mutant indeed fails to synthesize vitamin B12.
Figure 3. Correlation Between Vitamin B12 Biosynthesis and Repression of the Dietary Sensor.
(A) Ado-Cbl content measured by mass spectrometry in indicated bacterial strains. The red line indicates background levels of Ado-Cbl in the bacteria-free control.
(B) Correlation between vitamin B12 biosynthesis pathway presence and dietary sensor repression. Images of Pacdh-1::GFP animals fed various bacterial strains are located below a cartoon that indicates pathway status. Differences in exposure times for the inset images are indicated in yellow.
See also Figure S2.
We fed additional bacterial species to Pacdh-1::GFP dietary sensor animals and correlated their effect on GFP expression with the presence or absence of a vitamin B12 biosynthesis pathway. Four of these seven bacterial species have the capacity to synthesize vitamin B12 and three do not (Figure 3B). We found that the presence or absence of a vitamin B12 biosynthetic pathway in the bacterial diet correlates perfectly with the repression or activation of the acdh-1 promoter in C. elegans, respectively (Figure 3B).
Requirement of C. elegans Vitamin B12 Processing and Utilization
Vitamin B12 repressed GFP expression equally well whether supplemented to live or UV-killed E. coli OP50 diets, indicating that its effects do not depend on E. coli modification or metabolism (Figure S2A). We wondered why Ado-Cbl and Me-Cbl repress the dietary sensor equally well (Figure 2). From studies of human cobalamin deficiency disorders, it is known that after vitamin B12 is imported into the cell, it is stripped of its upper axial ligand, undergoes several processing steps, is modified into Ado-Cbl and Me-Cbl, and distributed to the two enzymes that use it (Figure S2B). Our supplementation experiments likely cannot discriminate between individual effects of these two forms of vitamin B12 because they can be interconverted. We wondered whether vitamin B12 processing and/or distribution in C. elegans are required for its repressive effect on Pacdh-1::GFP. In our previous C. elegans genetic screens, we did not retrieve any vitamin B12 import and processing genes (Watson et al., 2013), potentially due to inherently high false negative rates in RNA interference (RNAi) screens (Kamath et al., 2003). We predicted C. elegans vitamin B12 processing genes based on homology with known human genes (Figure S2B). RNAi of each of two genes tested, mtrr-1 and Y76A2B.5, resulted in failure of both Me-Cbl and Ado-Cbl to repress the dietary sensor (Figure S2C). This indicates that supplemented vitamin B12 must be properly processed and regenerated into its active forms within C. elegans to repress Pacdh-1::GFP.
We previously found that enzymes in propionyl-CoA breakdown and the methionine/SAM cycle are required for the repressive effect of the Comamonas aq. DA1877 diet on the dietary sensor (Watson et al., 2013). Thus, one would predict that these enzymes are also required for the repressive effects of vitamin B12 supplementation. We used two deletion mutants in each of these pathways to test whether they are also involved in mediating the response to vitamin B12. Indeed supplemention of vitamin B12 to either propionyl-CoA breakdown or methionine/SAM cycle mutants failed to repress acdh-1 promoter activity. (Figure S2D). Taken together, vitamin B12 processing/distributing genes and intact vitamin B12-dependent metabolic pathways are required for the effect of this cofactor on C. elegans.
Propionic Acid Can Override Vitamin B12 to Activate the Sensor
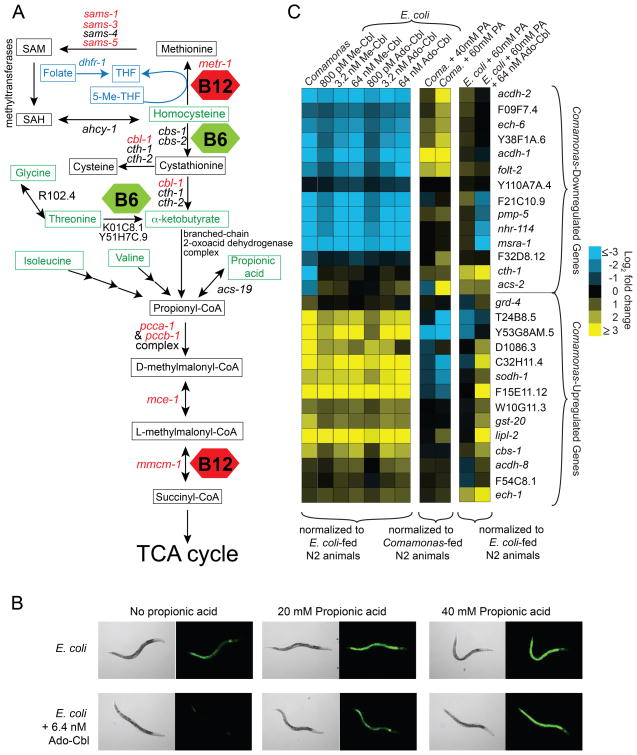
Vitamin B12 represses the dietary sensor, while vitamin B6, homocysteine, glycine, threonine, isoleucine, valine, α-ketobutyrate and propionic acid activate the acdh-1 promoter (Figure 2A). Interestingly, the breakdown of each of these amino acids as well as α-ketobutyrate and propionic acid involves conversion to propionyl-CoA (Figure 4A). Vitamin B6 functions as a cofactor in two reactions that lead to the production of propionyl-CoA (Figure 4A). Since vitamin B12 increases flux through the propionyl-CoA breakdown pathway, we hypothesized that the balance between vitamin B12 and propionyl-CoA levels may be the driving force controlling acdh-1 promoter activity. To test this hypothesis, we performed a chemical epistasis experiment and found that excess propionic acid can override the repressive effects of vitamin B12 on the dietary sensor in a dose-dependent manner (Figure 4B). This suggests that the effects of vitamin B12 on the acdh-1 promoter may depend on its capacity to repress the buildup of an activator, propionic acid.
Figure 4. Chemical Epistasis with Vitamin B12 and Propionic Acid on C. elegans Gene Expression.
(A) Network of the two vitamin B12-dependent pathways. Metabolites are indicated in rectangles. Green metabolites activate the dietary sensor when supplemented to the bacterial diet. Genes encoding the metabolic enzymes involved are indicated with arrows between metabolites. Genes that, when mutated, activate the dietary sensor are indicated in red. Red hexagons – vitamin B12; green hexagons – vitamin B6; blue indicates folate metabolism. 5-Me-THF = 5-methyltetrahydrofolate; THF = tetrahydrofolate, the biologically active form of folate.
(B) Chemical epistasis of combined supplementation of Ado-Cbl and propionic acid on Pacdh-1::GFP dietary sensor animals fed the E. coli OP50 diet.
(C) qRT-PCR of 14 Comamonas-upregulated and 14 Comamonas-downregulated genes in wild type animals fed indicated diets and supplemented metabolites. PA = propionic acid. N2 = wild type animals. Changes in expression for each gene are plotted as log2 fold change compared to respective mRNA levels in wild type animals fed E. coli OP50 (first and third columns) or Comamonas aq. DA1877 (middle column).
See also Table S4.
Vitamin B12 Mimics Broad Comamonas aq. DA1877 Mediated Gene Expression Changes
The Comamonas aq. DA1877 diet affects the expression of many C. elegans genes, including acdh-1 (MacNeil et al., 2013). To assess whether vitamin B12 can elicit similar broad effects on gene expression, we used quantitative RT-PCR (qRT-PCR) to determine relative expression levels of 14 representative Comamonas-downregulated genes and 14 representative Comamonas-upregulated genes in wild type animals fed E. coli OP50 with or without three doses of supplemented Me-Cbl or Ado-Cbl. All but one of the genes tested changed in expression on the Comamonas aq. DA1877 diet as described previously. Most of the Comamonas-downregulated genes were also downregulated when animals were fed E. coli OP50 supplemented with either form of vitamin B12 (Figure 4C). Likewise, most Comamonas-upregulated genes were also activated by supplementation of vitamin B12 (Figure 4C). Thus, vitamin B12 supplementation to the E. coli OP50 diet induces similar gene expression changes as those elicited by the Comamonas aq. DA1877 diet.
Six vitamin B12-downregulated genes, including acdh-1, were upregulated in response to propionic acid (Figure 4C). Further, propionic acid could override the repressive effect of vitamin B12 on these genes. Interestingly, a subset of the vitamin B12-downregulated genes did not respond to propionic acid. A similar trend was observed among the vitamin B12-activated genes; while some responded to propionic acid, others did not. Therefore, we determined that there are at least two classes of vitamin-B12 responsive genes: those that respond to propionic acid (hereafter referred to as type 1) and those that do not (type 2).
Vitamin B12 Accelerates C. elegans Developmental Rate and Egg Laying Timing and Requires the Methionine/SAM Cycle
Since vitamin B12 mimics the effects of the Comamonas aq. DA1877 diet on C. elegans gene expression, we next tested whether it also mimics the accelerated development, reduced fertility and accelerated aging induced by this bacterial diet (MacNeil et al., 2013). Vitamin B12 supplementation affected neither the mean nor the maximum lifespan of C. elegans (Figure S3A). This indicates that another factor must be responsible for the Comamonas aq. DA1877 effect on aging, which is in agreement with the observation that a diet consisting of E. coli OP50 supplemented with a small amount of Comamonas aq. DA1877 does not shorten lifespan (MacNeil et al., 2013).
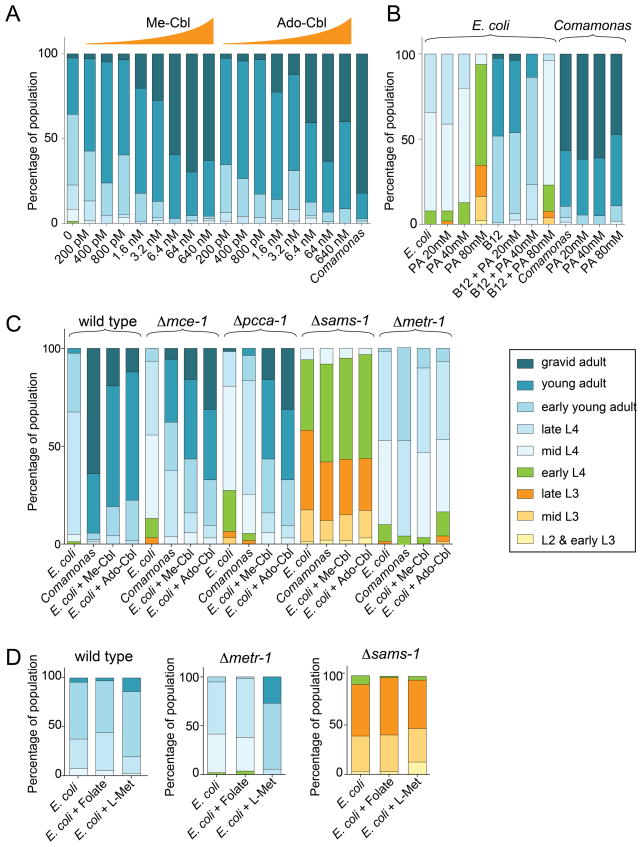
Addition of vitamin B12 to the E. coli OP50 diet did accelerate development, although it did not fully recapitulate the extent of developmental rate increase elicited by the Comamonas diet (Figure 5A). This indicates that additional growth enhancing bacterial factors may be provided by Comamonas. Propionic acid supplementation to the E. coli OP50 diet slowed C. elegans development, and supplementation of both propionic acid and vitamin B12 resulted in intermediate growth rates (Figure 5B). Based on this observation we hypothesized that propionic acid levels may dictate developmental rate: low concentrations caused by high vitamin B12 would accelerate development, whereas high propionic acid levels that occur when vitamin B12 is limiting would slow development. To test this hypothesis, we measured developmental timing in four metabolic gene mutants, two from each vitamin B12-dependent pathway: Δmce-1 and Δpcca-1 from propionyl-CoA metabolism and Δmetr-1 and Δsams-1 from the methionine/SAM cycle. When fed E. coli OP50, all four mutants exhibited slow growth compared to wild type animals (Figure 5C). Surprisingly, supplementation of vitamin B12 accelerated development in Δmce-1 and Δpcca-1 mutant animals, but failed to increase developmental rate in Δmetr-1 and Δsams-1 mutants (Figure 5C). Thus, vitamin B12-induced developmental acceleration requires a functional methionine/SAM cycle, but not the propionyl-CoA breakdown pathway. Δmce-1 and Δpcca-1 mutants fail to catabolize propionyl-CoA (Chandler et al., 2006), likely resulting in a buildup of propionic acid. Neither Δmce-1 nor Δpcca-1 mutants can break down propionic acid even when vitamin B12 is present; yet supplementation of this cofactor does accelerate development in these mutants. These data suggest that the levels of propionic acid are not the driving force behind developmental rate, but rather that development is accelerated via the methionine/SAM cycle, as a result of buildup or lack of specific metabolites therein.
Figure 5. Effects of Metabolite Supplementation on C. elegans Development.
(A) Developmental progression of synchronized wild type populations of animals fed indicated diets and supplemented metabolites. The parental generation was cultivated on E. coli OP50. (B) Developmental progression of synchronized wild type populations of animals fed indicated diets and supplemented metabolites. PA = propionic acid. Vitamin B12 = 64 nM Ado-Cbl. The parental generation was cultivated on E. coli OP50.
(C) Developmental progression of synchronized wild type and mutant populations of animals fed indicated diets and supplemented metabolites. 64 nM of each variant of vitamin B12 was used.
(D) Developmental progression of synchronized wild type and mutant populations of animals fed indicated diets and supplemented metabolites. L-Met = 4 mM methionine. 56 μM folate was used.
See also Figure S3.
To further test this model, we first focused on metr-1 (methionine synthase), which converts homocysteine to methionine by using vitamin B12 as a cofactor. In this reaction, 5-methyl-tetrahydrofolate is also converted to the biologically active form of folate, tetrahydrofolate (Figure 4A). Loss of METR-1 function can be caused either by a mutation in the corresponding gene or as a result of vitamin B12 deficiency. In humans, loss of the corresponding enzyme leads to depletion of tetrahydrofolate and methionine, and buildup of homocysteine. Folate supplementation (which is converted to tetrahydrofolate in vivo) did not rescue the slow growth of Δmetr-1 mutants (Figure 5D). Thus, tetrahydrofolate depletion is not a primary cause of developmental rate reduction. Methionine supplementation, on the other hand, largely rescued the developmental rate defect of Δmetr-1 mutants (Figure 5D). Interestingly, methionine supplementation did not accelerate development of Δsams-1 mutant animals, which are impaired in converting methionine to SAM (Figure 5D)(Walker et al., 2011). This suggests that the developmental acceleration by methionine depends on its conversion to SAM. Taken together, these data suggest that vitamin B12 accelerates C. elegans development primarily through its role as a cofactor in the methionine/SAM cycle, rather than in propionyl-CoA breakdown. However, excess propionic acid slows development, which is likely due to toxic effects.
Supplementing E. coli with vitamin B12 reduced total brood size and therefore mimics the Comamonas aq. DA1877 diet in regard to this phenotype as well (Figure S3B). Both the Comamonas aq. DA1877 diet and vitamin B12 supplementation altered the dynamics of egg laying; animals laid almost all of their eggs (93% and 96% respectively) within the first two days. In contrast, animals fed E. coli OP50 laid only 70% in the first two days (Figure S3C). This vitamin B12-induced shift to early egg laying was still observed in Δpcca-1 mutants, but the effect was lost in Δmetr-1 mutant animals (Figure S3B). Together, these results indicate that vitamin B12 both accelerates development and shifts egg-laying dynamics via its role in the methionine/SAM cycle.
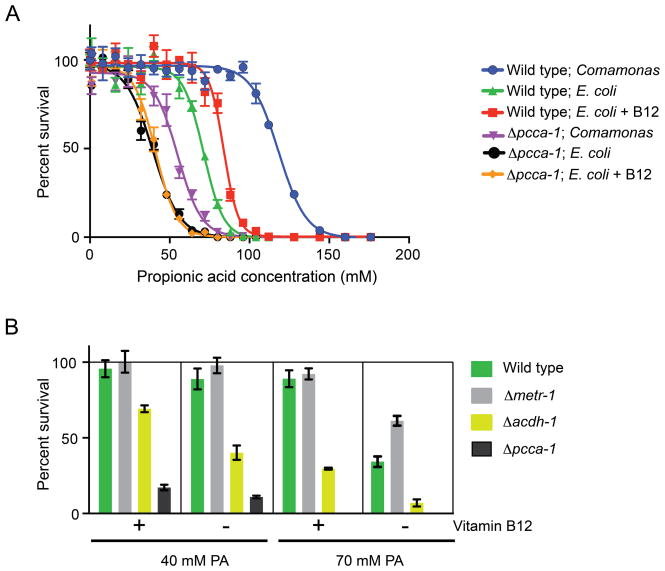
Vitamin B12 Protects Against Propionic Acid Toxicity and Requires the Propionyl-CoA Metabolism Pathway
The developmental acceleration induced by vitamin B12 did not rely on its role in propionyl-CoA breakdown, however vitamin B12 did prevent some of the developmental deceleration induced by high doses of propionic acid (Figure 5B). At such high doses, propionic acid is toxic to the animal (Figures 2A, 6A). We asked whether vitamin B12 supplementation could alleviate the toxic effects of excess dietary propionic acid in wild type and Δpcca-1 mutant animals. Vitamin B12 increased tolerance to propionic acid in wild type animals, but not in Δpcca-1 mutant animals, which are much more sensitive to propionic acid supplementation in general (Figure 6A). This is in agreement with the notion that animals lacking pcca-1 accumulate propionic acid and cannot utilize vitamin B12 to metabolize it. Interestingly, wild type animals fed Comamonas aq. DA1877 are much more resistant to propionic acid than those fed E. coli OP50 supplemented with 64 nM vitamin B12 (Figure 6A). This indicates that Comamonas aq. DA1877 may provide additional compounds that alleviate propionic acid toxicity, or that these bacteria metabolize more propionic acid, thus lowering the effective concentration delivered to the animal.
Figure 6. Effects of Vitamin B12 on Propionic Acid Toxicity.
(A) Toxicity analysis of incremental doses of propionic acid on wild type and Δpcca-1 mutant C. elegans in the absence and presence of 64 nM Ado-Cbl (B12). Mean +/− SEM is plotted from three combined experiments.
(B) Percent survival of different C. elegans strains fed E. coli OP50 at two concentrations of propionic acid (PA) in the absence or presence of 64 nM Ado-Cbl. Mean +/− SEM is plotted from four combined experiments.
We wondered whether loss of metr-1 would also increase sensitivity to propionic acid toxicity. Surprisingly, Δmetr-1 mutants tolerated excess propionic acid slightly better than wild type animals, although this difference was lost when vitamin B12 was in ample supply (Figure 6B). When vitamin B12 is limiting it is conceivable that MMCM-1 and METR-1 compete for this cofactor. As a result, MMCM-1 might be able to utilize more of the vitamin B12 pool for the breakdown of propionic acid when METR-1 is absent.
Transcription of acdh-1 is strongly activated in response to propionic acid supplementation, vitamin B12-deficient diets, or mutations in any of the four enzymes required for vitamin B12-dependent propionic acid breakdown. We wondered what the physiological role of this activation could be, and reasoned that acdh-1 may function in response to propionic acid buildup, perhaps to help dampen its toxic effects. We tested the sensitivity of Δacdh-1 mutants to propionic acid and found that they are much more sensitive than wild type animals (Figure 6B). Unlike Δpcca-1 mutants, however, Δacdh-1 mutants survived better on propionic acid when vitamin B12 was supplemented to the diet. This is likely because, unlike in Δpcca-1 mutants, the vitamin B12-dependent propionyl-CoA breakdown pathway is still intact in Δacdh-1 mutant animals. Taken together, type 1 vitamin B12 response genes that are activated by propionic acid may help to rewire flux through the metabolic network, thereby enabling the breakdown, excretion, or conversion of this metabolite, or alleviating its toxic effects indirectly.
DISCUSSION
Using an interspecies systems biology approach with C. elegans and two of its bacterial diets, we uncovered metabolites that affect gene expression and life history traits. We identified vitamin B12 as the major dilutable compound produced by Comamomas aq. DA1877 that regulates gene expression, accelerates development and affects fertility (MacNeil et al., 2013; this study). Further, we determined that the balance between vitamin B12 and propionyl-CoA is likely the main driving force in the regulation of type 1 vitamin B12-response genes. This finding may reconcile a puzzling observation, namely that both vitamin B12 supplementation and starvation repress the dietary sensor (MacNeil et al., 2013). How can supplementation of a compound and the absence of a compound have the same effect? In rats, starvation lowers levels of short chain fatty acids, including propionic acid, as animals catabolize their fat stores (Illman et al., 1986). In worms starvation and vitamin B12 supplementation may have the same repressive effect on the dietary sensor by reducing the levels of propionyl-CoA.
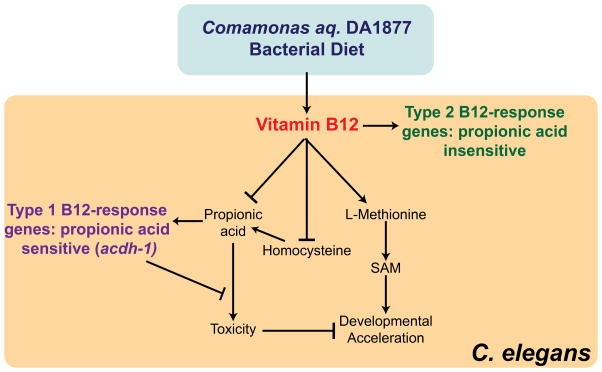
We identified different physiological functions for the two vitamin B12-dependent pathways in C. elegans: the methionine/SAM cycle affects development and fertility, whereas the propionyl-CoA breakdown pathway mitigates propionic acid-induced toxicity (Figure 7). The observation that the methionine/SAM cycle affects development is compatible with the notion that this is an anabolic pathway that generates SAM, the major methyl donor used to modify proteins such as histones (Liu et al., 2013), and to synthesize phosphatidylcholine (Walker et al., 2011), which is vital for the production of cell membranes during development.
Figure 7. Model.
Our findings demonstrate that the Comamonas aq. DA1877 diet provides high levels of vitamin B12, whereas the E. coli OP50 diet is low in this cofactor. Vitamin B12 acts through two different pathways: it lowers levels of propionyl-CoA, thereby preventing propionic acid toxicity, and enables the synthesis of methionine and SAM, thereby accelerating development and reducing fertility. Methionine synthesis reduces levels of homocysteine and likely propionyl-CoA levels. Our data suggest that vitamin B12 may affect these processes by regulating the expression of different sets of genes (type 1 and type 2); some that respond oppositely to propionyl-CoA and others that are insensitive to this metabolite.
Vitamin B12 represses acdh-1 expression at the level of transcription, though it is unclear whether it acts directly or indirectly. We previously identified several nuclear hormone receptors that control the response of the acdh-1 promoter to nutritional and metabolic conditions (MacNeil et al., 2013; Watson et al., 2013). Fat-soluble vitamins A and D are direct ligands for nuclear hormone receptors (Chawla et al., 2001). The C. elegans genome encodes 274 nuclear hormone receptors (Reece-Hoyes et al., 2005). It is tempting to speculate that vitamin B12 directly interacts with one or more of these nuclear hormone receptors. However, our data indicate that vitamin B12 can only repress Pacdh-1 when it can efficiently be used to break down propionyl-CoA because metabolic perturbations that block propionyl-CoA breakdown, or supplementation of excess propionic acid to wild type animals can override the repressive effect of vitamin B12 on acdh-1 expression. We speculate that propionic acid may directly interact with one or more nuclear hormone receptors to regulate type 1 vitamin B12-response genes.
C. elegans is likely to encounter many different bacterial food sources in the wild (Felix and Duveau, 2013). Not all bacterial species synthesize vitamin B12 and C. elegans may have evolved an optimal gene expression program to help the animal cope with diets low in vitamin B12-producing bacteria. One consequence of this gene expression program may be metabolic rewiring; the expression levels of several metabolic genes predicted to function in vitamin B12-dependent pathways are sensitive to dietary vitamin B12 content (MacNeil et al., 2013; Watson et al., 2013). Turning various enzymes on or off at the level of gene expression may help to optimize metabolic flux. acdh-1 expression is induced dramatically when vitamin B12 levels are low and propionic acid levels are high. While the exact metabolic functions of the ACDH-1 enzyme are unknown, we suspect that it may serve to re-route flux around propionyl-CoA within the metabolic network when vitamin B12 is limited to prevent toxic buildup of propionic acid in the animal. This notion is supported by our observation that Δacdh-1 mutants are sensitive to propionic acid toxicity.
Our findings suggest that C. elegans development, as it has been studied in the laboratory for decades using E. coli OP50, which is low in vitamin B12, only reflects one natural state. It will be interesting to study other characteristics of the animal such as behavior, mating and movement under ample vitamin B12 conditions.
Vitamin B12 deficiencies in humans can have severe impacts on health, causing anemias and neuropathies, as well as low birth weight, developmental retardation and defects in cognitive abilities in children of vitamin B12-deficient mothers (Pepper and Black, 2011). Such deficiencies can lead to propionic and methylmalonic acidemias, homocystinuria, methionine/SAM deficiency or a combination thereof, and it is not yet fully understood which of these metabolic imbalances lead to physiological and pathological consequences in humans. Each of these different diseases can also be caused by inborn errors in metabolic genes; for instance propionic acidemia is caused by mutations in the human ortholog of pcca-1 (Al-Lahham et al., 2010). Altogether, the interspecies paradigm we present provides a model to further characterize vitamin B12-dependent processes, and may provide a platform for the development of dietary or drug treatments to vitamin B12-related deficiencies.
Vitamin B12 is an essential cofactor in mammals and C. elegans (Bito et al., 2013), but is not used at all by plants, yeast or many insect species. The uneven distribution of vitamin B12 use among eukaryotes is interesting, as is the uneven distribution of vitamin B12 biosynthetic capability among bacterial species. Considering that many species of enteric bacteria can synthesize this essential vitamin suggests that the chemical outsourcing of vitamin B12 production may be an important keystone of a mutually beneficial relationship between gut microbiota and host (Albert et al., 1980; Goodman et al., 2009; LeBlanc et al., 2013). The microbiota greatly affects its host’s metabolism (Tremaroli and Backhed, 2012). For instance, bacteria in the gut break down dietary fiber, which results in the production of short-chain fatty acids such as propionic acid that are absorbed by intestinal epithelia and used to generate energy (Al-Lahham et al., 2010). Altogether, the microbiota facilitate digestion, affect xenobiotic response to drugs and other chemicals (Haiser et al., 2013; Maurice et al., 2013), and are critically important in the immunological defense (Kamada et al., 2013). The fact that both C. elegans and its bacterial diets are genetically tractable provides a powerful system to further unravel the complex interactions between diet, microbiota, metabolism, physiology and pathology, and the molecular mechanisms involved.
EXPERIMENTAL PROCEDURES
C. elegans Strains
N2 (Bristol) was used as the wild type strain and animals were maintained as described at 20°C (Brenner, 1974). sams-1(ok2946), mce-1(ok243), pcca-1(ok2282), metr-1(ok521) and acdh-1(ok1489) strains were provided by the C. elegans Gene Knockout Consortium and back-crossed as described (Watson et al., 2013). We previously described the Pacdh-1::GFP strain (VL749) (MacNeil et al., 2013).
Bacterial Strains
E. coli OP50, Serratia marcescens IGX2 and Comamonas aq. DA1877 were obtained from the CGC. Pseudomonas aeruginosa PA14 (gacA mutant) was a gift from Victor Ambros (UMMS), Corynebacterium glutamicum ATCC13032 and Comamonas testosteroni ATCC11996 were obtained from ATCC. Mycobacterium smegmatis MC2-155 was a gift from Chris Sassetti (UMMS). For the killed bacteria experiment, E. coli OP50 and Comamonas aq. DA1877 cultures were grown to log phase in LB, then harvested and washed, irradiated with UV (254nm, 5 J/cm2), concentrated, and added to NGM plates.
E. coli Deletion Collection Screen
Keio E. coli deletion collection clones (Thermo Scientific) were grown overnight in Luria Broth (LB) containing 50 μg/mL kanamyacin in 96-well deep well dishes. 15 μL of this overnight culture was seeded onto 96-well nematode growth media (NGM) agar plates containing 50 μg/mL kanamyacin. Pacdh-1::GFP gravid adults fed the Keio parent strain were bleached to harvest embryos, which were subsequently washed in M9 media and allowed to hatch overnight to synchronize the population by L1 arrest. Approximately 25 synchronized L1 animals were added to each well of the seeded plates. After incubating 48 hours at 20°C, animals were visually screened for GFP expression. Each plate from the Keio collection was screened twice, thus each E. coli gene was screened four times. Deletion mutants that affected GFP expression in at least two out of four experiments were considered hits.
Comamonas aq. DA1877 Transposon-Based Mutagenesis Screen
Comamonas aq. DA1877 Genome Sequencing, Assembly, and Annotation
Vitamin B12 Biosynthesis Pathway Analysis
To determine the presence or absence of vitamin B12 biosynthesis pathway in bacterial species used in this study, we first gathered EC numbers from KEGG (Kanehisa et al., 2012) and MetaCyc (Caspi et al., 2010) for all reactions comprising this pathway. For Comamamonas aq. DA1877, we cross-referenced these EC numbers against all EC numbers annotated in the draft metabolic network reconstruction from SEED and determined which vitamin B12 biosynthesis enzymes were encoded in the genome. For other bacterial species, we determined the presence or absence of vitamin B12 biosynthesis genes based on the same EC numbers, but used KEGG and Metacyc databases to guide annotation. If an EC number in the vitamin B12 biosynthesis pathway was annotated as present for a particular species in either or both of these databases, we called it present.
Metabolite Screen
Metabolites were purchased from Sigma-Aldrich and dissolved in water to the maximum soluble concentration. Dilutions were made in water as indicated in Figure 2. For each stock solution, pH was adjusted to 6.0 with sodium hydroxide or hydrochloric acid. Stock solutions were added to NGM in various doses just prior to plate pouring.
Mass-Spectrometry
RNAi Experiments
qRT-PCR
Embryos from N2 wild type gravid adults grown on E. coli OP50 were harvested, washed and hatched overnight in M9 to synchronize the population. Synchronized L1s were added to plates containing various concentrations of Me-Cbl or Ado-Cbl (both from Sigma), and/or propionic acid, seeded with E. coli OP50 or Comamonas DA1877. Animals were harvested for each condition when the majority of the population reached young adult stage. Total RNA was isolated using Trizol (Invitrogen), then purified of contaminating DNA by DNAseI treatment followed by cleanup using Qiagen RNeasy columns. cDNA was reverse-transcribed from RNA using oligo(dT)12–18 primer and Mu-MLV reverse transcriptase (NEB). Primer sequences for quantitative RT-PCR (qRT-PCR) were generated using the GETprime database (Gubelmann et al., 2011) and are listed in Table S4. qRT-PCR reactions were performed in triplicate using the Applied Biosystems StepOnePlus Real-Time PCR system and Fast Sybr Green Master Mix (Invitrogen). Relative transcript abundance was determined using the Δ ΔCt method, and normalized to averaged ama-1 and act-1 mRNA levels (Livak and Schmittgen, 2001).
Developmental Rate, Fertility and Lifespan Assays
For all experiments, animals were grown at 20°C. We measured the developmental state of a population by synchronizing animals by L1 arrest in M9 for 20 hr, then allowing animals to develop for 45 hours at 20°C under various dietary conditions. For L-methionine and folate supplementations, solutions (5 mg/mL L-Met in water or 0.1 mg/mL folate in 20% ethanol) were added on top of plain NGM plates and dried prior to bacterial seeding. After 45 hours, ~35–80 animals were mounted on agarose pads and visually categorized on a compound microscope into age groups based on the development of the vulva and germline as described (MacNeil et al., 2013).
Brood sizes were determined by transferring 30 L4 animals onto individual plates containing E. coli OP50, Comamonas aq. DA1877, or E. coli OP50 supplemented with 64 nM Ado-Cbl. Animals were transferred to new individual plates daily and the number of viable offspring on the plates was counted after hatching.
For the lifespan assays, ~125 L4 animals were transferred onto NGM plates containing 0.1 mg/ml FUDR, with or without vitamin B12, seeded with either E. coli OP50 or Comamonas aq. DA187. Animals were checked for body movement or pharyngeal pumping every two days. If no pumping was observed, animals were lightly tapped with a sterile platinum wire, and if animals failed to respond to the touch stimulus they were considered dead.
Propionic Acid Toxicity Assay
Approximately 100 starved L1 animals were placed on NGM media with increasing concentrations of propionic acid (pH 6.0). After 48 hours, live animals were counted. Each dose of propionic acid was analyzed in triplicate.
Supplementary Material
HIGHLIGHTS.
Interspecies systems biology paradigm: from genetics to metabolites
Vitamin B12 is produced by the Comamonas and not by the E. coli diet
Two types of vitamin B12 effects: development/fertility and propionic acid toxicity
Vitamin B12 accelerates development via SAM metabolism
Acknowledgments
We thank members of the Walhout laboratory, Amy Walker, Job Dekker and Chris Sassetti for helpful discussions. We thank Simon Dove and Bryan McGuffie for reagents and advice on transposon screening. This work was supported by the National Institute of Health (grant DK068429 to A.J.M.W.). A.D.R. was supported by NIH pre-doctoral fellowship AG041605. E.W. was supported by an AHA Founders Affiliate Predoctoral Fellowship. A.A.C is supported by the Ontario Early Researcher Award, by the Canadian Foundation for Innovation and the Ontario Leader’s Opportunity Fund, by the Canadian Institutes for Health Research, and by the Natural Sciences and Engineering Research Council of Canada. Some nematode strains used in this work were provided by the CGC, which is funded by the NIH National Center for Research Resources (NCRR).
REFERENCES CITED
- Al-Lahham SH, Peppelenbosch MP, Roelofsen H, Vonk RJ, Venema K. Biological effects of propionic acid in humans; metabolism, potential applications and underlying mechanisms. Biochimica et biophysica acta. 2010;1801:1175–1183. doi: 10.1016/j.bbalip.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Albert MJ, Mathan VI, Baker SJ. Vitamin B12 synthesis by human small intestinal bacteria. Nature. 1980;283:781–782. doi: 10.1038/283781a0. [DOI] [PubMed] [Google Scholar]
- Avery L, Shtonda BB. Food transport in the C. elegans pharynx. J Exp Biol. 2003;206:2441–2457. doi: 10.1242/jeb.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular Systems Biology. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassford PJ, Jr, Bradbeer C, Kadner RJ, Schnaitman CA. Transport of vitamin B12 in tonB mutants of Escherichia coli. J Bacteriol. 1976;128:242–247. doi: 10.1128/jb.128.1.242-247.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender DA. Nutritional biochemistry of the vitamins. Cambridge: Cambridge University Press; 2003. [Google Scholar]
- Bito T, Matsunaga Y, Yabuta Y, Kawano T, Watanabe F. Vitamin B12 deficiency in Caenorhabditis elegans results in loss of fertility, extended life cycle, and reduced lifespan. FEBS Open Bio. 2013;3:112–117. doi: 10.1016/j.fob.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic acids research. 2010;38:D473–479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler RJ, Aswani V, Tsai MS, Falk M, Wehrli N, Stabler S, Allen R, Sedensky M, Kazazian HH, Venditti CP. Propionyl-CoA and adenosylcobalamin metabolism in Caenorhabditis elegans: evidence for a role of methylmalonyl-CoA epimerase in intermediary metabolism. Mol Genet Metab. 2006;89:64–73. doi: 10.1016/j.ymgme.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla A, Repa JJ, Evans RM, Mangelsdorf DJ. Nuclear receptors and lipid physiology: opening the X-files. Science. 2001;294:1866–1870. doi: 10.1126/science.294.5548.1866. [DOI] [PubMed] [Google Scholar]
- Coolon JD, Jones KL, Todd TC, Carr BC, Herman MA. Caenorhabditis elegans genomic response to soil bacteria predicts environment-specific genetic effects on life history traits. PLoS genetics. 2009;5:e1000503. doi: 10.1371/journal.pgen.1000503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Duveau F. Population dynamics and habitat sharing of natural populations of Caenorhabditis elegans and C. briggsae. BMC Biology. 2013;10:59. doi: 10.1186/1741-7007-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, Lozupone CA, Knight R, Gordon JI. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gracida X, Eckmann CR. Fertility and germline stem cell maintenance under different diets requires nhr-114/HNF4 in C. elegans. Current biology: CB. 2013;23:607–613. doi: 10.1016/j.cub.2013.02.034. [DOI] [PubMed] [Google Scholar]
- Gubelmann C, Gattiker A, Massouras A, Hens K, David F, Decouttere F, Rougemont J, Deplancke B. GETPrime: a gene- or transcript-specific primter database for quantitative real-time PCR. Database bar040. 2011 doi: 10.1093/database/bar040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341:295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Midtvedt T, Gordon JI. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annual review of nutrition. 2002;22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- Illman RJ, Topping DL, Trimble RP. Effects of food restriction and starvation-refeeding on volatile fatty acid concentrations in the rat. J Nutr. 1986;116:1694–1700. doi: 10.1093/jn/116.9.1694. [DOI] [PubMed] [Google Scholar]
- Kadner RJ. Vitamin B12 transport in Escherichia coli: energy coupling between membranes. Mol Microbiol. 1990;4:2027–2033. doi: 10.1111/j.1365-2958.1990.tb00562.x. [DOI] [PubMed] [Google Scholar]
- Kamada N, Seo SU, Chen GY, Nunez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic acids research. 2012;40:D109–114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasseva V, Weiszfeiler JG, Lengyel Z. Synthesis of vitamin B12 by various species of mycobacteria. Zentralbl Bakteriol Orig A. 1977;239:514–520. [PubMed] [Google Scholar]
- LeBlanc JG, Milani C, de Giori GS, Sesma F, van Sinderen D, Ventura M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. 2013;24:160–168. doi: 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Lee KM, Go J, Yoon MY, Park Y, Kim SC, Yong DE, Yoon SS. Vitamin B12-mediated restoration of defective anaerobic growth leads to reduced biofilm formation in Pseudomonas aeruginosa. Infect Immun. 2012;80:1639–1649. doi: 10.1128/IAI.06161-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Galka M, Mori E, Liu X, Lin YF, Wei R, Pittock P, Voss C, Dhami G, Li X, et al. A method for systematic mapping of protein lysine methylation identifies functions for HP1beta in DNA damage response. Molecular cell. 2013;50:723–735. doi: 10.1016/j.molcel.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- MacNeil LT, Watson E, Arda HE, Zhu LJ, Walhout AJM. Diet-induced developmental acceleration independent of TOR and insulin in C. elegans. Cell. 2013;153:240–252. doi: 10.1016/j.cell.2013.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper MR, Black MM. B12 in fetal development. Semin Cell Dev Biol. 2011;22:619–623. doi: 10.1016/j.semcdb.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes JS, Deplancke B, Shingles J, Grove CA, Hope IA, Walhout AJM. A compendium of C. elegans regulatory transcription factors: a resource for mapping transcription regulatory networks. Genome Biol. 2005;6:R110. doi: 10.1186/gb-2005-6-13-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes Dev. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaroli V, Backhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, et al. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biology. 2012;10:67. doi: 10.1186/1741-7007-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Jacobs RL, Watts JL, Rottiers V, Jiang K, Finnegan DM, Shioda T, Hansen M, Yang F, Niebergall LJ, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, MacNeil LT, Arda HE, Zhu LJ, Walhout AJM. Integration of metabolic and gene regulatory networks modulates the C. elegans dietary response. Cell. 2013;153:253–266. doi: 10.1016/j.cell.2013.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.