Abstract
This article describes a method to quantify the movements of larval zebrafish in multi-well plates, using the open-source MATLAB® applications LSRtrack and LSRanalyze. The protocol comprises four stages: generation of high-quality, flatly-illuminated video recordings with exposure settings that facilitate object recognition; analysis of the resulting recordings using tools provided in LSRtrack to optimize tracking accuracy and motion detection; analysis of tracking data using LSRanalyze or custom MATLAB® scripts; implementation of validation controls. The method is reliable, automated and flexible, requires less than one hour of hands-on work for completion once optimized, and shows excellent signal:noise characteristics. The resulting data can be analyzed to determine: positional preference; displacement, velocity and acceleration; duration and frequency of movement events and rest periods. This approach is widely applicable to analyze spontaneous or stimulus-evoked zebrafish larval neurobehavioral phenotypes resulting from a broad array of genetic and environmental manipulations, in a multi-well plate format suitable for high-throughput applications.
Keywords: Zebrafish, Motor function, Neurobehavioral phenotyping, Video tracking, Neurological disease model, Software, Screening assay, Thigmotaxis, Infrared videography, LSRtrack
INTRODUCTION
Evaluation of zebrafish larval movement is a key component of the analysis of phenotypes resulting from genetic mutations1, gene knockdown approaches2, chemogenetic lesions3, drugs4 and toxins5 that target nervous system development, structure or function. Abnormal motor responses can provide evidence regarding the functional importance of biochemical or cellular abnormalities. The demonstration of motor phenotypes can support the relevance of zebrafish neurological disease models to their corresponding human disorders. Furthermore, high-throughput assays of motor function have been proposed as a means to screen chemical libraries for compounds that have neuropharmacological actions4,6, or for novel chemical structures that rescue pathological phenotypes7,8. Reliable and well-validated methods to evaluate larval motor function are central to these applications.
Motor function can be analyzed in multiple zebrafish simultaneously by acquiring video images showing larvae swimming in the wells of multi-well plates, and then analyzing the resulting video files using software tools that detect and quantify larval motion. Three basic approaches of increasing complexity (and computational intensity) have been reported for automated analysis of zebrafish movement from video recordings. Pixel quantification determines whether, in each region of interest (corresponding to the wells), the number of pixels whose value changes at each frame transition exceeds a threshold. This simple and robust form of motion detection enables analysis of motor activity patterns over time9. Larval tracking identifies, locates and assigns positional coordinates to a zebrafish in each region of interest, in each video frame. These data enable evaluation of positional preference and calculation of displacement, velocity and other kinetic parameters by determining how larval centroid coordinates change at frame transitions8. Kinematic analysis identifies anatomical landmarks on the zebrafish and then determines how their spatial relationships change in successive video frames. This allows measurement of trunk curvature, tail beating frequency and other details of individual movements, and has been used to distinguish turning and propulsive movements10. Here we describe a method to carry out pixel quantification and larval tracking.
Development of the protocol
LSRtrack is an open-source MATLAB® function that we developed for automated neurobehavioral phenotyping of zebrafish neurological disease models11. The software was designed to provide a reliable tracking algorithm, although the most recent version also includes a pixel quantification function. The tracking algorithm of LSRtrack has been validated extensively, both manually, by verifying its performance over many hours of video recording, and also by employing controls such as tricaine-immobilized animals to determine signal:noise characteristics over many millions of frame transitions11. The software includes automated error-reporting tools, which ensure that the user can reject wells from analysis if pre-determined criteria are not met for tracking accuracy. The interface of LSRtrack provides tools for live adjustment of tracking and pixel quantification thresholds, with a video stream showing real-time performance facilitating straightforward empiric optimization.
After initial development and validation of this method, we used LSRtrack to analyze motor responses of larval zebrafish following exposure to the dopaminergic neurotoxin MPP+ and the dopamine receptor drugs haloperidol, chlorpromazine, ropinirole and apomorphine8. We have also employed LSRtrack to investigate the role of endogenous zebrafish tor1, the homologue of the human DYT1 dystonia gene, during early motor development12. The current protocol, which was developed in the context of feedback from other research groups implementing LSRtrack in their work, has been extensively tested and should enable experimenters to implement this method with minimal difficulty.
Applications of the protocol
Video tracking using LSRtrack can be used to analyze a broad array of phenotypes resulting from diverse biological manipulations. The examples below demonstrate measurement of spontaneous motor activity8 and responses to changes in ambient illumination10,13. However, responses to other environmental cues or sensory stimuli could be analyzed using this method, provided the stimulus provokes a change in propulsive movement or positional preference and the response can be elicited in multiwell plates. The video tracking algorithm of LSRtrack generates data matrices that include information about zebrafish location and displacement at frame transitions. These data can be further analyzed in numerous ways to yield information about individual movement events, cumulative motor performance or patterns of motor activity. The tools provided in LSRanalyze automatically calculate many commonly measured parameters. However, additional custom algorithms are straightforward to design and execute using MATLAB®, so that a wide range of indices can be analyzed. Applications of this approach include characterization of neurobehavioral phenotypes resulting from gene mutations, transgene expression, morpholino gene targeting, chemogenetic lesions, toxin exposure and pharmacological manipulations, with assays encompassing both spontaneous and evoked behaviors. The multiwell format for which the method was optimized is also amenable to discovery-driven approaches, such as chemical modifier or genetic screens.
Comparison with other methods
Several proprietary software packages and hardware/software solutions are available for measuring zebrafish larval movement in multiwell plates, using similar centroid tracking and pixel quantification approaches as LSRtrack. The principal advantages of using commercial packages, which include the popular Zebralab® (Viewpoint Life Sciences, Montreal, QC) and Ethovision® (Noldus Information Technology, Leesburg, VA) applications, are: (i) the software is designed to work with standardized hardware that is provided by the same vendor, so that initial set up is straightforward; (ii) a live video stream can be analyzed in real time, so it is unnecessary to make a video recording and then analyze it offline, potentially increasing experimental throughput; and (iii) these applications are compatible with a broad range of plate formats, since well alignment is carried out manually.
LSRtrack presents other possible advantages. First, LSRtrack is freely available, can run on any computer (Windows, Mac OS or Linux) running MATLAB®, and the source code is published and can be modified or adapted for new applications. Second, experimental data are written directly into MATLAB® matrices, facilitating subsequent analysis using powerful MATLAB® tools for manipulating large datasets, or custom scripts using the high-level MATLAB® programming language (LSRanalyze, an open-source MATLAB® application that was designed to analyze data archives written by LSRtrack, is an example)11. Third, provided that specific standards are met for the video recording, LSRtrack can be used to analyze zebrafish larval movement regardless of the experimental hardware. This flexibility means that data can be acquired using a simple camcorder or more complex equipment (for example a high-speed camera to capture details of individual movements, or an infrared camera with an IR illumination source to enable visible illumination to be controlled independently). Finally, LSRtrack has inbuilt tools for automated well detection and alignment, tracking optimization and error reporting. These tools are powerful and easy to use, ensuring that reliable data can be generated under a wide range of experimental conditions.
Other published software applications enable researchers to perform kinematic analysis of high frame-rate recordings10,14, or high spatial resolution analysis of position and orientation in low frame-rate recordings15. It would be necessary to employ one of these methods instead of LSRtrack to detect abnormalities of trunk curvature angle, trunk bending frequency, or to detect a response characterized by changes in larval orientation. A recent review article provides a detailed overview of the theoretical basis for videographic analysis of zebrafish motor function and the currently available algorithms16.
In summary, for characterization of motor activity patterns and measurement of displacement, velocity and duration of movement and rest events, LSRtrack might be considered in situations where validation, hardware and platform flexibility, access to source code, and cost are key considerations, whereas proprietary packages offer comparatively rapid setup, increased plate format flexibility, and advantages in throughput, especially for assays with a prolonged time course. To measure details of individual movements such as trunk curvature, or to determine larval orientation, a different program should be selected according to the details of the proposed assay10,14,15,17.
Experimental design
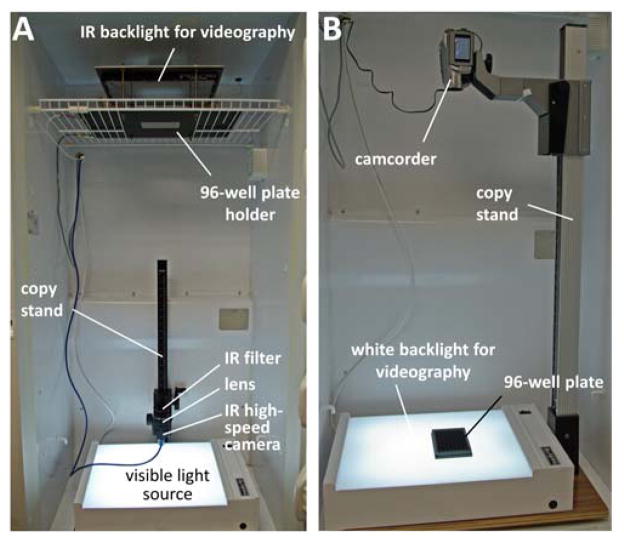
The experiment is carried out in four phases: (i) video capture; (ii) video analysis; (iii) data analysis; (iv) validation (figure 1). Video capture is the most critical stage. The objective is to generate a video recording showing zebrafish larvae swimming in the trans-illuminated wells of a multi-well plate. The video recording must meet the criteria shown in Box 1 in order to be analyzed using LSRtrack. This can be accomplished in practice using a variety of experimental arrangements, for example by positioning the camera below (inverted configuration; figure 2A) or above (upright configuration; figure 2B) the plate, and by employing infrared (figure 2A) or visible (figure 2B) back-illumination. Video analysis is carried out offline using LSRtrack and the resulting data are written into a series of MATLAB® matrices that can be further analyzed to compare motor function of different experimental groups or responses to different stimuli. A second MATLAB® function, LSRanalyze, automates the most common types of analysis and draws data figures.
Figure 1. Experimental design.
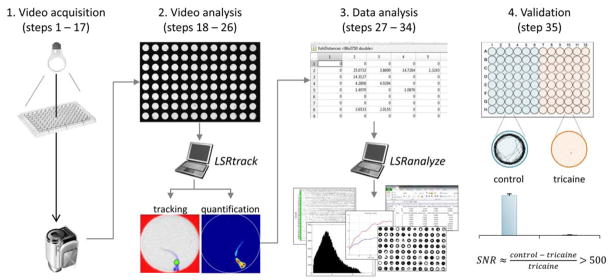
The four stages of the protocol: 1. Backlit video recordings are acquired of zebrafish swimming in the wells of multi-well plates; 2. The resulting recordings are analyzed using LSRtrack to locate zebrafish in each well, in every video frame; 3. The data matrices generated by LSRtrack are analyzed using MATLAB® functions, such as LSRanalyze; 4. The method is validated by measuring the motor activity of control and anesthetized zebrafish.
Box 1. Properties of video files that can be analyzed using LSRtrack.
The video file is a format that can be read by MATLAB® (a list of supported formats is available on http://www.mathworks.com/help/matlab/)
Dark zebrafish are shown on a light background
The wells are the only light areas in the image; the spaces between the wells and around the edge of the plate are dark
The wells are round and the number of wells is in the set {3 x 2n}, where n is a positive integer (this includes most common formats such as 6, 12, 24, 48 and 96-well plates)
Illumination is flat (the pixel value of a light area in the center of the image is close to the pixel value of a similar area at the edge of the image)
The camera is sufficiently far from the plate, or the wells sufficiently shallow, that the walls of the well are not visible
Figure 2. Equipment.
The photographs illustrate two different configurations for obtaining video recordings suitable for analysis. A: Inverted infrared (IR) videography. The camera is positioned below the plate, which is trans-illuminated using an IR light source. A visible light source at the bottom of the incubator enables ambient illumination to be controlled separately. B: Upright videography. A simple camcorder is positioned above the plate, which rests on a flat, white light source. Approval to conduct these experiments was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Details of experimental design vary according to the neurobehavioral response being studied and the aim of the experiment. However, some general points apply to many types of assays implementing this protocol. First, after setting up new equipment or changing any aspect of the apparatus or data acquisition, it is advisable to run an experiment comparing control zebrafish with tricaine-immobilized larvae, to establish that larval movement is being measured and not pixel noise or another artifact (see Validation below). Second, since spontaneous behavior and responses to stimuli can vary quantitatively between experimental sessions, it is desirable to include all experimental groups and controls in a single plate, and to run replicate experiments on different days to ensure that differences between groups are reproducible. Inclusion of appropriate controls can be accomplished by using 96-well plates, which can accommodate multiple groups of larvae, each of adequate size to ensure sufficient statistical power to detect differences between groups. For example, a transgenic line can be tested by including transgenic larvae, non-transgenic siblings from the same clutch and age-matched wild-type larvae; an assay to test neurobehavioral effects of a drug or toxin can include no-compound controls, different concentrations of test compound, and control compounds with known actions. Finally, in many cases the length of an assay is determined by the type of response being elicited; for example, acoustic startle responses can be measured in less than a second17, whereas evaluation of circadian variations in motor activity requires prolonged recordings over days9. To evaluate spontaneous motor function in white light, an hour of stable recording (see below) is sufficient to obtain reliable measurements of basic kinetic parameters such as distance, velocity and movement duration.
Limitations
LSRtrack was developed and optimized for high-throughput assays; its principal limitation is the requirement to use multiwell plates with a restricted set of specifications (Box 1). Only multiwell plates satisfying these specifications are compatible with the automated plate recognition and well alignment functions of LSRtrack. These functions make it unnecessary to draw regions of interest around each well, which can be a laborious task for 96-well plates. Automated positional recalibration also ensures that tracking accuracy is maintained during prolonged recordings. In addition, the plate format and well size is used by LSRanalyze to calculate a pixel:mm scaling factor, which permits calculation of distances in mm during data analysis steps of the protocol, regardless of image magnification. However, if the plate restrictions are problematic for a particular application, the source code of LSRtrack is provided and could be modified to accommodate a different format. Alternatively, a different software application could be employed (see Comparison with other methods above).
LSRtrack can only analyze a single animal within a region of interest; this limitation is shared with most other tracking algorithms. The detection of more than one larval object within a well generates a “too many objects” error for an individual frame (the cumulative error report at the end of the assay is used as a quality control measure to ensure that a single larval object has been tracked throughout the assay in each well). However, the activity of multiple larvae within each well can be measured using pixel quantification instead of tracking2, since the quantification approach does not rely on identification of discrete larval objects. In this case, the centroid tracking function could potentially be removed from LSRtrack to make the program run faster.
Finally, LSRtrack only reports displacement of larvae in the plane of the plate, and is insensitive to the depth at which the larvae are swimming in the wells. Relatively little has been published regarding this aspect of larval zebrafish behavior and the importance of this limitation is unclear at present.
MATERIALS
Reagents
E3 zebrafish embryo buffer: 5 mM NaCl (#S7653, Sigma, St. Louis, MO), 0.17 mM KCl (#P9541, Sigma), 0.33 mM CaCl2 (#C3881, Sigma), 0.33 mM MgSO4 (#M63, Fisher, Waltham, MA). 1 x working solution is made by diluting a 10 x stock in ddH20 and adjusting the pH to 7.4 using NaOH.
Tricaine (MS-222; Sigma #A5040). 0.04% solution is made by diluting 4% stock in 1 x E3 buffer.
-
Zebrafish larvae.
CAUTION: Experiments using zebrafish should be carried out in accordance with relevant guidelines and regulations. In the USA, this necessitates compliance with the NIH Guide for the Care and Use of Laboratory Animals, and approval by an Institutional Animal Care and Use Committee that all proposed procedures meet the standards for humane animal care specified by the US Animal Welfare Act.
Platinum Grade 0 brine shrimp eggs (F-ARGE-PTL, Argent Laboratories, Redmond, WA, USA)
Equipment
Essential
Video camera and lens. We use a Flea3 camera (#FL3-U3-13Y3M-C, Point Grey Research, Richmond, BC, Canada) and Fujifilm 50mm lens (#HF50SA1, B&H, New York, NY, USA); however, the method is compatible with a wide range of simple and more complex equipment. For measurement of displacement, or pixel quantification, equipment capable of a minimum resolution of 600 x 800 pixels at 2–8 frames/second (fps) is adequate for 96-well plates, although higher resolutions improve signal:noise ratio. For measurement of peak velocity or acceleration, a higher frame rate is necessary. High frame rates generally require better pixel resolution to enable movements to be adequately distinguished from pixel noise8. At the magnification used to record a 96-well plate, 1280 x 1024 pixels @ 60 frames/s is adequate for basic measurements of velocity and duration of individual movement events.
Camera support. We use a copy stand (Kaiser RS2XA, #205411, B&H, New York, NY, USA), although a small tripod or custom bracket work equally effectively.
-
Illumination source.
CRITICAL: illumination must be completely flat across the video frame for LSRtrack to work effectively. We use a high-quality infrared backlight (#BL812-880, Spectrum Illumination, Montague, MI, USA) supported on a custom-made holder to allow inverted videography (see figure 2A and Supplementary Methods). However, inexpensive equipment, such as a medical X-ray illumination box, works effectively, as long as illumination is even.
-
Multiwell plates. We use plates with optical glass bottoms (Costar #3720, Corning, Corning, NY, USA).
CRITICAL: the plates must have round wells separated by opaque/black areas, and the number of wells must be in the set {3x2n} where n is a positive integer (see section on ‘Limitations’ and Box 1). Plates with clear regions separating the wells can be used if a mask is applied to the bottom of the plate to ensure that only the wells appear light. This can be accomplished by printing a full size plate map with shaded inter-well areas on transparency film and applying it to the underside of plate. Alternatively, the video can be processed to apply a mask digitally, prior to tracking.
Stereozoom microscope (#S6E, Leica Microsystems, Wetzlar, Germany)
Pasteur pipettes (flame polished aperture >4 mm)
Eyelash (#113, Ted Pella, Redding, CA, USA)
Computer able to run MATLAB® and acquire digital video recordings
MATLAB® (MathWorks, Natick, Massachusetts, USA)
LSRtrack11. The source code and installation files are provided in supplementary files; the latest version can be obtained from the authors.
Software to capture digital video. We use FlyCapture 2 (Point Grey Research, Richmond, BC, Canada). MATLAB® is compatible with a broad range of video file formats; we have found that MPEG-4 with H.264 encoding works well with LSRtrack. If video captured from the chosen hardware is not compatible with MATLAB®, a number of free software tools are available that allow conversion to a compatible file format. Examples include Handbrake (http://handbrake.fr/) and Avidemux (http://avidemux.sourceforge.net/).
Optional
Infrared filter (Hoya #R72, B&H, New York, NY, USA)
Incubator to house experiment (#3721, Thermo Fisher, Waltham, Massachusetts, USA)
Ambient light source. Figure 2A shows a light box modified to use LEDs (#LSM-CW3X3, SuperBright LEDs, Earth City, MO, USA) for illumination (details of the modification are provided in Supplementary Methods)
USB relay board (EtherTek Circuits, Princeton, B.C., Canada)
Light meter (#PLMT56, Pyle Audio, Brooklyn, NY, USA)
Software to run USB relay for separate control of ambient illumination (EtherTek Circuits, Princeton, B.C., Canada)
Spreadsheet application (e.g. Microsoft Excel®, Corel Quattro Pro®, OpenOffice Calc, LibreOffice)
LSRanalyze11. The source code and installation files are provided in Supplementary Information; the latest version can be obtained from the authors.
Reagent setup
Zebrafish
After fertilization, raise embryos and larvae in E3 buffer at pH 7.4, in 100 x 20mm petri dishes with a maximum housing density of 30 larvae per dish in 25 ml of medium. Change media daily. House dishes in an incubator at 28.5 °C in a 14:10 hour light:dark cycle (light cycle begins at 8:00 am; the luminance is calibrated to 200 Lux and color temperature is 5500 K). Feed zebrafish older than 5 days post-fertilization (dpf) with Platinum Grade 0 brine shrimp eggs twice daily.
Larval age strongly influences responses and variability8. For tracking assays, it is recommended that larvae older than 14 dpf are recorded in 48-well or 24-well plates, depending on the types of responses that are being elicited.
CAUTION: Experiments using zebrafish should be carried out in accordance with relevant guidelines and regulations. In the USA this necessitates compliance with the NIH Guide for the Care and Use of Laboratory Animals, and approval by an Institutional Animal Care and Use Committee that all proposed procedures meet the standards for humane animal care specified by the US Animal Welfare Act.
Equipment setup
Camera and illumination
The quality of video images can be improved by housing the camera beneath the 96-well plate, which is trans-illuminated from above; the images are then acquired through the optical glass bottom of the plate rather than through the buffer meniscus. The data shown below were acquired using this configuration with an infrared backlight (figure 2A). However, in some situations it may be easier to house the camera above the plate with the light source below (figure 2B). In this upright configuration, care must be taken to ensure that the light source does not cause significant heating of the plate during the assay.
Location
The experiment should be shielded from extraneous stimuli, such as moving visual stimuli, changes in ambient illumination, noise, and mechanical stimuli, since these will evoke responses in the zebrafish larvae that add to variability and may mask differences between experimental groups or responses to experimental stimuli. We house the apparatus inside a low-temperature incubator kept at 28.5 °C to ensure that temperature is stable throughout the experiment and to allow manipulation of ambient illumination within the incubator. We also carry out experiments in a quiet room with restricted access, to prevent exposure of the zebrafish to unintended stimuli.
Optional control of ambient illumination
In the examples shown below, a light box containing white light LEDs (color temperature 5500K) was positioned at the bottom of the incubator and adjusted so that the 96-well plate was illuminated at an intensity of 200 Lux. The light box was controlled by a USB relay to allow independent control of illumination during the experiment.
PROCEDURE
Loading zebrafish larvae into a multi-well plate
Timing 20–30 min
-
1
Rinse the larvae in the petri dish with by changing the E3 buffer (pH 7.4) 2–3 times. Ensure buffer in the petri dish is free of debris after rinsing.
CRITICAL STEP: Avoid debris in the wells as this can interfere with tracking.
-
2
Put three drops of E3 buffer pH 7.4 into each well of a 96-well plate using a 10-ml serological pipette. Use an eyelash to remove bubbles from the walls and base of each well, using a dissecting microscope to visualize.
CRITICAL STEP: Avoid bubbles as they can interfere with tracking.
-
3
Transfer one zebrafish larva into each well, using a Pasteur pipette with a flame-polished aperture, taking great care not to damage the larva. Transfer as little buffer as possible from the petri dish to the 96-well plate. It is generally a good idea to determine in advance how the experimental groups will be positioned within the plate. LSRtrack numbers wells in the video image starting top left and progressing down each column of wells in turn, left to right; remember that the positions of the wells are inverted if the video is recorded from below.
CRITICAL STEP: Make an accurate record of which larvae are in which wells. For example, the range of well numbers housing each experimental group (e.g. transgene, mutant, control, [drug concentration], etc.) should be noted.
Troubleshooting
-
4
After loading the zebrafish, fill each well with fresh E3 buffer pH 7.4, such that there is a slightly convex meniscus at the top of the well.
-
5
Place the plate in the 28.5 °C incubator.
Setting up the recording
Timing 10–15 min
-
6
Turn on the illumination source and camera; connect the camera to the computer; allow the illumination source to reach stable intensity if appropriate.
-
7
Launch FlyCapture2 and open ‘camera settings’. Alternatively, exposure settings and focus can be adjusted using the equivalent settings dialogue in another video capture application, or camcorder controls that allow manual adjustment of exposure, frame rate and focus.
-
8
Set the desired frame rate. Simple recordings of motor activity patterns require a frame rate of 2–8 frames/s. Higher frame rates are required for measurement of velocity or acceleration, but the resulting video files are larger and take more time to analyze. Higher frame rate recordings also contain more frame transitions that contribute to stochastic pixel noise.
-
9
Adjust the plate and camera position so that the camera is directly below the center of the plate, the optical axis is perpendicular to the plate, and the video frame is filled by the wells. Ensure that all the wells fit completely within the frame. The wells should be the only light areas visible within the frame.
-
10
Focus the camera carefully on the larvae at maximum aperture (minimum f number).
Troubleshooting
-
11
Adjust buffer volume if any of the wells show meniscus shadow artifacts during framing and focusing in steps 9–10.
-
12
Check the exposure settings. In FlyCapture2, exposure adjustments are made with the assistance of the ‘histogram’ tool. Recommended pixel values for different parts of the image are as follows: black areas around the wells <30; white areas within the wells 220–250; darkest parts of the zebrafish 60–100. Use the lens aperture and camera shutter speed controls to ensure that these values are within the desired ranges. If an application other than FlyCapture is used for video capture, pixel values can be checked using ImageJ or equivalent image processing software off line.
CRITICAL STEP: Correct exposure settings greatly enhance the performance of LSRtrack.
CRITICAL STEP: Flat illumination is essential for LSRtrack to perform well. All parts of each well (which appear white) should have similar pixel values, in both the center and edges of the video frame.
Troubleshooting
-
13
(OPTIONAL) Since evaporation can alter the volume of buffer, potentially generating meniscus artifacts that interfere with tracking, we recommend housing the plate in a moist chamber when acquiring prolonged recordings. Significant evaporation could also reduce the temperature of the buffer which would, in turn, alter behavior.
-
14
Allow the larvae to habituate to the experimental conditions. If the recording is carried out under visible light illumination, a 30-min habituation period is usually adequate. If independent control of visible illumination is available through use of infrared videography, cycling light and dark conditions for 30 min [light on (5 min), light off (5 min), light on (5 min), light off (5 min), light on (10 min)] usually results in more stable results.
CRITICAL STEP: Failure to allow larvae to habituate to the experimental conditions increases the variability of responses and may make detection of small differences between experimental groups impossible.
-
15
Choose an output file location and name, and video format, in the video capture application.
Data management is a significant consideration; we employ a file naming convention that includes the initials of the person carrying out the experiment, reference to a lab book and page, and the date. Video compression can be used to minimize file size without significantly affecting tracking results. We have found that H.264/AVC compression works well with LSRtrack. It is important to note that some compression algorithms are incompatible with the pixel quantification function in LSRtrack and generate significant artifacts.
-
16
Start video recording
-
17
(OPTIONAL) Start the script running the USB relay to control the visible lights for the experiment.
Analyzing the video recording
Timing 10–15 min
-
18
(OPTIONAL) If necessary, transfer the video file to the computer that will be used for analysis using LSRtrack, for example if video capture and analysis are carried out using computers that run different operating systems.
-
19
Open MATLAB®, navigate to the LSRtrack folder in the MATLAB® browser window, and enter the command:
≫ LSRtrack <enter>
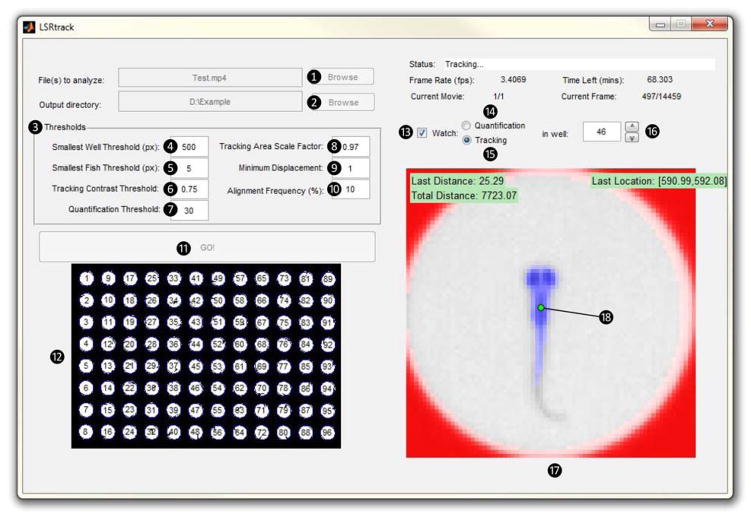
This launches the LSRtrack user interface (figure 3; components of the interface are numbered ①– ⑱ to correspond with the text below).
-
20
Click the ‘browse’ button ① next to the ‘file(s) to analyze’ prompt and navigate to the video file from the experiment that will be analyzed. Click the ‘Browse’ button ② next to the ‘output directory’ prompt and select the directory to which the MATLAB® data matrices will be written.
-
21
Set starting values for the seven parameters in the ‘thresholds’ panel ③ to optimize tracking accuracy (see Box 2). For recordings meeting the exposure and illumination criteria described in steps 6–12 and Box 1, the default values are usually adequate. Optimal settings enhance performance and signal:noise characteristics; these settings can be determined empirically as described in step 23 below.
CRITICAL STEP: Adjustments to these parameters do not compensate for defects in the illumination, focus or framing of video recordings in steps 9–12.
-
22
Press ‘GO!’ ⑪. The program starts by detecting the wells in the first frame of the video and then displays an image of the plate in the panel below the ‘GO!’ button ⑫. The wells are assigned numbers that correspond to the data in the output files. Well re-alignment can be manually triggered during the recording by clicking in the ‘tracking area scale factor’ box ⑧ and pressing <enter>. Re-alignment can improve tracking performance if there is movement of the plate or camera early in the experiment, or if initial plate alignment appears sub-optimal in the tracking window.
Troubleshooting
-
23
Check the ‘watch’ box ⑬ and select the ‘tracking’ option ⑮. Live tracking performance is shown in individual wells in the panel below ⑰. The outline of the well is colored red, the zebrafish colored blue and the larval centroid location is shown by a circle ⑱, which is colored green when the zebrafish is moving and red when it is stationary (figure 4A and B). The well that is shown is specified by the number in the box on the right ⑯. Scan through the wells by clicking the up and down buttons, to ensure that the tracking algorithm is performing appropriately.
CRITICAL STEP: It is important to check wells at the center, edges and corners of the plate to ensure that tracking is proceeding correctly. For each well, check that: there is a single zebrafish that is recognized by the software and colored blue; the walls of the well are recognized correctly by the software and colored red; and the centroid locator ⑱ is over the zebrafish (usually just caudal to the eyes) and changes from red to green when the zebrafish moves.
Troubleshooting
-
24
Click the ‘quantification’ button ⑭ to view the performance of pixel quantification live in individual wells. The well outline is colored black, and each pixel within the well is colored according to the amount by which its value decreased (i.e. the amount by which it became darker) from the previous frame, using a grey scale from white (no change) to black (−255). Changes of pixel value that are less than the ‘quantification threshold’ are not shown or included in analysis. Adjust the ‘quantification threshold’ ⑦ to a level where the only pixels that show as grey or black in the tracking window are within the larval outline during larval movement.
-
25
Once the best thresholds have been determined, uncheck the ‘watch’ box ⑬, which will enable the algorithm to run faster, and leave the program to complete tracking. If a large proportion of the recording was spent adjusting thresholds, it is best to re-start tracking at this point using the optimized settings.
-
26
When tracking is complete, LSRtrack writes a .mat archive that holds MATLAB® matrices containing tracking and quantification data, error reports, and other experimental data (see Anticipated results and Table 1) into the destination folder.
Figure 3. LSRtrack user interface.
The user interface of LSRtrack is annotated to show the controls and dynamic outputs employed in the protocol. Key features of the interface are numbered ①– ⑱ for cross-reference with the text in steps 18–26 and troubleshooting. Approval to conduct this experiment was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Box 2. Summary of adjustable thresholds in LSRtrack.
Smallest well threshold (pixels): Sets the minimum area used to determine which lighter objects are wells; any object smaller than the threshold will not be counted as a well during initial plate alignment.
Smallest fish threshold (pixels): The minimum size of a dark object within a well that is analyzed as a zebrafish larva. The value can be increased if small debris in the wells gives rise to ‘too many objects’ errors, or reduced if there are a large number of ‘no object’ errors.
Tracking contrast threshold: This parameter is set between 0 and 1 to specify the black/white threshold used to binarize the video image prior to zebrafish tracking. Decreasing the threshold makes the program analyze only darker pixels as part of the larval profile.
Quantization threshold: Pixels whose grey scale value decreases at a frame transition by more than this amount are included in the pixel quantification calculation.
Tracking area scale factor: The value is set between 0 and 1 to determine the proportion of the well area in which the program analyzes dark objects as zebrafish larvae.
Minimum displacement (pixels): The displacement in larval location that must be exceeded for a movement to be registered at a frame transition. This simple noise filter discards small displacements in larval coordinates that are caused by pixel noise in the video recording
Alignment frequency (%): The % of frames in the recording after which the program runs the automated well detection and alignment function. For example, a value of 5% triggers realignment at 20 evenly spaced intervals over the recording.
Figure 4. Troubleshooting LSRtrack.
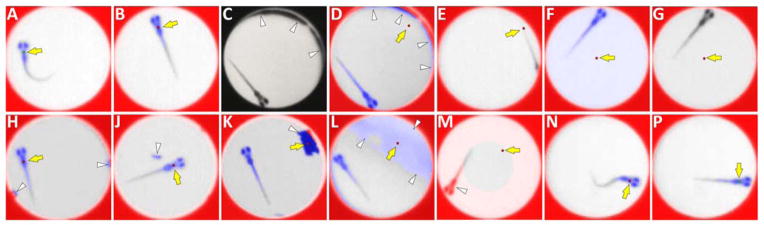
The panels show a range of common tracking errors that are discussed in troubleshooting. In each case, the position of the larval centroid marker is indicated by a yellow arrow; experimental artifacts, if present, are indicated by white arrowheads. A, B: Normal tracking. The well margins are red, the zebrafish is blue and the centroid marker is over the zebrafish. The marker is red when the zebrafish is stationary and green when its position moves. C, D: Well edge artifact shown in the video recording (C) and in the LSRtrack interface (D). This results from misalignment of plate and camera, insufficient plate-to-camera distance, or excessive well depth and is usually most prominent at the edges of the plate. E, F, G: Object assignment errors. E: The zebrafish is hidden by the well wall; F: The ‘tracking contrast threshold’ is too high; G: The ‘tracking contrast threshold’ is too low. H, J, K, L: ‘Too many objects’ errors. H: Bubbles in the water; J: small debris; K: debris sufficiently large to cause misidentification of larval object; L: uneven illumination or mark on bottom of plate. M: The ‘Tracking area scale factor’ is too low causing misidentification of the zebrafish as part of the well margin. N, P: Errors in detection of movement caused by the ‘minimum displacement’ threshold being too high (N) or too low (P). Approval to conduct these experiments was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Table 1.
Output matrices written by LSRtrack.
| Matrix name | Matrix dimensions | Description of data stored within the matrix |
|---|---|---|
| fishAreas | N × 4 | Contains the positional coordinates of the four extreme edges of each well (pixels) |
| fishCoords | N × F x 2 | Contains the positional coordinates of each larva in each frame (pixels) |
| fishDistances | N × F | Contains the distance moved by each larva at each frame transition (pixels) |
| fishQuants | N × F | Contains the sum of pixel values that decreased in each well by more than quantification threshold at each frame transition (Σ[change in pixel grey scale value]) |
| frameRate | 1 | Number of frames/second in video recording; enables subsequent data calculations to be reported in seconds |
| heatMap | W × L | Contains a pixel map of positional preference; each pixel is given a value between 0 and 255, according to the proportion of video frames that the pixel was occupied by a larval centroid |
| noObjectError | 1 × N | Contains the number of ‘no object’ errors in each well over the course of the video |
| noObjectErrorByFrame | N × F | For each well, each frame is scored as 0 (no error) or 1 (‘no object’ error) |
| radius | 1 | Contains the radius of each well in which zebrafish objects are analyzed after application of the ‘tracking area scale factor’ |
| tooManyObjectError | 1 × N | Contains the number of ‘too many objects’ errors in each well over the course of the video |
| tooManyObjectErrorByFrame | N × F | For each well, each frame is scored as 0 (no error) or 1 (‘too many objects’ error) |
| unscaledRadius | 1 | Contains the radius of wells (pixels); used to scale distances to mm in subsequent calculations |
Key: N = number of wells; F = number of video frames; W = width of plate (pixels); L = length of plate (pixels).
Analyzing the data
Timing <5 min to run LSRanalyze; additional time to carry out any further analysis
-
27
LSRanalyze is an open-source MATLAB® application enabling calculation of commonly used summary data from the MATLAB® archive that is output by LSRtrack. To employ LSRanalyze, follow steps 28–33. Additional analyses can be carried out using the MATLAB® programming language and inbuilt tools for manipulation of large data matrices; definitions and details of the data written by LSRtrack are provided in Table 1 to facilitate development of custom algorithms.
-
28
Navigate to the folder containing LSRanalyze using the MATLAB® interface and enter the command:
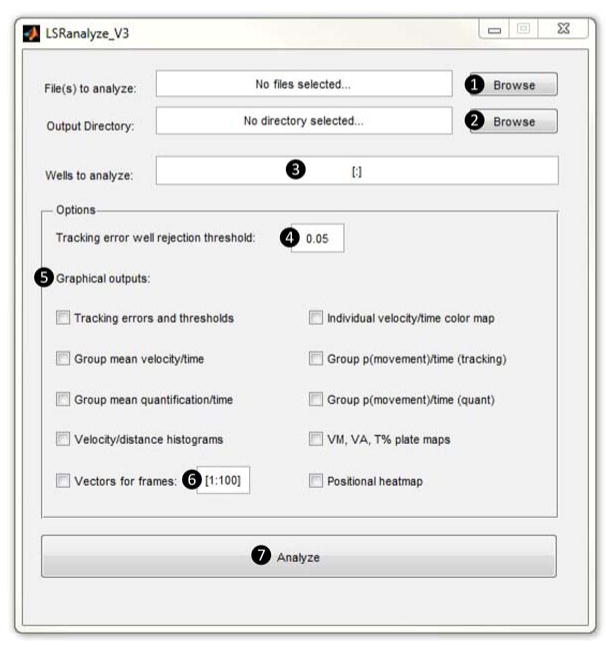
≫ LSRanalyze <enter>
This command opens the LSRanalyze interface (figure 5; components of the interface are numbered ①–⑦ to correspond with the text below).
-
29
Click the ‘browse’ button ① next to the ‘file(s) to analyze’ prompt and navigate to the .mat archive from the experiment that will be analyzed. Click the ‘browse’ button ② next to the ‘output directory’ prompt and select the directory in which the summary spreadsheet will be written.
-
30
Specify which wells will be included in analysis, in the ‘wells to analyze’ box ③. Results can be analyzed by individual larvae or in experimental groups (if groups are selected, the program outputs both group and individual data). The syntax for groups is [a:b],[c:d,e,f],[g,h,i:j], where square brackets specify experimental groups, and lower-case letters specify well numbers. Within groups, single wells are separated by commas, ranges of wells are indicated by colons. For example, [1:22],[23,30,32],[35:39,40,48] analyzes the data as three groups, the first containing wells 1–22, the second containing wells 23, 30 and 32, and the third containing wells 35–39, 40 and 48.
-
31
(OPTIONAL) By default, any well with >5% no object errors is deemed to be empty and excluded from analysis. In addition, the ‘tracking error well rejection threshold’ ④ can be set to exclude wells with total tracking errors (no object errors + too many objects errors) exceeding a user-defined proportion of frames. This option can be employed to adjust quality control criteria for the methodology, depending on application.
-
32
Select graphical outputs ⑤ by checking boxes next to each option. A description of each type of graphical output is shown in Box 3; examples are shown in figures 6, 7 and 8. If a vector plot output is selected, specify the range of video frames for which vectors will be drawn ⑥.
-
33
Press ‘analyze’ ⑦. LSRanalyze calculates summary data for individual larvae and for experimental groups, and generates the figures selected. Group data and tracking accuracy quality control data are shown in the MATLAB® interface.
Troubleshooting
-
34
Individual and group data are written into a spreadsheet for further quantitative analysis. The file contains four worksheets, showing centroid tracking and pixel quantification data, each analyzed by group and by individual larvae. Details of the output indices and their calculation are shown in Table 2.
Figure 5. LSRanalyze user interface.
The user interface of LSRanalyze is annotated to show the dialogue boxes and options employed in the protocol. Features of the interface are numbered ①– ⑦ for cross-reference with the text in steps 27–33.
Box 3. Optional graphical outputs from LSRanalyze.
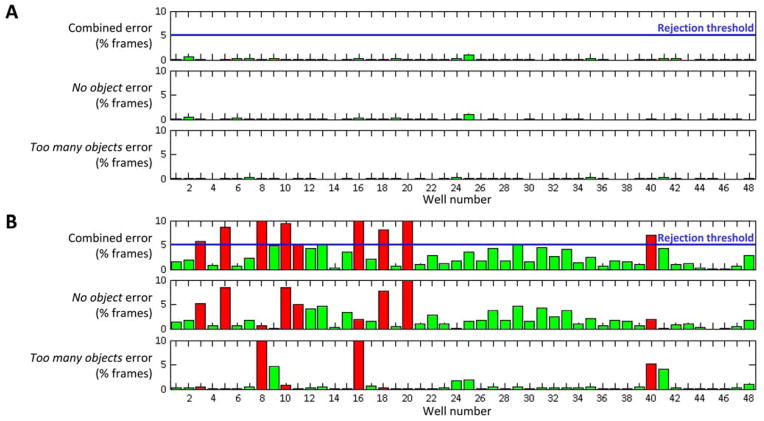
Tracking errors and thresholds: Histograms showing total tracking errors, ‘no object errors’ and ‘too many objects’ errors for each well. Wells discarded from analysis by exceeding ‘tracking error well rejection threshold’ (or empty well threshold) are shown in red (figure 7)
Individual velocity/time color map: Graph showing each individual well (y-axis) against time (x-axis). Each successive frame transition is shaded according to displacement of the larval centroid (figure 6D)
Group mean velocity/time: Mean centroid displacement at each frame transition is shown for each experimental group (figure 8)
Group p(movement)/time (tracking): The proportion of larvae whose centroid coordinates were displaced by an amount exceeding ‘minimum displacement’ is shown at each frame transition for each experimental group
Group mean quantification/time: Mean pixel quantification is shown at each frame transition for each experimental group
Group p(movement)/time (quant): The proportion of wells whose pixel quantification was greater than zero is shown at each frame transition for each experimental group.
Velocity/distance histograms: Histograms show total centroid displacement during recording (y-axis) in each instantaneous velocity bin (x-axis; calculated from the distribution of centroid displacements at each frame transition) for each experimental group (figure 6E)
Plate maps: Plate images; each well is shaded according to mean velocity, active velocity or % frame transitions showing displacement, over entire recording (figure 6C).
Positional heatmap: Plate image; each pixel is shaded according to the proportion of frames a larval centroid occupied the relevant coordinates over entire recording (figure 6B).
Vectors for frames [a:b]: Plate image; the position of each larval centroid is plotted over the range of video frames specified by a and b (figure 6A)
Figure 6. Typical results from protocol.
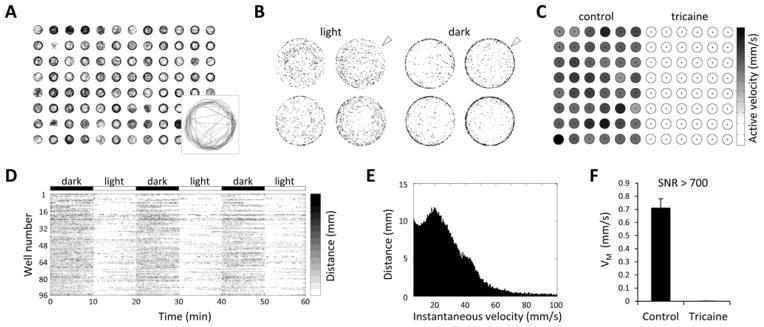
A: Vector plot showing swimming paths of 96 zebrafish larvae over the first 200 frames of a recording. The inset panel shows the vector plot from a single well enlarged for clarity. B: Heat map plot illustrating changes in larval positional preference following light-dark transition. Each point within the four wells (magnified from a 24-well plate) is shaded according to its average occupancy by the larval centroid over the course of the assay. In darkness, larvae tend to move in close proximity to the walls of the well (‘thigmotaxis’; white arrowheads). C: Well summary plot. A map of a 96-well plate is shown to illustrate an experiment in which 48 larvae were anesthetized with tricaine. Each well is shaded according to the grey scale on the right to indicate the mean active velocity of the zebrafish that occupied the well. D: Time-activity plot for 96 individual wells illustrating motor responses to changes in ambient illumination. Each row shows an individual zebrafish; points are shaded according to centroid displacement in each time bin, using the grey scale to the right. E: Instantaneous velocity histogram. Centroid displacements at frame transitions were used to estimate instantaneous velocities in a video segment captured at 60 frames/s. The histogram relates the distribution of instantaneous velocity measurements to the total displacement at each velocity over the course of the assay. Large displacements were registered infrequently and contributed relatively little to total displacement over the course of the assay; in contrast, small displacements were much more frequent and accounted for the majority of movement. F: Mean velocity was calculated for 48 control larvae and 48 tricaine-immobilized larvae over 60 min of spontaneous movement in white light. The signal:noise ratio for the assay , was >700 in this experiment. Values are typically >500 for high resolution, low frame rate video recordings with optimal illumination. Approval to conduct these experiments was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Figure 7. LSRtrack error reports.
Tracking error reports are shown for two different video recordings made and analyzed with (A) optimal or (B) suboptimal settings. For clarity, only the first 48 wells from each 96-well plate experiment are shown. The panels show the % frames in which ‘no object’ or ‘too many objects’ errors were reported for each well, and the combined error. The user-defined threshold for excluding a well from analysis is shown in blue. Wells included in analysis are shaded green; those rejected are shown in red. A: A plate with optimal settings for video capture and analysis shows very low error rates. B: Frequent ‘no object’ (wells 3, 5, 10, 11, 18, 20) and ‘too many objects’ (wells 8, 16, 40) errors led to exclusion of 9/48 wells from analysis of this plate. Approval to conduct these experiments was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Figure 8. Stability of group motor activity.
The graphs show group mean instantaneous velocity (y-axis) against time (x-axis). The mean displacement of 48 zebrafish was plotted for each successive frame transition (grey line). The average of this value over a moving 30-frame window was superimposed (black line) to illustrate the underlying trend. A: Consistent motor activity over an hour of recording; stable recordings facilitate numerical analysis and comparison of experimental groups. B: Insufficient habituation at the start of the assay results in changes to baseline activity as zebrafish acclimatize to experimental conditions. C: Extraneous stimuli such as temperature variations, noise, and visual stimuli cause instability in baseline activity that can obscure responses to experimental stimuli or differences between experimental samples. D: Group mean velocity/time graphs are also helpful for illustrating responses to stimuli, such as motor responses to changes in ambient illumination shown here. Approval to conduct these experiments was obtained from the University of Pittsburgh Institutional Animal Care and Use Committee.
Table 2.
Summary data outputs calculated by LSRanalyze
| Type of data | Name | Definition |
|---|---|---|
| Centroid tracking | Mean velocity | VM = total larval centroid displacement (mm)/duration of recording (s) |
| Active velocity | VA = VM/T% (an approximation of velocity during movement events) | |
| % Time moving | T% = number of frame transitions showing larval centroid displacement/number of frames in recording | |
| Active duration | TA = mean duration of contiguous video frames showing displacement of larval centroid (s) | |
| Rest duration | TR = mean duration of contiguous video frames showing no displacement of larval centroid (s) | |
| Pixel quantification | Mean quantification | QM = total pixel quantification (greyscale units)/duration of recording (s) |
| Active quantification | QA = QM/Q% | |
| % Time moving (quant) | TQ% = number of frame transitions with quantification > 0/number of frames in recording | |
| Active duration (quant) | TQA = mean duration of contiguous frames showing quantification > 0 (s) | |
| Rest duration (quant) | TQR = mean duration of contiguous frames showing quantification = 0 (s) |
Validation
Timing 2–3 hours
CRITICAL: Once the hardware and software components are running smoothly, it is necessary to conduct a validation experiment to ensure that the method measures zebrafish movement, and not apparent displacements in larval centroid location caused by pixel noise or other artifacts in the video recording or analysis. We recommend analyzing the movements of untreated control larvae in comparison with tricaine-immobilized larvae to confirm that the method reports larval movement and to enable calculation of the signal:noise ratio for the experimental conditions.
-
35
Carry out steps 1–34 using 48 control larvae and 48 larvae that are immobilized with tricaine. For tricaine treatment, replace E3 buffer with 0.04% tricaine in E3 in the last rinse of step 1, and leave the larvae in 0.04% tricaine when they are transferred to the 96-well plate. Use the method shown in the Anticipated results section to determine the signal:noise ratio for the experimental configuration and thresholds used. Once hardware components and software settings are set up to ensure reliable tracking and optimal signal:noise ratio, the protocol can be significantly shortened by using the same hardware configuration and exposure settings for each experiment, and changing the default tracking values in the source code. However, we recommend carefully checking framing, focus, pixel greyscale values and tracking accuracy for every experiment and running validation controls periodically.
Troubleshooting
TIMING
Steps 1–5: Loading larvae carefully into a 96-well plate takes 20–30 minutes.
Steps 6–17: Setting up the camera, plate and software takes 10–15 minutes of hands-on time; this is followed by a recording period that does not require supervision and is determined by the length of the assay.
Steps 18–26: The time required by LSRtrack to analyze a video recording depends on the duration of the experiment, the frame rate of the recording, the number of wells, and the specifications of the computer. For 96 wells, analysis rates of 4–8 frames/second are usual with a desktop PC, so that a 2 hour duration video at 2 frames/second takes 30–60 minutes to analyze. Longer recordings or higher frame rate videos can be batch processed unsupervised overnight.
Steps 27–34: Analysis of data matrices takes from < 1 minute to run LSRanalyze, to several hours or longer to design and execute custom MATLAB® scripts.
Step 35: Validation entails running the entire protocol with control and anesthetized larvae; this can generally be completed in 2–3 hours
TROUBLESHOOTING
| Step | Problem | Possible reasons | Solutions |
|---|---|---|---|
| 3 | Larval morphology abnormal | Larval damage during transfer | Replace larva Pipette gently and avoid trapping larva between the pipette and the |
| 10 | Circular shadow apparent in video image during focusing and framing | Buffer meniscus artifact caused by incorrect buffer volume | Adjust buffer volume |
| 10 | Outside walls of wells are visible within well area, problem worse at edges of plate (figure 4C) | ‘Well edge artifact’ is observed when the camera is too close to plate, or the wells are too deep | Move camera further away from the plate and use longer focal length lens |
| 10 | Wall on same side of all wells is visible within well area | Camera is not centered above/below the plate and the optical axis is not perpendicular to plate | Adjust camera/plate position |
| 12 | Light colored pixels in well area have lower value (i.e. are darker) at edge of image than in center | Illumination source is too small or too far from the
plate Optical defect in lens (‘vignetting’) Incorrect lens type for the sensor size |
Use light source with larger area of illumination or
move source closer to plate Try a smaller aperture (larger f number) or replace the lens Use a lens that is designed to project an image larger than the camera sensor |
| 22 | LSRtrack reports an error in well number | Incorrect plate format Light areas other than wells apparent in video image |
See Box 1
for plate format requirements Light areas visible around the edge of the plate can be removed by cropping video Try increasing ‘smallest well threshold’ ④ if unintended light areas are smaller than wells |
| 23 | Part of well boundary appears blue (figure 4D) | Well edge artifact, see above Tracking area scale factor’ too high ‘Tracking contrast threshold’ too high |
See troubleshooting advice for step 10 Try realigning wells by clicking in tracking area scale factor box ⑧ and pressing <enter> Try reducing ‘tracking area scale factor’ ⑧ Try reducing ‘tracking contrast threshold’ ⑥ if well edge artifact is brighter than the zebrafish |
| 23 | Zebrafish not visible in well or only partially visible and centroid locator not over zebrafish (figure 4E) | Empty well Zebrafish is hidden by being close to the well wall; this can typically occur on the opposite side of a well to significant well edge artifact (see troubleshooting advice for step 10) |
Check the experimental record to determine whether the
well was empty or contained a zebrafish ‘Disappearing fish’ artifact can be minimized by increasing distance between camera and plate or using shallower wells Examine error reporting; it may be necessary to discard well from analysis if no object errors are very prominent |
| 23 | Zebrafish visible in well but centroid marker not over zebrafish (figure 4F, G) |
‘Smallest fish
threshold’ too high ‘Tracking contrast threshold’ too high or too low |
Try reducing ‘smallest fish
threshold’ ⑤ Try adjusting ‘tracking contrast threshold’ ⑥ |
| 23 | Multiple objects in well colored blue; centroid marker is over zebrafish (figure 4H, J) | Presence of debris, bubbles or other well artifact | Try increasing ‘smallest fish threshold’ ⑤ or increasing ‘tracking contrast threshold’ ⑥. If smaller non-zebrafish object remains blue, data from well may still be accurate if centroid marker remains over zebrafish throughout the assay. |
| 23 | Multiple objects in well colored blue; centroid marker is not over zebrafish (figure 4K) | Presence of debris or other well artifact | If there is an artifact larger than the zebrafish, the well should be excluded from analysis |
| 23 | Part of well appears blue (figure 4L) | Uneven illumination or mark on underside of plate; ‘tracking contrast factor’ too high | Try reducing the ‘tracking contrast factor’; always use clean plates and ensure illumination is even throughout plate |
| 23 | Part of zebrafish appears red (figure 4M) | The ‘tracking area scale factor’ is too low | Try increasing the ‘tracking area scale factor’ ⑧ |
| 23 | Centroid marker is over the zebrafish; color stays red when zebrafish moves (figure 4N) | The ‘minimum displacement’ threshold is too high | Decrease value for the ‘minimum displacement’ threshold ⑨ |
| 23 | Centroid marker is over zebrafish; color changes from red to green without the zebrafish moving (figure 4P) | Pixel noise artifact causing apparent centroid displacement | Increase value for the ‘minimum displacement’ threshold ⑨ |
| 33 | High frequency of ‘Too many objects’ errors in tracking data | Presence of debris or bubbles (figure 4H, J, K) More than one zebrafish in the well Well edge artifact and tracking area scale factor too high (figure 4D, L) |
Watch well while tracking; artifact may be eliminated by adjusting tracking thresholds depending on type. If centroid marker over larva for duration of assay this error can be ignored, otherwise exclude well from analysis |
| 33 | High frequency of ‘no object’ errors | The well is empty Well edge artifact so larva not visible to camera (figure 4E) The ‘tracking contrast factor’ is too high or too low (figure 4F, G) |
Check experimental record to ensure a zebrafish was
present in the well Well edge artifact may be minimized by moving camera away from plate. Increasing ‘tracking area scale factor’ or reducing ‘minimum fish size’ can also mitigate this problem Adjust the ‘tracking contrast factor’ |
| 33 | Tracking errors become more frequent during recording | Buffer evaporation leading to meniscus artifact
Drift in alignment of plate and camera |
Use a moist chamber, see step 13 Increase the ‘alignment frequency’ ⑩ |
| 33 | Motor activity not stable during recording | Inadvertent extraneous stimuli | House experiment inside incubator in a quiet room |
| 35 | Poor signal:noise performance | Tracking errors The ‘minimum displacement’ threshold is too low Pixel resolution of the image is too low The video frame rate is too high Tricaine animals not immobilized |
Detect and correct tracking errors in step 33
Adjust the ‘minimum displacement’ threshold ⑨ Increase pixel resolution of camera Decrease temporal resolution of camera Use fresh tricaine, or increase its concentration |
ANTICIPATED RESULTS
LSRtrack writes data into a MATLAB® archive comprising a series of matrices (table 1), which contain: tracking data (coordinates in each frame, displacement at each frame transition, and positional preference over the course of the assay); pixel quantification data; error reports; information about scaling and well sizes; and video frame rate. The data in these matrices can be analyzed in a large number of different ways to ask specific questions about movements of the larvae during the experiment. LSRanalyze calculates commonly measured summary data, writes a spreadsheet containing summary indices (table 2), and generates a set of MATLAB® figures to illustrate the data, assist in analysis and provide essential quality controls. Examples of typical data and quality control parameters output by LSRtrack and LSRanalyze are shown in figures 6–8.
One key feature of LSRtrack is that a record is kept of all tracking errors (individual frames in which the algorithm detects 0 or >1 object in a well; figure 7). For positional calculations, LSRtrack assigns the last known coordinates when there is a ‘no object’ error and assigns coordinates to the largest object when there is a ‘too many objects’ error. We previously showed by manual verification that, in most situations, these assignments do not introduce significant errors11. If the quality of videography is good and care was taken to minimize or eliminate well edge artifacts (see troubleshooting), there are limited locations within a well where the zebrafish cannot be tracked; to remain undetected for an appreciable length of time the animal is typically stationary. Conversely, when there is a ‘too many objects’ error, the largest object is most frequently the zebrafish (unless there are two larvae or an unusually large item of debris in the well). Consequently, even wells with a significant number of reported tracking errors can often be used in analysis. However, for most experiments, it is reasonable to expect the tracking error rate to be below 5% in most wells.
Tricaine-immobilized controls can be used to verify that reported displacements are not caused by tracking errors or pixel noise; LSRtrack should ideally report little or no movement for tricaine-immobilized animals (small passive displacements of anesthetized larvae through mechanisms such as drift are removed by the ‘minimal displacement’ threshold). By analyzing movement in 48 control animals and 48 tricaine-immobilized animals, the assay signal:noise ratio (SNR) can be estimated. For example, for mean velocity (VM), . SNR for should be greater than 100 and is frequently above 500 for recordings obtained using this protocol (figure 6F). For pixel quantification data, SNR for % time active (TQ%) estimated using the same approach is often higher than 2000, depending on the threshold chosen for determining how much pixel value change is necessary for inclusion in analysis.
In the absence of external stimuli, the activity of a group of larvae illuminated in white light is usually stable over an hour of recording, but slowly declines over several hours. The stability of motor function over time can be evaluated using the ‘Group mean velocity/time’ option in LSRanalyze, which plots mean larval displacement (calculated from the fishDistances matrix) for each experimental group in successive video frames, thereby providing a population mean instantaneous velocity trace (figure 8A). Stable motor activity during a spontaneous movement assay is desirable, since reduced variability enhances statistical power to detect quantitative differences between experimental groups in parameters such as VM, VA, and T%. The mean instantaneous velocity graph also enables the detection of artifacts caused by inadequate habituation (figure 8B) and unintentional extraneous stimuli (figure 8C) and can be used to identify and evaluate responses to sensory stimuli, such as the typical phases of the behavioral responses to abrupt changes in illumination (figure 8D).
Supplementary Material
Acknowledgments
Development of LSRtrack and the current protocol was supported by research grants from NINDS (NS080881, NS058369), NIEHS (ES022644), the Pittsburgh Foundation (M2005-0071), and the Society for Progressive Supranuclear Palsy (468-08). YZ is a Tsinghua Scholar at the University of Pittsburgh.
Footnotes
AUTHOR CONTRIBUTION STATEMENTS:
YZ, CLC, EAB designed and wrote software; YZ, RC, QB, EAB developed the experimental protocol; YZ, RC, QB carried out experiments; YC, RC, EAB wrote the manuscript.
COMPETING FINANCIAL INTERESTS:
The authors declare no competing financial interests.
Supplementary information:
Supplementary methods: Instructions and drawings are shown for construction of optional equipment used in the protocol: (i) plate holder for inverted infrared videography (shown in figure 2A); and (ii) USB relay-controlled LED light box, employed to elicit responses to changes in ambient illumination (shown in figures 6D and 8D).
Software files: The .zip archive contains current versions of LSRtrack and LSRanalyze. After downloading, navigate MATLAB® to the directory containing the uncompressed files to run the software as indicated in the protocol. The source code files can also be edited to adapt the software for custom applications.
References
- 1.Mahmood F, Fu S, Cooke J, et al. A zebrafish model of CLN2 disease is deficient in tripeptidyl peptidase 1 and displays progressive neurodegeneration accompanied by a reduction in proliferation. Brain. 2013;136(Pt 5):1488–1507. doi: 10.1093/brain/awt043. [DOI] [PubMed] [Google Scholar]
- 2.Milanese C, Sager JJ, Bai Q, et al. Hypokinesia and reduced dopamine levels in zebrafish lacking beta- and gamma1-synucleins. J Biol Chem. 2012;287(5):2971–2983. doi: 10.1074/jbc.M111.308312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elbaz I, Yelin-Bekerman L, Nicenboim J, et al. Genetic ablation of hypocretin neurons alters behavioral state transitions in zebrafish. J Neurosci. 2012;32(37):12961–12972. doi: 10.1523/JNEUROSCI.1284-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rihel J, Prober DA, Arvanites A, et al. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327(5963):348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sallinen V, Torkko V, Sundvik M, et al. MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J Neurochem. 2009;108(3):719–731. doi: 10.1111/j.1471-4159.2008.05793.x. [DOI] [PubMed] [Google Scholar]
- 6.Kokel D, Bryan J, Laggner C, et al. Rapid behavior-based identification of neuroactive small molecules in the zebrafish. Nat Chem Biol. 2010;6(3):231–237. doi: 10.1038/nchembio.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4(1):35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 8.Farrell TC, Cario CL, Milanese C, et al. Evaluation of spontaneous propulsive movement as a screening tool to detect rescue of Parkinsonism phenotypes in zebrafish models. Neurobiol Dis. 2011;44(1):9–18. doi: 10.1016/j.nbd.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prober DA, Rihel J, Onah AA, et al. Hypocretin/orexin overexpression induces an insomnia-like phenotype in zebrafish. J Neurosci. 2006;26(51):13400–13410. doi: 10.1523/JNEUROSCI.4332-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burgess HA, Granato M. Modulation of locomotor activity in larval zebrafish during light adaptation. J Exp Biol. 2007;210(Pt 14):2526–2539. doi: 10.1242/jeb.003939. [DOI] [PubMed] [Google Scholar]
- 11.Cario CL, Farrell TC, Milanese C, et al. Automated measurement of zebrafish larval movement. J Physiol. 2011;589(Pt 15):3703–3708. doi: 10.1113/jphysiol.2011.207308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sager JJ, Torres GE, Burton EA. The zebrafish homologue of the human DYT1 dystonia gene Is widely expressed in CNS neurons but non-essential for early motor system development. PLoS ONE. 2012;7(9):e45175. doi: 10.1371/journal.pone.0045175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richendrfer H, Pelkowski SD, Colwill RM, et al. On the edge: pharmacological evidence for anxiety-related behavior in zebrafish larvae. Behav Brain Res. 2012;228(1):99–106. doi: 10.1016/j.bbr.2011.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontaine E, Lentink D, Kranenbarg S, et al. Automated visual tracking for studying the ontogeny of zebrafish swimming. J Exp Biol. 2008;211(Pt 8):1305–1316. doi: 10.1242/jeb.010272. [DOI] [PubMed] [Google Scholar]
- 15.Creton R. Automated analysis of behavior in zebrafish larvae. Behav Brain Res. 2009;203(1):127–136. doi: 10.1016/j.bbr.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 16.Martineau PR, Mourrain P. Tracking zebrafish larvae in group--status and perspectives. Methods. 2013;62(3):292–303. doi: 10.1016/j.ymeth.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgess HA, Granato M. Sensorimotor gating in larval zebrafish. J Neurosci. 2007;27(18):4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.