Abstract
Methionine adenosyltransferase (MAT) is a family of enzymes that utilizes ATP and methionine to produce S-adenosylmethionine (AdoMet), the most crucial methyl donor in the biological methylation of biomolecules and bioactive natural products. Here, we report that the MAT from Sulfolobus solfataricus (sMAT), an enzyme from a poorly explored class of the MAT family, has the ability to produce a range of differentially alkylated AdoMet analogs in the presence of non-native methionine analogs and ATP. To investigate the molecular basis for AdoMet analog production, we have crystallized the sMAT in the AdoMet bound, S-adenosylethionine (AdoMet) bound, and unbound forms. Notably, among these structures, the AdoEth-bound form offers the first MAT structure containing a non-native product and cumulatively, these structures add new structural insight into the MAT family and allow for detailed active site comparison with its homologs in E. coli and human. As a thermostable MAT structure from archaea, the structures herein also provide as a basis for future engineering to potentially broaden AdoMet analog production as reagents for methyltransferase-catalyzed ‘alkylrandomization’ and/or the study of methylation in the context of biological processes.
Keywords: natural product, X-ray diffraction, S-adenosylmethionine, enzyme engineering, Methionine adenosyltransferase
Introduction
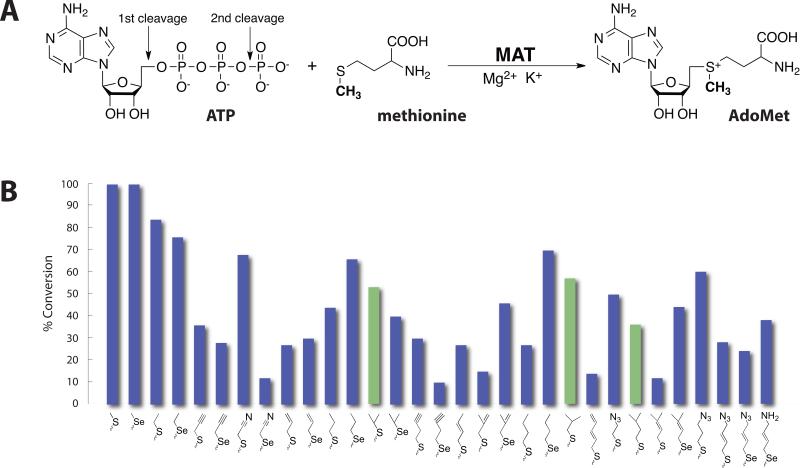
AdoMet is the most widely used methyl donor in biological systems, and the only known family of enzymes that synthesize AdoMet is methionine adenosyltransferase (MAT), which utilizes methionine and ATP as substrates (Fig. 1A) [1]. The reaction occurs in an unusual two-step mechanism, in which the adenosyl group is transferred from ATP to the sulfur atom of methionine cleaving the triphosphate and the triphosphate is subsequently hydrolyzed to PPi and Pi [2]. MAT is present in all living organisms from bacteria to mammals. To date, MAT structures from E. coli [3, 4], Campylobacter jejuni [5], Burkholderia pseudomallei (PDB code 3IML), Entamoeba histolytica (PDB code 3SO4), Mycobacterium marinum (PDB code 3RV2), Mycobacterium avium (PDB code 3S82), Mycobacterium tuberculosis (PDB code 3TDE), Thermococcus kodakarensis [6], Rattus norvegicus [7] and Homo sapiens [8] have been solved. They are more than 50% identical to one other and share several common features: (i) they usually appear as oligomeric proteins with a highly conserved three-domain fold [9]; (ii) divalent cations such as Mg2+ are required for activity and monovalent cations such as K+ are able to enhance the reaction rate [10]; (iii) a flexible loop suspended above the active site, serves as a “gate” and is involved in the catalytic activity of the enzyme [11]. In addition, some members of the MAT family have recently been demonstrated to produce non-native AdoMet analogs from non-native methionine analogs [12-14]. And in the context of coupled reactions containing permissive methyltransferases, those MATs enable the generation of natural product ‘alkylrandomizations’ [15]. Thus, there is renewed interest in assessing the substrate tolerance of additional MATs and extending the molecular level understanding of MAT-substrate interactions.
Fig. 1.
(A) The reaction catalyzed by methionine adenosyltransferase. (B) Turnover of S/Se-Met analogs to the corresponding AdoMet analogs catalyzed by sMAT based upon RP-HPLC (average percent error ≤4%, see Table 2 and Fig. 4). Bars colored green (referring to branched L-alkyl-substituted analogs) denote analogs for which only trace turnover was observed with MATs studied to date [15]. As controls, no product formation was observed in the absence of sMAT, S/Se-Met analogs or ATP.
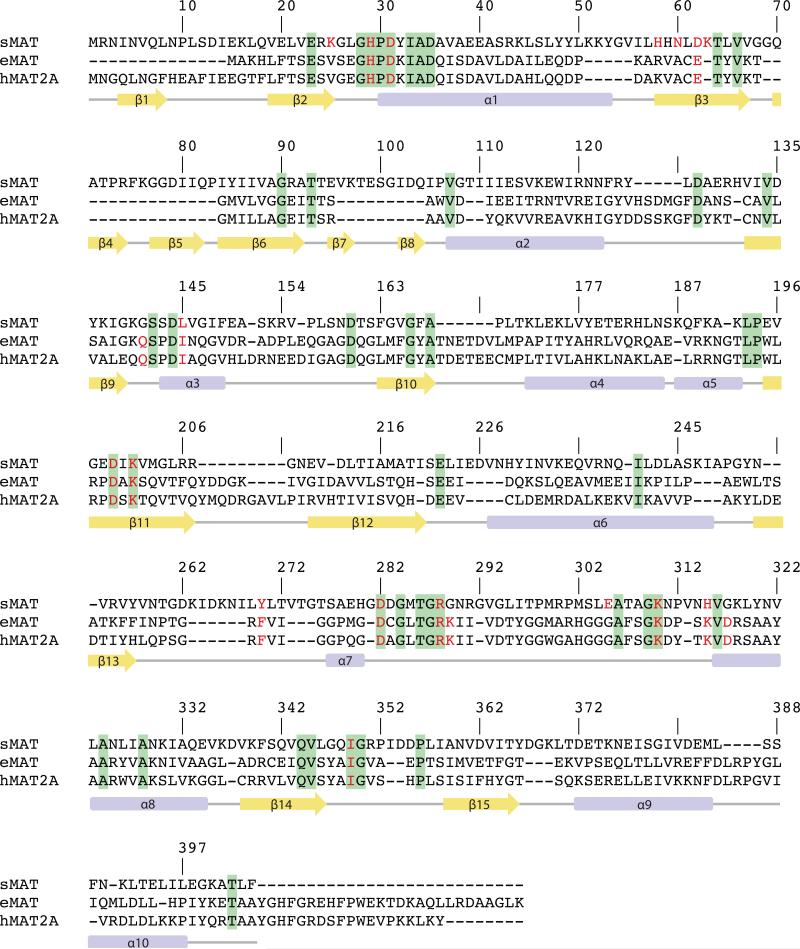
In contrast to well-characterized MATs in bacteria and eukaryotes, studies of a sequence divergent class of MATs in archaea has been limited. The first archaeal MAT was identified from Sulfolobus solfataricus by Porcelli et. al. [16] in 1988, having only ~20% sequence identity with E.coli and human MATs (Fig. 2). Later on, another archaeal MAT from Methanococcus jannaschii (mjMAT) was characterized in detail in terms of kinetics parameters, substrate specificity, and folding [12, 17]. More recently, an apo crystal structure of MAT from Thermococcus kodakarensis (tkMAT) has been reported, which provides new structural insights on archaeal MAT [5]. The improved thermostability but comparable kinetics parameters with MATs in bacteria and eukaryotes show archaeal MATs to be of great interest for enzyme engineering applications. However, structural information of active site contents and broad substrate specificity assessment for archaeal MATs has been lacking. Here we report Sulfolobus solfataricus MAT (sMAT) to enable the cumulative synthesis of a broad panel of unnatural AdoMet analogs (31 analogs detected) starting from synthetic S/Se-alkylated Met analogs (42 analogs) or commercial sources (3 analogs). In addition, this study highlights the crystal structures of a thermostable MAT (sMAT), in three different forms: AdoMet bound, a non-native product S-adenosylethionine (AdoEth) bound, and the unbound form. Interestingly, in contrast to its low sequence similarity to other MATs, sMAT displays the typical three-domain fold and partly conserved active site architecture. Unlike other known MAT structures, the activity of sMAT cannot be stimulated by ionic potassium [16]. This can be supported structurally by the presence of a lysine side chain (K63) in sMAT, which likely has a similar function to potassium ion in other MATs. Further, the capture of the first atypical ligand bound structure of MAT provides insights on the nature of sMAT broad substrate specificity and a potential template for future engineering toward expanding the substrate scope. Cumulatively, the results in this study provide the first atomic view of the poorly explored class of MATs from archaea and expose sMAT as an efficient catalyst for AdoMet analog production that is amendable to downstream AdoMet-utilizing processes.
Fig. 2.
The structure-based sequence alignment of MAT from Sulfolobus solfataricus (sMAT), MAT from E.coli (eMAT) and human MAT (hMAT2A). Secondary structural features of sMAT are shown at the bottom. The numbering of the amino acids in the figure corresponds to sMAT. Identical residues between all three sequences are shown in green; identical residues between two sequences are shown in yellow. The critical residues involved in substrates binding are highlighted in red letters.
Results and Discussion
Overall Structural Organization
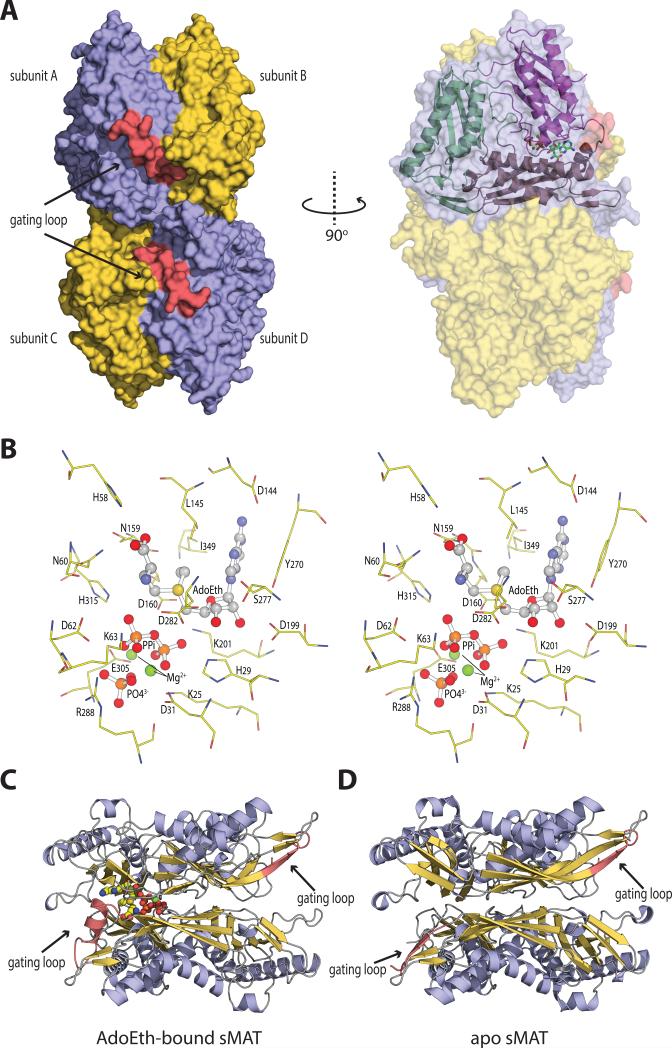
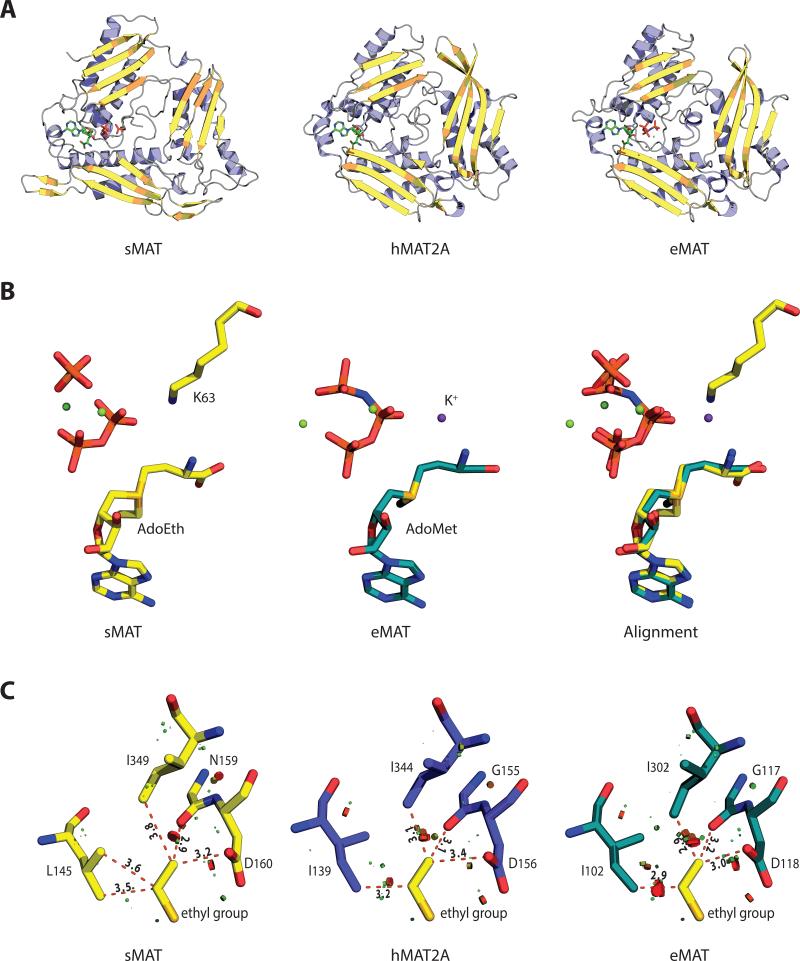
The crystal structures of sMAT have been determined successfully at 2.19 Å or 2.39 Å resolution for the AdoMet-liganded form, 2.49 Å for the AdoEth-liganded form and 2.21 Å for the unliganded form (Table 1). Similar to E.coli MAT (eMAT) [18], Rat liver MAT (rlMAT) [19], human MATs (hMAT1A and hMAT2A) [8], sMAT packs as a tetramer (Fig. 3A). All four sMAT structures contain two subunits, A and B, in the asymmetric unit and the tetramer is formed by a 2-fold crystallographic symmetry axis. The buried surface interface between the two subunits A and B, and the two dimers AB and CD are calculated to be 2570 and 1870 Å2 respectively [20]. These areas are similar to those from other bacterial MATs [3-5]; but larger than rat liver MAT [7] and slightly smaller than human MAT [8]. In the tetramer, there are four potential ligand-binding sites: two sites sandwiched by A and B, and the other two sites between C and D. Compared with other known MAT structures [9], the sMAT monomer adopts a similar 3-domain architecture with secondary structure variants (Fig. 3A). Interestingly, unlike other MATs, structural variations were observed between subunits A and B in sMAT with an average rmsd of 0.51 Å in both the liganded and unliganded structures. In addition, the maximum rmsd between all the A subunits and the maximum rmsd between all the B subunits in all sMAT structures are calculated as 0.29 Å and 0.21 Å, respectively. Similar observation was also reported in a recently solved archaeal MAT structure [6]. As a result, half of the active sites within sMAT have a more open conformation than the others. Consistently, in all the ligand-bound sMAT structures, only half of the active sites within the sMAT tetramer are occupied while the other half are unoccupied. In addition, only the gating loops outside the occupied active site become ordered (Fig. 3C, 3D).
Table 1.
Data Collection and Refinement Statistics for sMAT in the apo form and ligand-bound forms
| sMAT | sMAT+AdoMet | sMAT+AdoMet | sMAT+AdoEth | |
|---|---|---|---|---|
| Added in crystallization | --- | SAM | ADP, Met | ADP, Eth |
| Modeled in structure | PPi | AdoMet, PPi | AdoMet, PPi | AdoEth, PPi |
| Data collection | ||||
| Resolution range (Å) | 50-2.21 | 50-2.19 | 50-2.39 | 50-2.49 |
| Wavelength (Å) | 0.98 | 0.98 | 0.98 | 0.98 |
| Space group | P6522 | P6522 | P6522 | P6522 |
| a, b, c (Å) | 151.3, 151.3, 221.2 | 151.6, 151.6, 226.1 | 150.1, 150.1, 222.6 | 151.6, 151.6, 226.1 |
| No. of molecules per asymmetric unit | 2 | 2 | 2 | 2 |
| Measured reflections | 1,478,624 | 1,541,606 | 1,854,076 | 1,101,136 |
| Unique reflections | 74,368 | 78,995 | 59,745 | 53,034 |
| R merge | 0.118 (0.909) | 0.142 (0.918) | 0.165 (0.606) | 0.184 (0.225) |
| Completeness | 99.61 (96.38) | 99.75 (97.50) | 100.0 (100.0) | 98.31 (82.97) |
| Redundancy | 19.9 (9.2) | 19.5 (14.3) | 26.0 (22.2) | 20.7 (21.0) |
| Mean I/sigma | 14.55 (1.41) | 14.94 (3.08) | 27.01 (5.14) | 11.94 (3.85) |
| Refinement | ||||
| Rcryst/Rfreea | 0.1990/0.2201 | 0.1639/0.1888 | 0.1710/0.1978 | 0.1595/0.1843 |
| No. of protein atoms | 6301 | 6,356 | 6,396 | 6,410 |
| No. of ligand atoms | --- | 48 | 43 | 44 |
| No. of solvent atoms | 304 | 707 | 596 | 589 |
| Average B factor (Å2) | ||||
| Protein | 38.20 | 31.6 | 37.5 | 40.1 |
| Ligands | --- | 28.7 | 30.3 | 38.2 |
| Ligands RSCCb | --- | 0.93 | 0.98 | 0.96 |
| RMSD from ideal | ||||
| Bond length (Å) | 0.002 | 0.007 | 0.006 | 0.007 |
| Bond angles (deg) | 0.69 | 1.33 | 0.70 | 0.97 |
| Ramachandran plot (%) | ||||
| Favored regions | 99 | 98 | 98 | 99 |
| Outliers | 0.25 | 0.12 | 0.12 | 0 |
| PDB ID | 4HPV | 4L7I | 4K0B | 4L2Z |
Values in parentheses are for the highest resolution shell.
Rfree was calculated as Rcryst using 5.0% of randomly selected unique reflections (in thin resolution shells) that were omitted from the structure refinement.
RSCC is the real-space correlation to electron density calculated by Phenix.
Fig. 3. The overall molecular structure and active site contents of sMAT.
(A) Tetramer assembly of sMAT in the crystal structure as calculated by PISA. The surface of the protein is displayed: four protein monomers are shown in light blue or yellow and the gating loops are shown in red. One protein monomer is displayed on the right showing the three intertwined domains as cartoon and the ligands as sticks.
(B) Stereoview of sMAT-ligands interactions. The stick model of AdoEth, PPi, PO43- and Mg2+ is depicted in spheres and the interacting sMAT residues are labeled and illustrated in green.
(C) Side view of sMAT dimer with AdoEth bound. The gating loop region is highlighted in red.
(D) Side view of apo sMAT dimer. The gating loop region is highlighted in red.
The substrate specificity for sMAT based upon RP-HPLC is illustrated in Fig. 1B wherein observed 5’-methyl-thio(seleno)-5’-deoxyadenosine (MSeA) production (via RP-HPLC, Fig. 4) was interpreted as product based upon the well-established AdoMet decay pathways indicating MSeA to directly derive from AdoMet (not ATP). [21-23] The putative substrates tested were those recently reported to profile the substrate specificity of a range of MATs and these analogs were specifically designed to interrogate both steric and electronic contributions to turnover [15]. Of the 45 putative substrates (Table 2) tested with sMAT, 11 led to appreciable (>50%) AdoMet analog production, an additional 15 led to moderate (25-50%) conversion, while 5 offered detectable product (<25%) under the conditions described. In general, smaller alkyl substitutions were better tolerated, suggesting steric infringement to possibly prohibit larger substitutions. Interestingly, in the case where direct comparisons could be made, the degree of unsaturation correlated with a reduction in turnover (e.g., propyl > allyl > propargyl). Importantly, notable turnover was observed with branched analogs (Fig. 1B, highlighted in green) that previously led to only trace product with MATs studied to date.
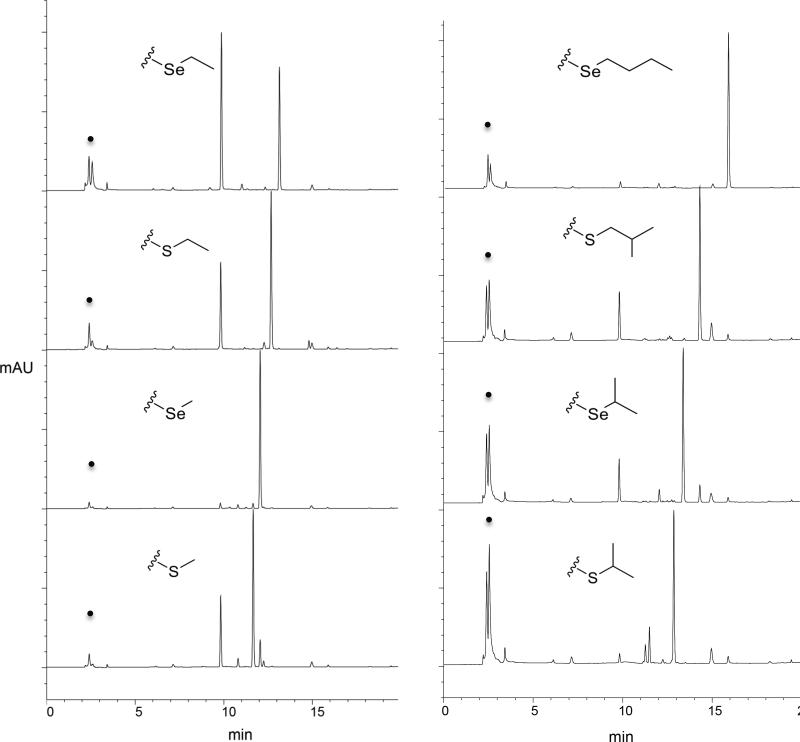
Fig. 4.
HPLC traces for representative sMAT reactions illustrating the production of AdoMet analogs and/or 5’-methyl-thio(seleno)-5’-deoxyadenosine (MSeA) in the presence of a select set of L-Met analogs. Starting material (ATP) is designated by a dot.
Table 2.
Summary of sMAT reactions
| Analog | Analog name | Percentage Turn-Over |
Theoretical mass | Observed mass |
|---|---|---|---|---|
|
|
Methionine | 100 | 399.1445, 297.0896[a] | 399.1444, 298.0965[b] |
|
|
Se-Methionine | 100 | 447.0890, 345.0340[a] | 447.0895, 346.0416[b] |
|
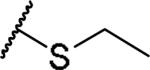
(2S)-2-amino-4-(ethylsulfanyl)butanoic acid | 84 | 413.1602, 311.1052[a] | 413.1599, 312.1121[b] |
|
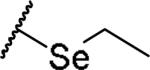
(2S)-2-amino-4-(ethylselanyl)butanoic acid | 76 | 461.1046, 359.0497[a] | 416.1040, 360.0565[b] |
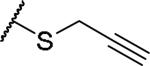
|
(2S)-2-amino-4-(prop-2-yn-1-ylsulfanyl)butanoic acid | 36 | 321.0896[a] | 322.1051[b] |
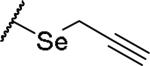
|
(2S)-2-amino-4-(prop-2-yn-1-ylselanyl)butanoic acid | 28 | 369.0340[a] | 370.0411[b] |
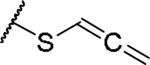
|
(2S)-2-amino-4-(propa-1,2-dien-1-ylsulfanyl)butanoic acid | ND | NA | NA |
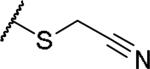
|
(2S)-2-amino-4-[(cyanomethyl)sulfanyl]butanoic acid | 68 | 322.0848[a] | 323.0919[b] |
|
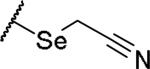
(2S)-2-amino-4-[(cyanomethyl)selanyl]butanoic acid | 12 | 370.0293[a] | 371.0371[b] |
|
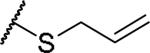
(2S)-2-amino-4-(prop-2-en-1-ylsulfanyl)butanoic acid | 27 | 425.1602, 323.1052[a] | 425.1609, 324.1129[b] |
|
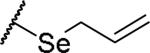
(2S)-2-amino-4-(prop-2-en-1-ylselanyl)butanoic acid | 30 | 371.0497[a] | 372.0568[b] |
|
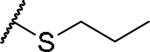
(2S)-2-amino-4-(propylsulfanyl)butanoic acid | 44 | 427.1758, 325.1209[a] | 427.1759, 326.1281[b] |
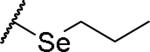
|
(2S)-2-amino-4-(propylselanyl)butanoic acid | 66 | 475.1203, 373.0653[a] | 475.1200, 374.0726[b] |
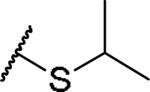
|
(2S)-2-amino-4-(propan-2-ylsulfanyl)butanoic acid | 53 | 427.1758, 325.1209[a] | 427.1762, 326.1282[b] |
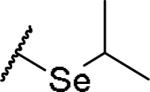
|
(2S)-2-amino-4-(propan-2-ylselanyl)butanoic acid | 40 | 373.0653[a] | 374.0743[b] |
|
|
(2S)-2-amino-4-(but-3-yn-1-ylsulfanyl)butanoic acid | 30 | 437.1602, 335.1052[a] | 437.1599, 336.1124[b] |
|
|
(2S)-2-amino-4-(but-3-yn-1-ylselanyl)butanoic acid | 10 | 383.0497[a] | 384.0566[b] |
|
|
(2S)-2-amino-4-[(2E)-but-2-en-1-ylsulfanyl]butanoic acid | 27 | 439.1758, 337.1209[a] | 439.1754, 338.1275[b] |
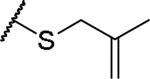
|
(2S)-2-amino-4-[(2-methylprop-2-en-1-yl)sulfanyl]butanoic acid | 15 | 439.1758, 337.1209[a] | 439.1754, 338.1282[b] |
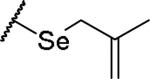
|
(2S)-2-amino-4-[(2-methylprop-2-en-1-yl)selanyl]butanoic acid | 46 | 385.0653[a] | 386.0730[b] |
|
|
(2S)-2-amino-4-(butylsulfanyl)butanoic acid | 27 | 441.1915, 339.1365[a] | 441.1966, 340.1477[b] |
|
|
(2S)-2-amino-4-(butylselanyl)butanoic acid | 70 | 489.1359, 387.0810[a] | 489.1358, 388.0882[b] |
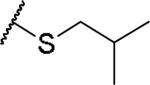
|
(2S)-2-amino-4-[(2-methylpropyl)sulfanyl]butanoic acid | 57 | 441.1915, 339.1365[a] | 441.1918, 340.1439[b] |
|
|
(2S)-2-amino-4-[(3-aminopropyl)sulfanyl]butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-[(3-aminopropyl)selanyl]butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-[(2E)-penta-2,4-dien-1-ylsulfanyl]butanoic acid | 14 | 451.1758 | 451.1756 |
|
|
(2S)-2-amino-4-[(2-azidoethyl)sulfanyl]butanoic acid | 50 | 454.1616, 352.1066[a] | 454.1615, 353.1140[b] |
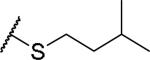
|
(2S)-2-amino-4-[(3-methylbutyl)sulfanyl]butanoic acid | 36 | 455.2071, 353.1522[a] | 455.2123, 354.1634[b] |
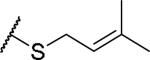
|
(2S)-2-amino-4-[(3-methylbut-2-en-1-yl)sulfanyl]butanoic acid | 12 | 453.1915 | 453.2044 |
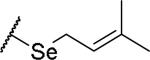
|
(2S)-2-amino-4-[(3-methylbut-2-en-1-yl)selanyl]butanoic acid | 44 | 399.0810[a] | 400.0878[b] |
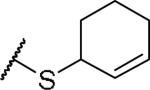
|
(2S)-2-amino-4-(cyclohex-2-en-1-ylsulfanyl)butanoic acid | ND | NA | NA |
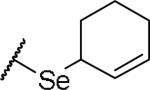
|
(2S)-2-amino-4-(cyclohex-2-en-1-ylselanyl)butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-[(3-azidopropyl)sulfanyl]butanoic acid | 60 | 468.1772, 366.1223[a] | 468.1772, 367.1296[b] |
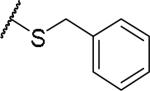
|
(2S)-2-amino-4-(benzylsulfanyl)butanoic acid | ND | NA | NA |
|
(2S)-2-amino-4-(benzylselanyl)butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-{[(2E)-4-azidobut-2-en-1-yl]sulfanyl}butanoic acid | 28 | 480.1772, 378.1223[a] | 480.1768, 379.1292[b] |
|
|
(2S)-2-amino-4-{[(2E)-4-azidobut-2-en-1-yl]selanyl}butanoic acid | 24 | 426.0667[a] | 427.0687[b] |
|
|
(2S)-2-amino-4-{[(2E)-4-aminobut-2-en-1-yl]selanyl}butanoic acid | 38 | 400.0762[a] | 401.0830[b] |
|
(2S)-2-amino-4-{[(4-methylphenyl)methyl]sulfanyl}butanoic acid | ND | NA | NA |
|
(2S)-2-amino-4-{[(2E)-3-phenylprop-2-en-1-yl]sulfanyl}butanoic acid | ND | NA | NA |
|
(2S)-2-amino-4-{[(4-nitrophenyl)methyl]sulfanyl}butanoic acid | ND | NA | NA |
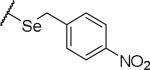
|
(2S)-2-amino-4-{[(4-nitrophenyl)methyl]selanyl}butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-[(2-{[(3S)-3-amino-3-carboxypropyl]sulfanyl}ethyl)sulfanyl]butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-[(3-{[(3S)-3-amino-3-carboxypropyl]sulfanyl}propyl)sulfanyl]butanoic acid | ND | NA | NA |
|
|
(2S)-2-amino-4-{[(2E)-4-{[(3S)-3-amino-3-carboxypropyl]sulfanyl}but-2-en-1-yl]sulfanyl}butanoic acid | ND | NA | NA |
theoretical mass of degraded 5’-alkyl-thio(seleno)-5’-deoxyadenosine (MSeA); observed mass of degraded 5’-alkyl thio(seleno)-5’-deoxyadenosine (MSeA)
[M+H]
NDnot detected
NAnot applicable.
Table 3 highlights a comparison of the kinetic parameters for L-methionine and the non-native substrate for which a ligand-bound structure is available (L-ethionine). The changes in kinetic parameters of the sMAT for both the substrates are moderate from 37 °C to 65 °C. Compared with Thermococcus kodakarensis MAT and Methanococcus jannaschii MAT, sMAT appears to be a somewhat better enzyme because sMAT has a 100 times smaller Km for methionine, a slightly smaller Km for ATP, and a similar kcat. More importantly, the data reflect that L-ethionine is kinetically competent and comparable to the native substrate L-methionine. At either temperature, the kcat values for the sMAT reaction with L-methionine or L-ethionine are similar and the reduced proficiency with L-ethionine when compared to the native substrate L-methionine derive from a combination of higher Km values for both L-ethionine and ATP.
Table 3.
Kinetic parameters of sMAT enzyme.
| Temperature (°C) | Constant Substrate | Varied Substrate | kcat (min−1) | Km (mM) | kcat / Km (mM−1min−1) |
|---|---|---|---|---|---|
| 37 | ATP | L-methionine | 2.06 ± 0.03 | 0.0023 ± 0.0003 | 895 ± 100 |
| ATP | L-ethionine | 2.34 ± 0.07 | 0.0056 ± 0.0015 | 417 ± 47 | |
| L-methionine | ATP | 1.97 ± 0.08 | 0.0686 ± 0.0110 | 29 ± 7 | |
| L-ethionine | ATP | 2.48 ± 0.09 | 0.0969 ± 0.0130 | 26 ± 7 | |
| 65 | ATP | L-methionine | 2.92 ± 0.07 | 0.0028 ± 0.0007 | 1042 ± 100 |
| ATP | L-ethionine | 2.99 ± 0.11 | 0.0070 ± 0.0016 | 427 ± 69 | |
| L-methionine | ATP | 2.806 ± 0.08 | 0.0145 ± 0.0025 | 194 ± 32 | |
| L-ethionine | ATP | 3.10 ± 0.14 | 0.0592 ± 0.0108 | 52 ± 13 | |
Active Site Contents
The MAT catalyzed AdoMet formation, as mentioned in the introduction, occurs via a sequential two-step mechanism. In the first step, AdoMet is formed by a direct SN2 reaction, in which the sulfur atom of methionine attacks the C5’ position of ATP and thus cleaves the polyphosphate chain from ATP. In the second step, the triphosphate is further hydrolyzed to diphosphate (PPi) and phosphate (Pi) [9]. Komoto et al. [3] identified two critical residues, lysine 165 and histidine 14, in eMAT for this proposed SN2 reaction based on their ligand-bound structures. Interestingly, even with significant sequential variations to eMAT and other MATs, several conserved residues were observed in sMAT, mainly located around the active site, including the two crucial residues, lysine (K201) and histidine (H29) for the proposed SN2 reaction (Table 4).
Table 4.
Ligand-based Alignment of Active Site Residues between sMAT and eMAT
| Interaction Partnera | sMAT | eMAT | DALI Alignment | Chain |
|---|---|---|---|---|
| O1, O2 in PPi, O2 in PO4 | K25 | K245 | not detected | B |
| O3 in PPi | H29 | H14 | detected | B |
| Mg2+ | D31 | D16 | detected | B |
| O7, O8 in AdoEth | H58 | Q98 | not detected | A |
| O7, O8 in AdoEth | N60 | K269 | not detected | A |
| K63 (K+ in eMAT) | D62 | E42 | detected | A |
| O5, O7 in PPi | K63 | K+ | - | A |
| ethyl group in AdoEth | L145 | I102 | detected | A |
| O26 in AdoEth | D199 | D163 | detected | B |
| O3 in PPi | K201 | K165 | detected | B |
| stacked with adenine ring | Y270 | F230 | detected | B |
| O27 in AdoEth | D282 | D238 | detected | B |
| O1, O3 in PO4 | R288 | R244 | detected | B |
| Mg2+ | E305 | D271 | not detected | A |
| O6 in PPi, O1 in PO4 | K310 | K265 | detected | A |
| O5 in PPi | H315 | - | - | A |
| ethyl group in AdoEth | I349 | I302 | detected | A |
The atom numbers used for interaction analysis here are the numbers from the AdoEth-bound structure (PDB code 4L2Z).
The interaction between sMAT and products in the active site are multifaceted as illustrated in Fig. 3B. The adenine ring of AdoMet is recognized by a hydrogen bond with the side chain of aspartate 144 and a stacking interaction with the aromatic ring of tyrosine 270. Several water molecules surrounding by the adenine ring also form a hydrogen bond network to the enzyme. The 2’-OH and 3’-OH of the adenosine ribose interact with the side chains of aspartate 199, aspartate 282 and serine 277. Similar interactions involving aspartic acids have been seen in other MAT structures bound with AdoMet, but not for serine [3, 8]. The methionine/ethionine moiety (of AdoMet and AdoEth, respectively) forms hydrogen bonds with four residues, in which the amino group interacts with the side chain of aspartate 282, and the carboxylate group interacts with the side chain of histidine 58, asparagine 60 and asparagine 159. The methyl or ethyl group is buried in a slightly hydrophobic pocket surrounded by asparagine 159, aspartate 160, isoleucine 349, leucine 145 and the adenine ring. As similar observations in eMAT [3], the PPi and Pi formed a U-shaped conformation with two magnesium ions closely stacked on both sides. Further, the two magnesium sites are formed with the side chains of aspartate 31, glutamate 305 and three water molecules. The phosphate groups are surrounded and stabilized by the side chains of several basic amino acids: lysine 25, histidine 29, lysine 201, arginine 288 and lysine 310.
As described, the ligands are solvent inaccessible and thus, the entrance of the active site requires a dynamic and flexible region. A flexible loop region was previously identified as the gate for the active site in MATs [24]. This gate loop feature has been confirmed by crystal structures of eMAT and hMAT2A, in which the loop becomes ordered when ligands are bound and it becomes disordered when the active site is empty [3, 8]. A similar gating loop (residues 141-155) region was identified in sMAT (Fig. 3), which has a similar pose to eMAT and hMAT2A and interacts with the adenine ring and the methionine or ethionine moiety. When the active site is empty, residues 141-144 form a β-sheet with residues 95-96 while the rest of the loop is poorly seen or unidentifiable in electron density. When ligands are bound in the active site, the loop region becomes ordered and is anchored above the active site with residues 141-144 shifting from the origin position, and with residues 145-149 forming a small α-helix.
In various structures of eMAT [3] and hMAT2A [8], the active site ligand occupation is correlated with an ordered active site gating loop. These studies are consistent with our observations in sMAT structures. In addition to this, ligand orientations in the eMAT and human MAT studies are similar to sMAT's. Different ligand orientations with an unordered gating loop have been reported before in rlMAT structures [7] and an earlier set of eMAT structures [4], but have some unusual features: (i) In rlMAT the temperature factors of most of the ligand coordinates are above 70 Å2, and in earlier eMAT structures, the temperature factors of ADP (or ADP mimic) are above 114 Å2; (ii) The flexible loop above the active site was undefined even though the active sites were reported to be occupied [25]; (iii) Even though rlMAT and eMAT have a 59% sequence identity and all critical residues are conserved, they show completely different ligand orientations in the active site; (iv) X-ray data for rlMAT and earlier eMAT structures are not available online, and thus ligand real-space correlation coefficients cannot be calculated. These unusual features suggest that the active site contents of rlMAT and earlier eMAT are questionable. In contrast to rlMAT and the earlier eMAT structures, all the hMAT2A, later eMAT and sMAT structures have reasonable temperature factors and a good ligand real-space correlation to electron density at the active site.
Unusual Product Formation During Crystallization
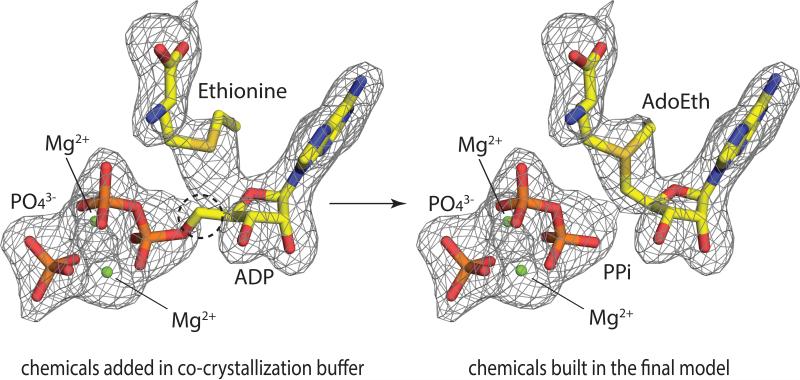
As described in the methods section, two sets of ligand bound crystals were obtained in the presence of 5 mM ADP, 10 mM ethionine (or methionine), 10 mM Mg2+ ion, and 1.4 M NaHPO4/K2HPO4. Thus based on the simulated annealing Fo-Fc omit map of the active site, one ADP, one PO4, two Mg2+ and one ethionine (or methionine) molecule were initially built in (Fig. 5, left). However, this model does not fit the electron density perfectly, because the Fo-Fc omit map does not agree with the placement of the crucial carbon atom circled in Fig. 5. Thus, it is very clear that the product has already formed and a model including PPi and AdoEth (or AdoMet) is more appropriate. The new model (Fig. 5, right) has a lower temperature factor and a better real-space correlation to electron density in the active site. MAT-catalyzed AdoMet/AdoEth formation via ADP and Met/Eth has not been previously observed. In addition, incubation of sMAT in the presence of ADP and methionine at 65 °C for 90 min under standard assay conditions led to less than 2% AdoMet formation. Thus, two explanations for this unusual product formation have been proposed: (i) The ADP stock solution is contaminated by a sufficient amount of ATP; (ii) The unusual reaction catalyzed by sMAT can actually occur in vitro, but may take as long as one month to complete, which corresponds to the time of crystal growth in this experiment.
Fig. 5.
The active site of sMAT with the simulated annealing Fo-Fc omit map (contoured at 3.0σ). Substrates including ADP, ethionine, PO43- and Mg2+ are modeled on the left. Products including AdoEth, PPi, PO43- and Mg2+ are modeled on the right.
Structure Homology
A DALI search [26] for structures similar to the sMAT monomer returned several hits, all of which are previously solved MAT structures with Z-scores between 23 and 29. Those MAT structures share a very high level of overall sequence identity (> 50%) and a high level of conservation among residues associated with substrates binding. Interestingly, sMAT only has a maximum sequence identity of 19% with these known MATs, but shares a similar three-domain (Fig. 6A).
Fig. 6.
Folding and active site comparisons between sMAT, hMAT2A and eMAT (A) Monomers of sMAT, hMAT2A and eMAT show a similar 3-domain fold. (B) Substrate-based alignment of sMAT and eMAT shows a space overlap between lysine in sMAT and K+ in eMAT.
(C) Substrate-based alignment of sMAT, hMAT2A and eMAT displays the clashes around the sMAT ethyl group. The clashes are calculated by “show bumps” in PyMOL and the serious/medium/minor clashes are shown in red/brown/green.
For the comparison of active site residues, crystal structures of sMAT, eMAT [3] and hMAT2A [8] were aligned by ligands as described in the methods section (Table 4). Surprisingly, 16 of 17 active site residues detected in sMAT have an identical or similar residue in eMAT and hMAT2A. The only extra residue sMAT has is H315, which forms hydrogen bond with O5 in di-phosphate. DALI-based sequence alignment was able to identify 11 pairs of residues (Fig. 2). Eight of them are conserved among sMAT and other MATs, including the crucial residues histidine and lysine for the proposed SN2-like mechanism [3]. The other three pairs are very similar residues at the same spot: for example, in sMAT tyrosine 270 forms stacking interactions with the adenine ring of AdoEth/AdoMet, while in eMAT it is phenylalanine 230. Intriguingly, there are another 5 pairs of residues that are not detectable via DALI search: the side chain of sMAT lysine 25 (eMAT lysine 245) helps stabilize the tri-phosphate group; the side chains of sMAT histidine 58 and asparagine 60 (eMAT glutamine 98 and lysine 269) form hydrogen bonds with the carboxyl group of methionine or ethionine; the side chain of sMAT glutamate 305 (eMAT aspartate 271) form ionic bonds with the magnesium ion; the side chain of sMAT lysine 63 occupies the same spot as the eMAT potassium ion and helps stabilize the di-phosphate ligand (Fig. 6B).
Unlike other known MATs, it has been previously reported that the activity of sMAT cannot be enhanced by K+ [16]. In the present study, all the crystals of sMAT were obtained from the crystallization condition containing more than 150 mM potassium, but electron density suitable for K+ was not observed in any datasets. In addition, potassium dependency has been previously reported in a close MAT homolog from Methanococcus jannaschii, which share the same active site residues with sMAT except for the lysine [17]. Combined with the active sites alignment evidence above (Fig. 6B), it is very likely that the catalytic activity of sMAT is not affected by K+, because the lysine in sMAT serves to present the requisite cation properties.
Interestingly, eMAT and hMAT2A also have some ability to incorporate ethionine. The ethionine turns over with sMAT and hMAT2A is near 100% whereas eMAT is just 10% [15]. A ligand-based alignment (Fig. 6C) shows that sMAT has a larger cavity around the ethyl/methyl group than either hMAT2A or eMAT. Placement of the ethyl group in eMAT will cause serious clashes with isoleucine 102 and 302, while in hMAT2A the ethionine causes moderate clashes with isoleucine 139 and 344. In sMAT a leucine (L145) is substituted for one of the conserved isoleucine in other MATs (isoleucine 102 in eMAT), which provides more active site flexibility for ethyl group binding. The ethyl group in sMAT only has minor clashes with leucine 145 and isoleucine 349. Therefore, it is very likely that the better proficiency of sMAT is caused by a larger cavity adjacent to the methyl/ethyl group. Also, branched analogs highlighted in Fig. 1B turnover significantly better with sMAT, comparing with eMAT, hMAT2A and mjMAT [15]. Interestingly, mjMAT is a thermophilic archaeal MAT that has all active site residues conserved with sMAT. However, further comparison between their active site cavities cannot be conducted, because mjMAT structure remains unknown. The current structural information suggests the better turnover rate of branched AdoMet analogs with sMAT is possibly mediated by some general orientation/dynamics of the gating loop and/or secondary shell variations. The specific residues contribute to this are currently unknown.
Conclusions
The crystal structures herein provide the atomic view of a clearly divergent class of MATs from archaea and add new active site architecture understanding to the MAT family. The sMAT has the characteristic fold and the typical tetramer assembly of known MATs, but it is the first structure in protein data bank for a thermostable MAT. In addition, the slightly expanded substrate scope of sMAT over other MATs studied to date highlight sMAT as a useful tool for the production of AdoMet analogs. In conjunction with the recent demonstration of coupled MAT-methyltransferase systems for differential alkylation [15], this chemoenzymatic strategy circumvents a major liability in the use of synthetic AdoMet analogs – namely, the dramatic instability of the AdoMet analogs. Thus, the structural insights regarding sMAT provided herein offer another blueprint from which to pursue future engineering to further broaden the catalyst promiscuity toward AdoMet analog production.
Further, the elucidation of the active site architecture with the atypical product bound, the characterization of the gating loop region, as well as the sMAT turnover reactions with different AdoMet analogs provide a blueprint for future AdoMet analog production. Since the two-step AdoMet synthesis process catalyzed by sMAT does not involve large conformational changes of ATP and methionine/ethionine, the cavity around the methyl/ethyl group is most likely the limiting factor for installing more function groups on the deoxyribose sugar. Since the cavity is formed by the adenine ring, asparagine 159, aspartate 160, isoleucine 349 and leucine 145 in the gating loop, mutations of these amino acids, especially on isoleucine 349 and leucine 145, or/and mutations on the gating loop residues can potentially increase the size of the cavity or change the local electrostatic field to accept more functional groups, such as the compounds with low or zero turnover rate described in Table 2.
Experimental Procedures
Expression and Purification of sMAT
The methionine adenosyltransferase (MAT) gene (UniProt accession: Q980S9) was cloned into NdeI/EcoRI-digested pET28a to enable production of recombinant N-His6- sMAT). For protein production, the corresponding pET28a-sMAT construct was transformed into the E.coli methionine auxotroph strain B834 (DE3) and auto-induction media [27] was used for expression at 37 °C. The cells were harvested by centrifugation at 5000 rpm for 30 min and resuspended in buffer 20 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole pH 7.8. The cells were lysed via lysozyme incubation followed by sonication on ice. Subsequently, N-His6-sMAT was purified via Ni-NTA chelating column (GE Healthcare) following a protocol with a linear imidazole (10–500 mM) elution gradient (50 mM NaH2PO4, 300 mM NaCl, pH 8.0) The His6-tag was removed by thrombin (Novagen) cleavage and the affinity tag was removed via a second round of Ni-NTA affinity chromatography. After the buffer exchange using PD-10 column (25 mM Tris-HCl, pH 8), the desired truncated SeMet-labeled sMAT was concentrated to 27 mg/mL, flash frozen in liquid nitrogen and stored at -80 °C. Protein concentrations were determined by Bradford assay (Bio-Rad, Hercules, CA, USA) using BSA as a standard.
Crystallization, diffraction, and structure determination
General screens were performed with PEGRx HT, Crystal Screen HT, Index HT, and SaltRx HT (Hampton Research) utilizing a Mosquito® dispenser (TTP labTech) by the sitting drop method. Crystal growth was monitored by Bruker Nonius Crystal Farms at 20°C. All sMAT crystals with or without substrates were obtained by mixing 2 μL of protein solution and 2 μL of reservoir solution, 1.40 M sodium phosphate monobasic monohydrate/potassium phosphate dibasic pH 5.6, using the sitting drop method. For unbound sMAT crystals, the protein solution contained 0.15-0.2 mM sMAT and 25 mM Tris pH 8.0. For sMAT:(AdoMet) condition, the protein solution contains 0.15-0.2 mM sMAT, 1 mM AdoMet and 25 mM Tris pH 8.0. For sMAT:(ADP+Met/Eth), the protein solution contained 0.2-0.3 mM sMAT, 5 mM ADP, 10 mM Met (or Eth), 10 mM MgCl2, 12.5 mM KCl, 7.5 mM DTT and 25 mM Tris pH 8. All crystals were cryoprotected by 25% DMSO or 25% ethylene glycol and flash frozen in liquid nitrogen.
X-ray diffraction data were collected at the Life Science Collaborative Access Team (LSCAT) with an X-ray wavelength of 0.98 Å for all sMAT crystals at the Advanced Photon Source at Argonne National Laboratory. Datasets were indexed and scaled by HKL2000 or XDS [28, 29]. For structure solution of apo sMAT, phenix.HySS was used for determination of selenium atom sub-structure, AutoSol for phasing and phenix.autobuild for model building [30]. For the other structures of sMAT with ligands, molecular replacement was utilized using the apo sMAT structure as a starting model. The structures including several double conformations were manually rebuilt in several rounds by Coot [31] and further refined by phenix.refine [30]. MolProbity was used to validate the quality of the coordinates [32]. All structural figures in this paper were generated using PyMOL [33].
Ligand-based Structural alignment of MAT homologs
In order to best compare the active site contents between structures with low sequence identity, four structures were aligned based on ligand positions by PyMOL. They are eMAT with AdoMet and PPNP (PDB code 1RG9) [3], hMAT2A with AdoMet (PDB code 2P02) [8], sMAT with AdoMet, PPi and PO4 (PDB code 4K0B) and sMAT with AdoEth, PPi and PO4 (4L2Z). 1RG9, 2P02 and 4K0B were first aligned by AdoMet molecule, and thus 4L2Z was aligned to 4K0B via several active site residues. Atom clashes were calculated in presence of hydrogen atoms using PyMOL and displayed as bumps. For clear display, hydrogen atoms were not illustrated in Fig. 6C.
In-vitro sMAT assay
In vitro sMAT reactions were conducted in a volume of 50 μl with 2 mM S/Se-alkylated analog, 1 mM ATP and 5 μM purified sMAT in 25 mM Tris buffer pH 8.0, 5 mM MgCl2, 50 mM KCl for 4 h at 65 °C. Reactions were quenched by adding an equal volume of methanol followed by centrifugation (10,000 xg for 15 min) to remove the precipitated protein and product formation for each reaction was subsequently analyzed by reverse phase HPLC (RP-HPLC) using method described below. For each reaction, percent yield was based upon the integration of species at 254 nm and calculated by dividing the integrated area of product and/or decomposed product by the sum of integrated area of product and/or decomposed product and the remaining substrate. The assays were repeated twice under identical conditions and Table 2 (and Fig. 4) represents an average value of two assays. The inclusion of adenine and 5’-R-thio-5’-adenosine in these calculations is based upon the established AdoMet and ATP chemical decomposition pathways which indicate adenine and 5’-R-thio-5’-adenosine to only derive from AdoMet (not ATP). All putative products were subsequently confirmed by high resolution electrospray ionization (ESI) mass spectra with positive (+) and/or negative (-) mode (see general methods).
RP-HPLC method
Reactions were quenched by the addition of equal volume of methanol followed by centrifugation at 10,000 g for 15 min to remove precipitated protein before applying on an analytical Varian ProStar HPLC [Luna C18 column, 5 μm, 4.6 mm × 250 mm; Phenomenex, Torrance, California, USA; gradient of 10% B to 40% B over 15 min, 40% B to 75% B over 5 min (A = 10 mM NaH2PO4, 5 mM octane sulfonic acid, pH was adjusted to 3.5 using phosphoric acid; B = acetonitrile) flow rate = 1 mL min-1; A254]. Reaction products were confirmed by High-resolution Mass Spectrometry.
Kinetic Measurements of sMAT Reactions
Pseudo first-order kinetics was assessed in triplicate under saturating ATP (2.5 mM) and variable L-methionine/L-ethionine (0.007, 0.013, 0.033, 0.066, 0.133, 0.333, 0.66, 1.33 mM) and saturating L-methionine/L-ethionine (2.5 mM) and variable ATP (0.013, 0.033, 0.066, 0.133, 0.333, 0.66, 1.33 mM) concentrations. Reactions were performed in buffer containing 25 mM Tris pH 8.0, 40 mM KCl and 5 mM MgCl2, and 0.5 μg purified sMAT was added and incubated at 37 °C or 65 °C for 30 min in a final volume of 20 μL. Reaction products were analyzed using RP-HPLC method above and product quantification was carried out using standard curve generated by commercial AdoMet. The kinetic curves were fit to the Michaelis-Menton equation using Prism software.
Acknowledgments
We want to thank the Center for Eukaryotic Structural Genomics for numerous contributions, Dr. Samuel E. Butcher and Dr. Jordan E. Burke (University of Wisconsin Madison) for the help of collecting SAXS data and Dr. Hongnan Cao (Rice University) for his helpful discussions on this paper. We also wish to thank the staff at the LS-CAT beamline at the Advanced Photo Source for help collecting the diffraction data. This work was supported by Protein Structure Initiative grants U01 GM098248, NIH RO1 CA84374 (JST) and the National Center for Advancing Translational Sciences (UL1TR000117).
Abbreviations
- PDB
Protein Data Bank
- MAT
Methionine adenosyltransferase
- AdoMet
S-adenosylmethionine
- AdoEth
S-adenosylethionine
Footnotes
Author Contributions
FW, JST and GNP planned experiments. FW, KEH, and KAH performed the X-ray crystallography experiments. SS, JZ and THD performed the biochemistry assays. FW, CAB, and GNP analyzed data. MS, RDG, AJM contributed essential methionine analogs. FW, JST and GNP wrote the paper.
References
- 1.Lu SC, Mato JM. S-Adenosylmethionine in cell growth, apoptosis and liver cancer. J Gastroenterol Hepatol. 2008;23(Suppl 1):S73–7. doi: 10.1111/j.1440-1746.2007.05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mudd SH, Cantoni GL. Activation of Methionine for Transmethylation III Methionine-Activating Enzyme of Bakers Yeast. J Biol Chem. 1958;231:481–492. [PubMed] [Google Scholar]
- 3.Komoto J, Yamada T, Takata Y, Markham GD, Takusagawa F. Crystal structure of the S-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of S-adenosylmethionine synthesis from ATP and Met. Biochemistry. 2004;43:1821–31. doi: 10.1021/bi035611t. [DOI] [PubMed] [Google Scholar]
- 4.Takusagawa F, Kamitori S, Markham GD. Structure and function of S-adenosylmethionine synthetase: crystal structures of S-adenosylmethionine synthetase with ADP, BrADP, and PPi at 28 angstroms resolution. Biochemistry. 1996;35:2586–96. doi: 10.1021/bi952604z. [DOI] [PubMed] [Google Scholar]
- 5.Zano SP, Pavlovsky AG, Viola RE. Structure of an unusual S-adenosylmethionine synthetase from Campylobacter jejuni. Acta Crystallogr D Biol Crystallogr. 2014;70:442–50. doi: 10.1107/S139900471303023X. [DOI] [PubMed] [Google Scholar]
- 6.Schlesier J, Siegrist J, Gerhardt S, Erb A, Blaesi S, Richter M, Einsle O, Andexer JN. Structural and functional characterisation of the methionine adenosyltransferase from Thermococcus kodakarensis. BMC Struct Biol. 2013;13:22. doi: 10.1186/1472-6807-13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez B, Pajares MA, Hermoso JA, Guillerm D, Guillerm G, Sanz-Aparicio J. Crystal structures of methionine adenosyltransferase complexed with substrates and products reveal the methionine-ATP recognition and give insights into the catalytic mechanism. J Mol Biol. 2003;331:407–16. doi: 10.1016/s0022-2836(03)00728-9. [DOI] [PubMed] [Google Scholar]
- 8.Shafqat N, Muniz JR, Pilka ES, Papagrigoriou E, von Delft F, Oppermann U, Yue WW. Insight into S-adenosylmethionine biosynthesis from the crystal structures of the human methionine adenosyltransferase catalytic and regulatory subunits. Biochem J. 2013;452:27–36. doi: 10.1042/BJ20121580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markham GD, Pajares MA. Structure-function relationships in methionine adenosyltransferases. Cell Mol Life Sci. 2009;66:636–48. doi: 10.1007/s00018-008-8516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mato JM, Alvarez L, Ortiz P, Pajares MA. S-adenosylmethionine synthesis: molecular mechanisms and clinical implications. Pharmacol Ther. 1997;73:265–80. doi: 10.1016/s0163-7258(96)00197-0. [DOI] [PubMed] [Google Scholar]
- 11.Fu Z, Hu Y, Markham GD, Takusagawa F. Flexible loop in the structure of S-adenosylmethionine synthetase crystallized in the tetragonal modification. J Biomol Struct Dyn. 1996;13:727–39. doi: 10.1080/07391102.1996.10508887. [DOI] [PubMed] [Google Scholar]
- 12.Lu ZJ, Markham GD. Enzymatic properties of S-adenosylmethionine synthetase from the archaeon Methanococcus jannaschii. J Biol Chem. 2002;277:16624–31. doi: 10.1074/jbc.M110456200. [DOI] [PubMed] [Google Scholar]
- 13.Ottink OM, Nelissen FH, Derks Y, Wijmenga SS, Heus HA. Enzymatic stereospecific preparation of fluorescent S-adenosyl-L-methionine analogs. Anal Biochem. 2010;396:280–3. doi: 10.1016/j.ab.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 14.Wang R, Islam K, Liu Y, Zheng W, Tang H, Lailler N, Blum G, Deng H, Luo M. Profiling genome-wide chromatin methylation with engineered posttranslation apparatus within living cells. J Am Chem Soc. 2013;135:1048–56. doi: 10.1021/ja309412s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh S, Zhang J, Huber TD, Sunkara M, Hurley KA, Goff RD, Wang G, Zhang W, Liu C, Rohr J, Van Lanen SG, Morris AJ, J.S., T Facile Strategies for the Synthesis and Utilization of S-Adenosyl-L-Methionine Analogs. Angewandte Chemie. 2014 doi: 10.1002/anie.201308272. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porcelli M, Cacciapuoti G, Carteni-Farina M, Gambacorta A. S-adenosylmethionine synthetase in the thermophilic archaebacterium Sulfolobus solfataricus. Purification and characterization of two isoforms. Eur J Biochem. 1988;177:273–80. doi: 10.1111/j.1432-1033.1988.tb14373.x. [DOI] [PubMed] [Google Scholar]
- 17.Graham DE, Bock CL, Schalk-Hihi C, Lu ZJ, Markham GD. Identification of a highly diverged class of S-adenosylmethionine synthetases in the archaea. J Biol Chem. 2000;275:4055–9. doi: 10.1074/jbc.275.6.4055. [DOI] [PubMed] [Google Scholar]
- 18.Takusagawa F, Kamitori S, Misaki S, Markham GD. Crystal structure of S-adenosylmethionine synthetase. J Biol Chem. 1996;271:136–47. [PubMed] [Google Scholar]
- 19.Gonzalez B, Pajares MA, Hermoso JA, Alvarez L, Garrido F, Sufrin JR, Sanz-Aparicio J. The crystal structure of tetrameric methionine adenosyltransferase from rat liver reveals the methionine-binding site. J Mol Biol. 2000;300:363–75. doi: 10.1006/jmbi.2000.3858. [DOI] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–97. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman JL. Chromatographic analysis of the chiral and covalent instability of S-adenosyl-L-methionine. Biochemistry. 1986;25:4444–9. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- 22.Iwig DF, Booker SJ. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine. Biochemistry. 2004;43:13496–509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- 23.Willnow S, Martin M, Luscher B, Weinhold E. A selenium-based click AdoMet analogue for versatile substrate labeling with wild-type protein methyltransferases. Chembiochem. 2012;13:1167–73. doi: 10.1002/cbic.201100781. [DOI] [PubMed] [Google Scholar]
- 24.Taylor JC, Markham GD. Conformational dynamics of the active site loop of S-adenosylmethionine synthetase illuminated by site-directed spin labeling. Arch Biochem Biophys. 2003;415:164–71. doi: 10.1016/s0003-9861(03)00277-7. [DOI] [PubMed] [Google Scholar]
- 25.Taylor JC, Takusagawa F, Markham GD. The active site loop of S-adenosylmethionine synthetase modulates catalytic efficiency. Biochemistry. 2002;41:9358–69. doi: 10.1021/bi025851t. [DOI] [PubMed] [Google Scholar]
- 26.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr., Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40:256–67. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrodinger, LLC The PyMOL Molecular Graphics System, Version 1.3r1 in. 2010.