Abstract
Analysis of the polar lipids of many pathogenic and non-pathogenic clostridia has revealed the presence of plasmalogens, alk-1′-enyl ether-containing phospholipids and glycolipids. An exception to this finding so far has been Clostridium difficile, an important human pathogen which is the cause of antibiotic-associated diarrhea and other more serious complications. We have examined the polar lipids of three strains of C. difficile by thin-layer chromatography and have found acid-labile polar lipids indicative of the presence of plasmalogens. The lipids from one of these strains were subjected to further analysis by liquid chromatography coupled to electrospray ionization-mass spectrometry (LC/ESI-MS), which revealed the presence of phosphatidylglycerol, cardiolipin, monohexosyldiradylglycerol, dihexosyldiradylglycerol, and two unusual glycolipids identified as an aminohexosyl-hexosyldiradylglycerol, and a trihexosyldiradylglycerol. High resolution tandem mass spectrometry determined that monohexosyldiradylglycerol, cardiolipin and phosphatidylglycerol contained significant amounts of plasmalogens. C. difficile thus joins the growing list of clostridia that have plasmalogens. Since plasmalogens in clostridia are formed by an anaerobic pathway distinct from that in animal cells, their formation represents a potential novel target for antibiotic action.
Supplementary key words: Clostridium difficile, glycolipid, mass spectrometry, phospholipid, plasmalogen
1. Introduction
Plasmalogens, alk-1′-enyl ether lipids, have a unique distribution in nature. They are found in anaerobic, but not in aerobic or facultative bacteria, and they are present in many animals. Indeed these lipids appear to have evolved twice since they are made anaerobically in bacteria and by an oxygen-dependent desaturation of a saturated ether lipid in animal cells [1,2]. A major group of anaerobic bacteria in which plasmalogens have been found is the genus Clostridium. With one exception, Clostridium difficile [3,4] all examined species have been found to contain plasmalogens [5–11]. Since the biosynthesis of plasmalogens by an anaerobic pathway not found in animal cells offers a potential target for novel antibacterial compounds, it is important to know if C. difficile contains these ether lipids. C. difficile is a major cause of antibiotic-associated intestinal disease, including pseudomembranous colitis, a severe inflammation of the lower intestine. C. difficile overgrows the normal flora after many of the latter have been removed by antibiotic treatment. It produces toxins and is difficult to treat owing to its resistance to antibiotics.
Acid-lability distinguishes plasmalogens from diacylglycerolipids; the presence of an acid labile lipid-like material in animal cells was key to the discovery of this class of lipids [12]. We have used thin-layer chromatography (TLC) combined with acid-treatment to demonstrate the presence of acid-labile lipids in C. difficile. A more precise tool in the study of microbial plasmalogens is high resolution mass spectrometry [5–7][5–7] which can distinguish plasmalogen molecular species from diacyl glycerolipids having essentially the same molecular mass. In this paper we show that C. difficile has both phospholipids and glycolipids that consist of diacyl and plasmalogen species. In addition to the previously reported phosphatidylglycerol (PG), we have found cardiolipin in the tetraacyl and plasmalogen forms, monohexosyldiradylglycerol (MHDRG), dihexosyldiradylglycerol (DHDRG) and two unusual glycosyl diradylglycerols. Plasmalogens were most abundant in the phospholipids.
2. Methods
2.1. Bacterial strain and growth conditions
C. difficile strains CD630, UK1 (from J. Sorg, Texas A&M University), and VPI10463 (ATCC 43255) were grown as described previously [13] in an anaerobic workstation (Coy Laboratory Productions, USA) at 37°C in pre-reduced BHIS, Brain-Heart Infusion (Bacto, USA) medium supplemented with yeast extract (5 mg/mL; Bacto, USA) and L-cysteine (0.1% wt/vol; Sigma-Aldrich, St. Louis, MO). An anaerobic atmosphere was maintained with 5% H2, 5% CO2, and 90% N2. Cultures were incubated overnight without shaking, then removed from the anaerobic hood and immediately centrifuged at 6000 × g for 10 min at 4°C. Cells were washed once in 1× PBS and transferred to Corex® 25 mL glass centrifuge tubes. The cell pellets were stored at − 80°C.
2.2. Lipid extraction and thin-layer chromatography (TLC)
Total lipids were extracted from the wet cell pellets with chloroform-methanol [14] with minor modifications[15]. TLC was done on silica gel 60, 10 ×10 cm, thin-layer plates using the following solvents: System A, chloroform/methanol/concentrated ammonia/water, 65:30:2.5:2.5 (by vol.) in the first dimension and System B, chloroform/methanol/acetic acid/water, 80:18:12:5 (by vol.) in the second dimension. Acid treatment of plates was performed by exposing the lipids to HCl fumes for 20–25 sec at the origin for one-dimensional TLC or the strip containing the lipids above the origin for 2D-TLC. The plates were then air dried in a fume hood and after the HCl evaporated, moisture was removed under vacuum. The plates were then immediately placed in a TLC tank for the second dimension run. Phospholipids or amine-containing lipids were detected with 0.3% (w/v) molybdenum blue (Sigma-Aldrich) or 0.3% ninhydrin in ethanol, respectively, followed by heating at 120 °C for 10 min, respectively. Glycolipids were detected by α-naphthol staining [16].
Preparative TLC was performed on a 10 × 10 cm silica gel 60 TLC plate with the lipids extracted from a 500 ml culture of strain CD630 spread on a line at the origin. The lipids were chromatographed in Solvent A and all but the furthest left material was scraped in 1 cm bands which were eluted as described above for extraction of cellular lipids. The leftmost material was stained for phospholipids with molybdate reagent and charred.
2.3. Quantification of lipid compositions
C. difficile CD630 was grown overnight as described above in 10 ml cultures containing 10 μCi [1-14C] acetate (60 mCi/mol, Perkin Elmer Life Sciences, Waltham, MA). The lipids were extracted, carrier lipids from the same species were added and ~10,000 cpm of the radioactively labeled lipids were separated by 2D-TLC as described above. Radiolabeled lipids were visualized and quantified with Typhoon 9410 phosphorimager (Amersham Biosciences, Arlington Heights, IL), equipped with Image-Quant software.
2.4. Liquid chromatography/tandem mass spectrometry (LC/MS/MS)
Normal phase LC was performed on an Agilent 1200 Quaternary LC system equipped with an Ascentis Silica HPLC column, 5 μm, 25 cm × 2.1 mm (Sigma-Aldrich, St. Louis, MO). Mobile phase A consisted of chloroform/methanol/aqueous ammonium hydroxide (800:195:5, v/v); mobile phase B consisted of chloroform/methanol/water/aqueous ammonium hydroxide (600:340:50:5, v/v); mobile phase C consisted of chloroform/methanol/water/aqueous ammonium hydroxide (450:450:95:5, v/v). The elution program consisted of the following: 100% mobile phase A was held isocratically for 2 min and then linearly increased to 100% mobile phase B over 14 min and held at 100% B for 11 min. The LC gradient was then changed to 100% mobile phase C over 3 min and held at 100% C for 3 min, then returned to 100% A over 0.5 min and held at 100% A for 5 min. The LC eluent (with a total flow rate of 300 μl/min) was introduced into the ESI source of a high resolution TripleTOF5600 mass spectrometer (Applied Biosystem, Foster City, CA). Instrumental settings for negative ion ESI and MS/MS analysis of lipid species were as follows: IS= −4500 V; CUR= 20 psi; GSI= 20 psi; DP= −55 V; and FP= −150 V. The MS/MS analysis used nitrogen as the collision gas. Data analysis was performed using Analyst TF1.5 software (Applied Biosystems, Foster City, CA).
2.5 Acid hydrolysis of total lipids
To perform the acid treatment, the dried lipids were dissolved in a single-phase Bligh-Dyer mixture (methanol-chloroform-water, 2:1:0.8, v/v/v), containing hydrochloric acid at a final concentration of 0.2 N, and incubated at room temperature for 20 min. Next, the mixture was converted to a two-phase Bligh-Dyer system by adding chloroform and H2O. After centrifugation, the lower phase was dried under N2, and then re-suspended into a chloroform/methanol mixture (2:1 v/v) before being subjected to LC/MS analysis.
3. Results
3.1. Identification and quantification of C. difficile polar lipids
Total lipids from C. difficile CD630, VPI and UKI were first subjected to one-dimensional TLC with and without prior treatment with fumes of hydrochloric acid. We detected acid-labile lipids by charring all the lipids, which revealed accumulation of neutral lipids, presumably aldehydes, at the solvent front after acid hydrolysis (Supplementary Fig. 1A) and more clearly by staining the lipids for phosphorus with molybdenum blue which revealed the formation of polar phospholipids, presumably lyso lipids produced by hydrolysis of the alk-1-enyl ether (Supplementary Fig. 1B). The three strains gave similar results.
C. difficile CD630 cells were radioactively labeled by overnight growth with [1-14C] acetate. The extracted total lipids were separated by 2D-TLC. A representative chromatogram is shown in Fig. 1. Radioactivity was quantified on a phosphorimager and the results are presented in Table 1. Four lipids: MHDRG (spot 1), cardiolipin (spot 4), PG (spot 5) and DHDRG (spot 6) were identified by their positions relative to standards (not shown) and by staining with phosphomolybdate for phosphorus and by α-naphthol for sugars. Their identities were confirmed by liquid chromatography/mass spectrometry (see below). Spot 2, which migrated ahead of cardiolipin in the first dimension, was identified as PG by preparative TLC, elution of the lipid from silica gel and MS of the eluted lipid (see below). These four lipids represent 74.4% of the total radioactivity. Two unknown glycolipids (spots 7 and 8 in Fig. 1) represent 21.7% of the total radioactivity.
Fig. 1. Analysis by 2D-TLC of [14C]-labeled lipids from C. difficile CD630.
Based on their positions relative to standards and specific staining the major lipids were tentatively identified as (1) MHDRG, (2) phosphatidylglycerol, (3) unknown artifact, probably acetic acid, (4) cardiolipin, (5) phosphatidylglycerol (6) DHDRG, (7 and 8) unknown glycolipids. The symbols indicate * α-naphthol positive, † molybdenum blue-positive and # ninhydrin-positive. The arrows indicate the first and second dimensions of the chromatograms.
Table 1.
Polar lipid composition of Clostridium difficile CD630 as judged by radiolabeling (% cpm ± S.D., n = 4).
| Monohexosyldiradylglycerol (1)* | 14.2 ± 3.3 |
| Cardiolipin (4) | 16.1 ± 3.8 |
| Phosphatidylglycerol (5) | 4.5 ± 1.0 |
| Second phosphatidylglycerol spot (2) | 24.7 ± 4.7 |
| Dihexosyldiradylglycerol (6) | 14.9 ± 2.2 |
| Aminohexosyl-hexosyldiradylglycerol (7) | 16.2 ± 3.8 |
| Trihexosyldiradylglycerol (8) | 5.5 ± 1.6 |
3.2. Mass spectrometry
In order to confirm the identities of lipids detected by TLC, the total lipid extract of C. difficile CD630 was subjected to normal phase LC-ESIMS/MS using a silica column for lipid separation. Diacylglycerol emerged in a peak at 2.36 min (Fig. 2, Suppl. Fig. 2). The peak emerging at 3.2 min showed the characteristic molecular masses of a monohexosyldiacylglycerol (MHDAG) corresponding to molecular species 30:0, 32:1, 32:0, 34:1 and 34:0 (Fig. 3). Small amounts of the corresponding plasmalogens corresponding to the major diacyl species seen at m/z -16 from the diacyl forms are designated with arrows. The presence of plasmalogens in the MGDRG was confirmed by HCl hydrolysis on a 2D-TLC plate as described under Methods (Supplementary Fig. 3, spots 1 and 2). The peak at 7.4 min (see Fig. 2) contained phosphatidylglycerol (PG) with the principal molecular species 32:1 and 34:1 and plasmenylglycerol (PlaG) 32:1, and 34:1 (Fig. 4). Based on FAB-MS, the m/z 703.5 and 731.5 ions were previously interpreted as PG (31:2) and (33:2), presumably containing 15:1 and/or 17:1 acyl chains [3]. However, the exact masses show 15.996 Da differences between the diacyl and plasmalogen forms not the 16.031 Da differences expected for two diacyl lipids (Supplementary figure 4). MS/MS of the m/z 703.5 ion showed that the major acyl chain was 16:1. An alk-1-enyl 16:0 fragment was observed at m/z 239.224 corresponding to the anion of hexadecenal as shown in supplementary Fig. 5. The peak emerging at 11.8 min (Fig. 2) also contained PG molecular species, however, the diacyl species 32:1 and 34:1 were replaced by 32:2 and 34:2, with m/z 717.507 and 745.539, respectively. The plasmalogen species remained the same, m/z 703.492 and 731.523 (data not shown). PG may be separating into two fractions on this normal phase column as the less polar protonated form and the more polar salt form. Separation of PG into two spots was also seen on 2D-TLC (Fig. 1).
Fig. 2. Total ion chromatogram of LC/MS of C. difficile total lipids.
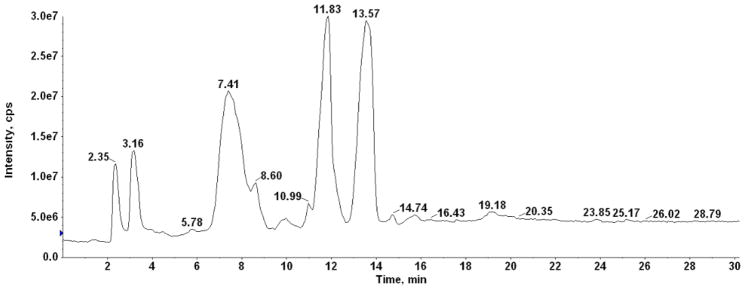
The chromatogram of total lipids indicates the peaks of emergence at which the mass spectra were averaged.
Fig. 3. LC/MS identification of monohexosyldiradylglycerol from C. difficile CD630.
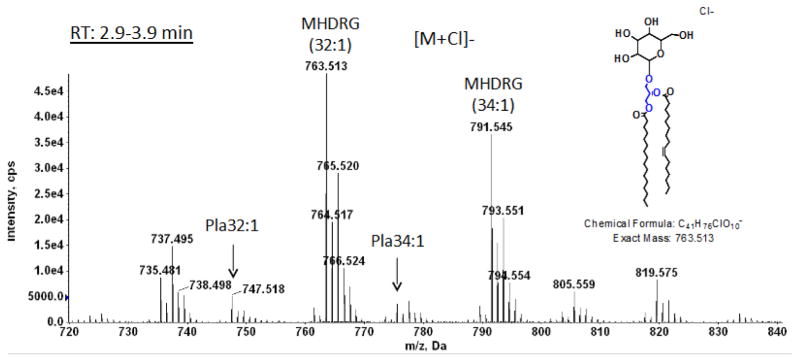
Mass spectrum of the material emerging between 3 and 4 min (see Fig. 2) showing the [M + Cl]− ions of MHDRG. The arrows indicate plasmalogen species.
Fig. 4. LC/MS identification of phosphatidylglycerol from C. difficile CD630.
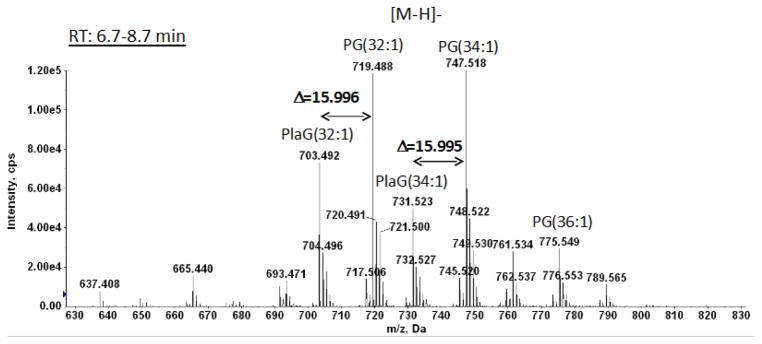
Mass spectrum showing the [M – H]− ions of PG. Two of the major species of PG, 32:1 and 34:1, are found to be in both diacyl and plasmalogen (PlaG) forms. The exact difference of 15.996 Da between the diacyl and alk-1′-enyl, acyl forms is consistent with the plasmalogen structure (see Supplementary figure 4).
A peak emerging between 9.0 and 10.5 min was identified as dihexosyldiacylglcyerol (Suppl. Fig. 6). Cardiolipin, which emerged with a peak at 13.57 min displayed, as expected from the numerous molecular species of PG and PlaG, a complex pattern of mass peaks with major species from 60:1 to 70:0. There were major tetra-acyl, alk-1′-enyl, triacyl, and di- alk-1′-enyl, diacyl species (Fig. 5 and supplementary material Table 1).
Fig. 5. LC/MS identification of cardiolipin from C. difficile CD630.
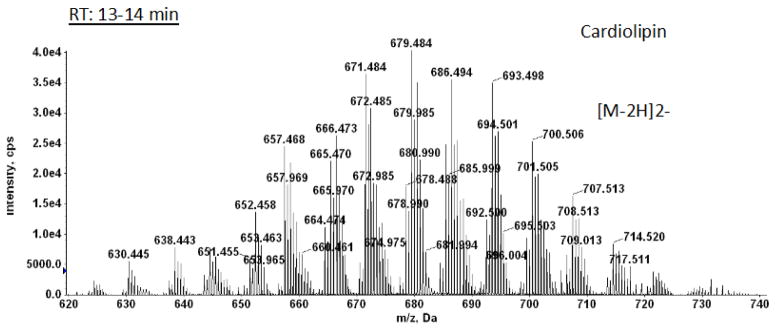
Mass spectrum showing the [M – 2H]2− ions of cardiolipin molecular species. Preliminary identifications are given in Supplementary Material Table 1.
We have checked for the presence of alkyl ether analogs of the polar lipids by carrying out acid hydrolysis of total C. difficile lipids followed by MS analysis. In supplementary figures 7 and 8 we focus on the results with phosphatidylglycerol and its plasmalogen. After hydrolysis, the plasmalogens at m/z 703.492 and 731.523 disappear, as expected (Suppl. Fig. 7). We see no signal at m/z 705.5 where the alkyl ether (32:1) would appear. We do see a signal at 733.500 which could be the alkyl ether (34:1). However, MS/MS of this peak shows that it consists of diacyl PG (33:0) with the possible acyl chain combinations of 16:0/17:1, 16:1/17:0, and some 18:0/15:1 (Suppl. Fig. 8).
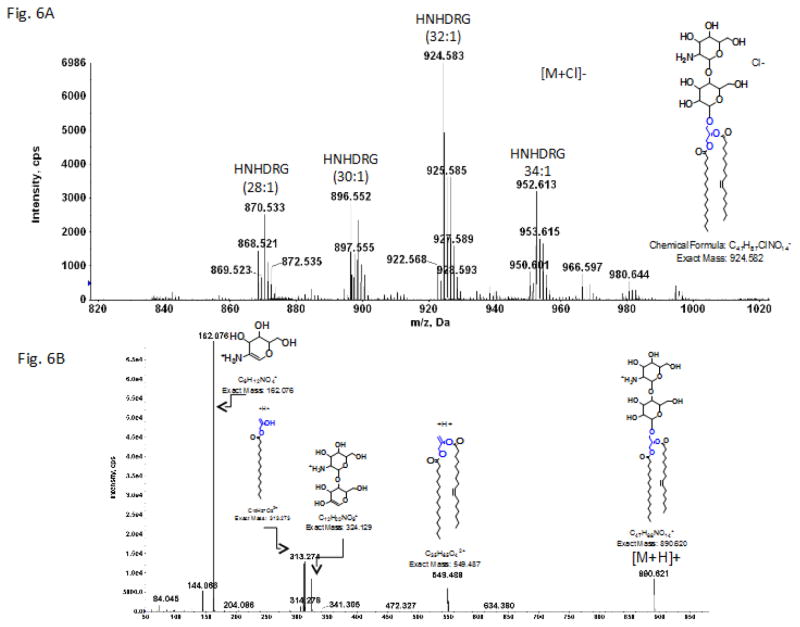
3.3 Identification of the two unknown glycolipids
One of two remaining glycolipids was identified as an aminohexosyl-hexosyldiradylglycerol (HNHDRG). The mass spectrum shown in Figure 6 was generated from lipid obtained by preparative TLC in a band corresponding to ninhydrin-positive spot 7 (Fig. 1). Positive ion MS/MS of the m/z 890.6 [M+H]+ ion of HNHDRG (32:1) (corresponding to m/z 924.583 [M+Cl]− seen in the negative ion mode) gave the fragmentation pattern shown in Fig. 6B. Characteristic fragments corresponding to a protonated hexosamine-hexose, m/z 324.13, and a hexosamine, m/z 162.076, in addition to fragments derived from the diacylglycerol are also seen.
Fig. 6.
LC/MS identification of hexoasaminylhexosylDRG from C. difficile CD630.
A. Mass spectrum showing the [M + Cl]− ions of a hexosaminylhexosylDRG. B. MS/MS of [M+H]+ ion at m/z 890.6. The structure is for illustrative purposes only. The positions of the glycosidic bonds have not been determined.
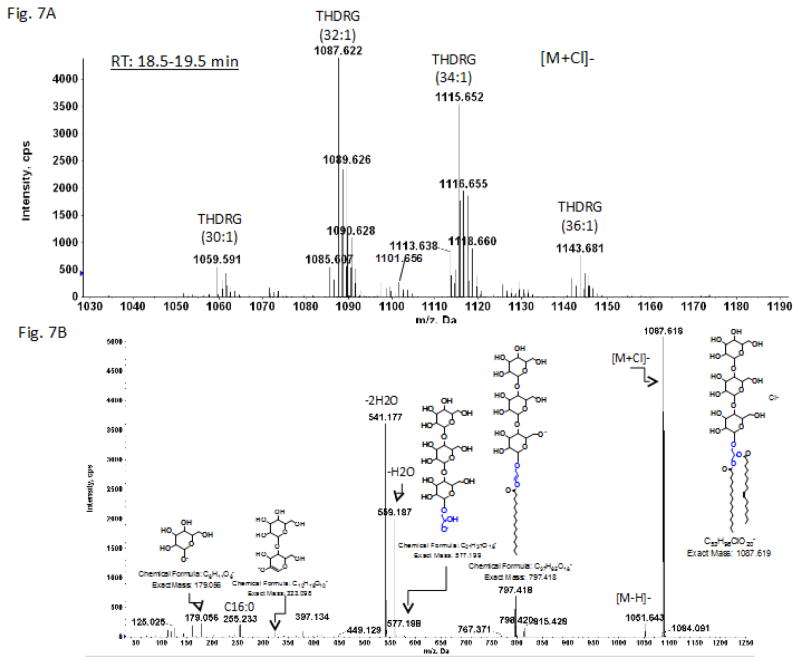
The most polar glycolipid corresponding to spot 8 (Fig. 1) isolated by preparative TLC gave the spectrum shown in Fig. 7. The exact masses correspond to a series of trihexosyldiacylglycerols (THDRG). MS/MS of the m/z 1087.6 THDRG (32:1) gave the fragmentation pattern shown in Fig. 7B. A fragment corresponding to trihexosylglycerol at m/z 577.198, whose H2O-loss ions are seen at m/z 559.187 and m/z 541.177. In addition, a small signal for a deprotonated dihexose at m/z 323.1 and a single deprotonated hexose at m/z 179.056 were also evident. The signal at m/z 797.418 corresponds to the product ion after the loss of one acyl chain from the parent ion. The presence of small amounts of plasmalogens in both the HNHDRG and the THDRG was confirmed by HCl hydrolysis on a 2D-TLC plate as described under Methods (Supplementary Fig. 3, spots 10 and 11 and corresponding aldehydes and lyso-lipids at the solvent front and trailing the diacyl compounds, respectively). The ion peak at m/z 1101.656 corresponds to a 34:0 plasmalogen species (Fig. 7A).
Fig. 7.
LC/MS identification of trihexosylDRG from C. difficile CD630.
A. Mass spectrum showing the [M + Cl]− ions of a trihexosylDRG. B. MS/MS of [M+Cl]− at m/z 1087.6. The structure is for illustrative purposes only. The positions of the glycosidic bonds have not been determined.
3. Discussion
In this paper we show that C. difficile contains glycerophospholipids and glycosyldiradyl-glycerols. The major identified glycerophospholipids are cardiolipin and PG, both of which are found in the plasmalogen as well as the tetraacyl and diacyl forms, respectively. Plasmalogen forms were not found in previous studies [3,4]. The previous assignment of specific PG molecular species to those containing an odd chain, is understandable because low-resolution fast atom bombardment FAB/MS used in previous work could not make the distinction between the exact masses of these molecules which only differ by 36 mDA (Fig. 4 and Supplementary Fig. 4). In agreement with past studies, we have not found phosphatidylethanolamine in C. difficile [3,4]. Examination of the published genome of C. difficile CD630 reveals that the genes needed for phosphatidylethanolamine biosynthesis, pss and psd, are absent and no pss orthologs were found in C. difficile strains when BLAST searches were done using the DNA sequence of Clostridium beijerinckii pss.
The anaerobic pathway to plasmalogens starts with the acylation of glycerol-3-P to form phosphatidic acid continues through CDP-diacylglycerol, which then branches to form phosphatidylserine and phosphatidylethanolamine on one branch and PG and cardiolipin on the other. The formation of plasmalogens in previously studied clostridia ends with the conversion of PE and PG to their respective plasmalogens [17–19]. The intermediates: phosphatidic acid, CDP-diacylglycerol, phosphatidylserine and phosphatidylglycerophosphate were found to have little or no plasmalogen species [6,7]. The plasmalogen form of MHDAG is presumably formed from the corresponding diacyl lipid.
Although PG was previously shown to be a major phospholipid in C. difficile, cardiolipin was not found [3]. Since PG is a precursor of cardiolipin, it is possible that the ratio of these phospholipids undergoes changes during growth. Since the objective of our study was to explore the total lipidome of this organism for plasmalogens, we have not studied changes during growth or in response to changes in the environment, which can be significant and worthy of future studies.
A glucosyl-N-acylglucosaminyldiacylglycerol was previously reported in Bacillus acidocaldarius [20]. We propose that the hexosamine residue in the HNHDRG of C. difficile is distal to the hexose because there is a hexosylDRG, but not a hexosaminyDRG. Given the lack of zwitterionic phosphatidylethanolamine (PE) in C. difficile, this amino-containing glycolipid, which represents 16% of the total, provides a means for balancing the negative charges of PG and cardiolipin. TriglycosylDAG has been reported in several Gram-positive bacteria [21].
C. difficile is distantly related to most of the clostridia studied previously. Based on 16S rRNA sequencing the genus Clostridium was divided into 14 clusters and it was proposed that the genus be divided into five new genera and eleven new combinations. C. difficile was assigned to cluster XI, which in itself is taxonomically heterogeneous. Most of the clostridia species previously studied for their lipid composition were placed in cluster I. Clostridium innocuum, which like C. difficile has no PE and has lipids unique to that species [22], was assigned to the narrow cluster XVI [23].
C. difficile is a growing health concern because C. difficile-associated diarrhea (CDAD) often progresses to intestinal lesions which can lead to pseudomembranous colitis with raised yellow plaques throughout the colon mucosa. This chronic condition can in some case ultimately result in death [24]. CDAD can be a significant outcome of antibiotic treatment, which may provide specific changes in the host flora that provide an environment favorable to C. difficile growth and toxin production [24]. There is clearly a need for new antibiotics that specifically inhibit clostridial growth. Since the anaerobic pathway to plasmalogens differs from the oxygen-dependent pathway in animal cells [2], the enzyme(s) responsible for plasmalogen formation in C. difficile and in other anaerobes should provide a new target for specific inhibitors once the proteins are known. The search for the genes responsible for plasmalogen biosynthesis in anaerobes should have a high priority. Once the genes have been identified, the study of the biological and pathological roles of these lipids in C. difficile could be approached through genetic manipulation.
Supplementary Material
Highlights.
Clostridium difficile is a major human pathogen
Clostridium difficile glycolipids and phospholipids are characterized
Both glycolipids and phospholipids have plasmalogen species
An anaerobic pathway to plasmalogens represents a potential target for antibiotics
Acknowledgments
This work was partially supported by the Lipid Maps Collaborative Grant GM-069338 from the National Institutes of Health (to Z.G.) and NIH U54 AI 057168 (to J.Z).We wish to thank Sharon Ng for help with the preparative TLC.
Abbreviations
- CL
cardiolipin
- DHDRG
dihexosyldiradylglycerol
- HNHDRG
aminohexosyl-hexosyldiradylglycerol
- MHDRG
monohexosyldiradylglycerol
- PG
phosphatidylglycerol
- PlaG
plasmenylglycerol
- THDRG
trihexosyldiradylglycerol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.McIntyre TM, Snyder F, Marathe GK. Ether-Linked Lipids and Their Bioactive Species. In: Vance DE, Vance JE, editors. Biochemistry of Lipids, Lipoproteins and Membranes. Elsevier; Amsterdam: 2008. pp. 245–276. [Google Scholar]
- 2.Goldfine H. The appearance, disappearance and reappearance of plasmalogens in evolution. Prog Lipid Res. 2010;49:493–498. doi: 10.1016/j.plipres.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 3.Drucker DB, Wardle HM, Boote V. Phospholipid profiles of Clostridium difficile. J Bacteriol. 1996;178:5844–5846. doi: 10.1128/jb.178.19.5844-5846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Korachi M, Rupnik M, Blinkhorn AS, Boote V, Drucker DB. Comparison of polar lipid profiles of Clostridium difficile isolates from different geographical locations. Anaerobe. 2002;8:35–39. [Google Scholar]
- 5.Guan Z, Johnston NC, Raetz CRH, Johnson EA, Goldfine H. Lipid diversity among botulinum neurotoxin-producing clostridia. Microbiology. 2012;158:2577–2584. doi: 10.1099/mic.0.060707-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan Z, Johnston NC, Aygun-Sunar S, Daldal F, Raetz CR, Goldfine H. Structural characterization of the polar lipids of Clostridium novyi NT. Further evidence for a novel anaerobic biosynthetic pathway to plasmalogens. Biochim Biophys Acta. 2011;1811:186–193. doi: 10.1016/j.bbalip.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston NC, Aygun-Sunar S, Guan Z, Ribeiro AA, Daldal F, Raetz CR, Goldfine H. A phosphoethanolamine-modified glycosyl diradylglycerol in the polar lipids of Clostridium tetani. J Lipid Res. 2010;51:1953–1961. doi: 10.1194/jlr.M004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldfine H. Membrane Biogenesis. In: Timmis KN, editor. Hydrocarbons, Oils and Lipids: Diversity, Properties and Formation, vol. 1, Handbook of Hydrocarbon and Lipid Microbiology. Springer; Berlin: 2010. pp. 417–424. [Google Scholar]
- 9.Goldfine H, Johnston NC. Membrane Lipids of Clostridia. In: Dürre P, editor. Handbook on clostridia. Taylor & Francis; Boca Raton, FL: 2005. pp. 297–310. [Google Scholar]
- 10.Oulevey J, Bahl H, Thiele OW. Novel alk-1-enyl ether lipids isolated from Clostridium acetobutylicum. Arch Microbiol. 1986;144:166–168. [Google Scholar]
- 11.Kamio Y, Kanegasaki S, Takahashi H. Occurrence of plasmalogens in anaerobic bacteria. J Gen Appl Microbiol. 1969;15:439–451. [Google Scholar]
- 12.Debuch H, Seng P. The History of Ether-Linked Lipids Through 1960. In: Snyder F, editor. Ether lipids. Academic Press; New York and London: 72 A.D. pp. 1–24. [Google Scholar]
- 13.Katzianer DS, Yano T, Rubin H, Zhu J. A high-throughput small-molecule screen to identify a novel chemical inhibotor of Clostridium difficile. Int J Antimicrob Agents. 2014 doi: 10.1016/j.ijantimicag.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 15.Kates M. Techniques of Lipidology. Isolation, Analysis and Identification of Lipids, Techniques of Lipidology. Isolation, Analysis and Identification of Lipids. 1990:106. [Google Scholar]
- 16.Siakotos AN, Rouser G. Analytical separation of nonlipid water soluble substances and gangliosides from other lipids by dextran gel column chromatography. J Am Chem Soc. 1965;42:913–919. doi: 10.1007/BF02632444. [DOI] [PubMed] [Google Scholar]
- 17.Baumann NA, Hagen PO, Goldfine H. Phospholipids of Clostridium butyricum: Studies on plasmalogen composition and biosynthesis. J Biol Chem. 1965;240:1559–1567. [PubMed] [Google Scholar]
- 18.Koga Y, Goldfine H. Biosynthesis of phospholipids in Clostridium butyricum: the kinetics of synthesis of plasmalogens and the glycerol acetal of ethanolamine plasmalogen. J Bacteriol. 1984;159:597–604. doi: 10.1128/jb.159.2.597-604.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacDonald DL, Goldfine H. Phosphatidylglycerol acetal of plasmenylethanolamine as an intermediate in ether lipid formation in Clostridium butyricum. Biochem Cell Biol. 1990;68:225–230. doi: 10.1139/o90-030. [DOI] [PubMed] [Google Scholar]
- 20.Langworthy TA, Mayberry WR, Smith PF. A sulfonolipid and novel glucosamidyl glycolipids from the extreme thermoacidophile Bacillus acidocaldarius. Biochim Biophys Acta. 1976;431:550–569. doi: 10.1016/0005-2760(76)90220-4. [DOI] [PubMed] [Google Scholar]
- 21.Kates M. Glyco-, Phospho-, and Sulfoglycoglycerolipids of Bacteria. In: Kates M, editor. Handbook of Lipid Research, Vol. 6. Glycolipids, phosphoglycolipids, and sulfoglycolipids. Plenum Press; New York: 1990. pp. 1–122. [Google Scholar]
- 22.Johnston NC, Goldfine H, Fischer W. Novel polar lipid composition of Clostridium innocuum as the basis for an assessment of its taxonomic status. Microbiology. 1994;140:105–111. doi: 10.1099/13500872-140-1-105. [DOI] [PubMed] [Google Scholar]
- 23.Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow JAE. The phylogeny of the Genus Clostridium; Proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 24.Giel JL, Sorg JA, Sonenshein AL, Zhu J. Metabolism of Bile Salts in Mice Influences Spore Germination in Clostridium difficile. Plos One. 2010;5 doi: 10.1371/journal.pone.0008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.