Abstract
Burkholderia cepacia complex (Bcc) is a set of closely related bacterial species that are notorious pathogens of cystic fibrosis patients, responsible for life-threatening lung infections. Expression of several virulence factors of Bcc is controlled by a mechanism known as quorum sensing (QS). QS is a means of bacterial communication used to coordinate gene expression in a cell-density–dependent manner. The system involves the production of diffusible signaling molecules (N-acyl-L-homoserine lactones, AHLs), that bind to cognate transcriptional regulators and influence their ability to regulate gene expression. One such system that is highly conserved in Bcc consists of CepI and CepR. CepI is AHL synthase, while CepR is an AHL-dependent transcription factor. In most members of the Bcc group, the cepI and cepR genes are divergently transcribed and separated by additional genes. One of them, bcam1869, encodes the BcRsaM protein, which was recently postulated to modulate the abundance or activity of CepI or CepR. Here we show the crystal structure of BcRsaM from B. cenocepacia J2315. It is a single-domain protein with unique topology and presents a novel fold. The protein is a dimer in the crystal and in solution. This regulator has no known DNA binding motifs and direct binding of BcRsaM to the cepI promoter could not be detected in in vitro assays. Therefore, we propose that the modulatory action of RsaM might result from interactions with other components of the QS machinery rather than from direct association with the DNA promoter.
Keywords: quorum sensing, bcam1869, BcRsaM, TofM, Burkholderia cenocepacia
Introduction
The Burkholderia species are betaproteobacteria that inhabit various ecosystems (reviewed in Vial et al. [1], where they play a wide variety of ecological roles. For instance, some species colonize the rhizosphere and engage in symbiosis that promotes plant growth. Some form nodules on the roots of legumes and are capable of fixing atmospheric nitrogen. They also occur naturally in water and soil, where they could potentially be exploited in bioremediation due to their ability to degrade a number of toxic compounds, including oil-derived pollutants and xenobiotics (reviewed in O’Sullivan et al. [2]. However, besides their positive relationships with other organisms and the environment, Burkholderia are also known as precarious human and plant pathogens, which raises safety concerns for their biotechnological applications.
Burkholderia cepacia complex (Bcc) is a group of at least eighteen species [3-6]. Some members are recognized as causative agents of pneumonia in immunocompromised patients with a preexisting lung disease, such as cystic fibrosis (CF) or chronic granulomatous disease (CGD). The most commonly identified pathogenic species in affected individuals are B. cenocepacia and B. multivorans [7]. In CF patients, Burkholderia is the second major pathogen responsible for chronic lung infection, with Pseudomonas aeruginosa holding the dishonorable first place. In extreme cases, Burkholderia-associated inflammation, induced by antibiotic resistant species, leads to the so-called “cepacia syndrome” – a condition manifesting in high fever, leukocytosis and progressive respiratory failure with poor prognosis of survival [8]. Molecular mechanisms of B. cenocepacia pathogenicity remain poorly understood. A detailed research of the Burkholderia species, and the B. cenocepacia epidemic lineage ET-12 in particular, enabled the identification of a number of virulence factors critical for bacterial invasion [9].
Expression of several virulence-related genes in B. cenocepacia is controlled by quorum sensing (QS) systems (reviewed by Subramoni and Sokol [10]. QS is a method of social communication that enables bacteria to coordinate behavior of their community in a cell-density–dependent manner. QS systems rely on the synthesis of small signaling molecules (autoinducers), their passive diffusion or active transport across the cell envelope [11-13] and their receptor-mediated detection by members of the same species. In proteobacteria, these signals are most often N-acyl-L-homoserine lactones (AHL). These compounds reprogram the gene transcription pattern to induce physiological processes that are beneficial for a larger bacterial population but could be nonproductive for individual cells. A typical QS system requires an AHL synthase that resembles LuxI of Vibrio fischeri and an AHL-dependent transcription factor that resembles the V. fischeri LuxR protein. As the bacterial population grows, AHL accumulates intra- and extracellularly until it reaches a critical level. At sufficiently high concentration, AHL binds to and activates the LuxR-type regulator, which subsequently affects expression of QS-controlled target genes, including QS genes themselves. A small number of LuxR-type proteins are active only as apoproteins and are functionally inactive when binding their cognate AHL [14]. Most LuxR-type proteins are transcription activators, but a few can function as transcriptional repressors.
All known species of Burkholderia encode proteins orthologous to the cepI and cepR genes of B. cenocepacia [15, 16]. These systems consist of a CepR transcription regulator and CepI autoinducer synthase that generates primarily N-octanoyl-L-homoserine lactone (OHL). CepI can also synthesize N-hexanoyl-L-homoserine lactone (HHL) when overexpressed in E. coli [17]. Besides CepIR, virulent B. cenocepacia strains encode the CciIR system that is found on the cenocepacia island (cci) [18]. CciI produces HHL and small amounts of OHL. For the most part, CepR operates as an activator while CciR acts as a repressor of the same pool of genes. In addition, some isolates of B. cenocepacia encode an additional LuxR-type protein called CepR2. CepR2 lacks a cognate AHL synthase and is therefore designated an orphan receptor. In one study, CepR2 was reported to function independently of AHLs [19], while a second study demonstrated that CepR2 functions only as an apoprotein and that its action is inhibited by OHL [20]. Apo-CepR2 represses several promoters located near the cepR2 gene [19, 20]. All B. cenocepacia QS mechanisms have complex regulatory connections and work together in an orchestrated manner. These relationships have been recently thoroughly reviewed by Subramoni and Sokol [10].
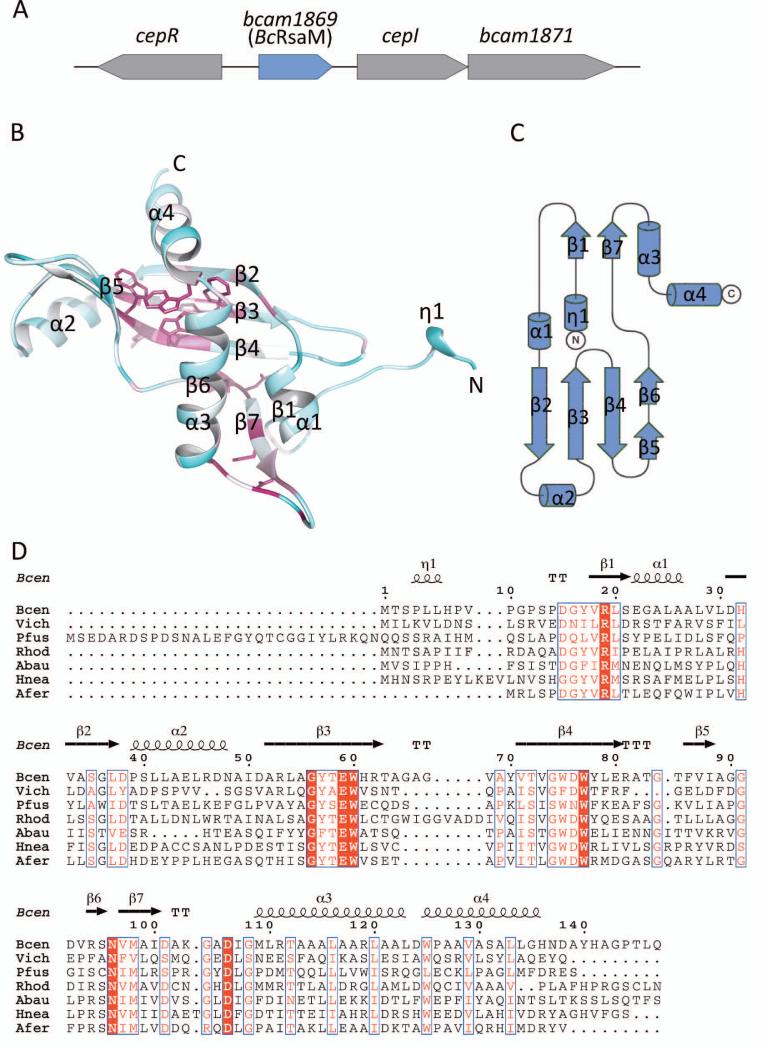
Recently, it became recognized that QS response might be further modulated by additional factors present on QS loci. luxIR modules show a number of different chromosomal arrangements within the QS loci. In the simplest scenarios, the two genes are located next to each other and are transcribed either divergently, or convergently, or in the same direction (for example cciIR) [21]. In more intricate cases, the lux genes are separated by one or more additional genes, multiplying the number of possible genetic architectures. QS loci may also contain other genes present outside of the luxIR-encoding region. The two latter situations may be illustrated by the B. cenocepacia cepIR locus (Fig. 1A), which contains bcam1869 and bcam1871 QS-related genes. bcam1869 is localized in between the divergent cepR and cepI genes and is transcribed in the same direction as cepI. bcam1871 is positioned downstream of cepI and is cotranscribed with cepI. Both these genes were recently shown to be activated by CepR-OHL complexes [22-25].
Fig. 1.
The bcam1869 gene of B. cenocepacia and its product, BcRsaM protein. A). Genetic organization of the B. cenocepacia bcam1869 locus. The bcam1869, cepI, and bcam1871 genes are transcriptionally induced by CepR-OHL complexes [25]. B). Overall fold of the B. cenocepacia BcRsaM monomer colored according to sequence conservation, with the most conserved residues shown in maroon (also in a stick representation) and the most variable ones shown in cyan. C). Topology diagram of BcRsaM. D). Multiple sequence alignment of B. cenocepacia BcRsaM with a subset of homologs: Bcen — B. cenocepacia J2315 (gi|206563717), Vich — V. ichthyoenteri (gi|493766304), Pfus — P. fuscovaginae (gi|290454886), Rhod — Rhodanobacter sp. 115 (gi|495487176), Abau — A. baumannii (gi|491275299), Hnea — H. neapolitanus c2 (gi|261855566), Afer — A. ferrooxidans ATCC 53993 (gi|198283773). The alignment was generated using the Muscle algorithm [59] and displayed using ESPript [60]. Strictly conserved residues are shown in white on a red background, moderately conserved ones are in a red font. The secondary structure elements for BcRsaM are shown above the alignment with α corresponding to α-helices, β to β-strands and η to helix 310. T indicates turns.
The bcam1869 product, here designated as BcRsaM, is the focus of the present study, which was undertaken because of reports suggesting that its homologs play modulatory roles in QS [22]. A homolog of BcRsaM, the RsaM protein from Pseudomonas fuscovaginae, was identified as part of the QS system of that organism [22]. P. fuscovaginae contains two QS systems, PfvIR and PfsIR, with the rsaM gene localized between pfsR and pfsI. Strikingly, the majority of cepR and cepI genes found in Burkholderia spp are separated by a gene that resembles rsaM [26]. Generally, genome-wide analyses indicate that the presence of rsaM-like genes is limited to β- and γ-proteobacteria with AHL-QS systems [27]. This group includes Burkholderia spp., Actinetobacter baumannii, Acidithiobacillus ferrooxidans, Halothiobacillus neapolitanus and, mentioned earlier, P. fuscovaginae [27]. In all genomes, the rsaM genes are found directly adjacent to the luxI homolog and are oriented in the same direction. Most commonly, it also neighbors the luxR–family gene, but occasionally these two genes are separated by additional ORFs. There are also examples of rsaM homologs that lack a nearby luxR gene (in B. cepacia AMMD and B. ambifaria MC40-6 strains) [26].
The RsaM protein from P. fuscovaginae, as well as its homolog from the B. cenocepacia H111 strain have been shown to down-regulate AHL production [22, 28]. This phenomenon has been attributed to repression of the luxI-like gene transcription, although posttranscriptional mechanisms have also been considered [28]. Remarkably, in the P. fuscovaginae study, RsaM influenced expression not only of its cognate AHL synthase, PfsI, but also inhibited PfvR-dependent pfvI transcription, despite the fact that the PfvIR system contains its own repressor, RsaL. A series of experiments with the B. cenocepacia H111 QS system also demonstrated that its BcRsaM protein influences expression or activity of the CepIR system as well as the Cep2R regulator [28]. Consequently, it affects transcription of the Cep-regulated downstream genes. Therefore, RsaM appears to be a major regulatory protein that fine-tunes the QS apparatus in β- and γ-proteobacteria.
As BcRsaM shares no sequence similarity with biochemically or structurally characterized proteins, the molecular basis of its modulatory action remains to be discovered. We have undertaken crystallographic studies and biochemical characterization of BcRsaM of B. cenocepacia strain J2315, hoping that structural information will provide some insights into the physiological function of BcRsaM in QS systems. Here we present the BcRsaM crystal structure determined at 2.3 Å resolution and biochemical characterization of the protein.
Results and discussion
Overall fold
BcRsaM is a one-domain protein (147 amino acid residues) with a melting temperature of 43.5° ± 0.5°. The core of the molecule consists of a five-stranded antiparallel β-sheet (strands ↑ -β2, ↓-β3, ↑ -β4, ↓-β5 and ↓-β6) that wraps around two α-helices (α3 and α4, Fig. 1B). The two pairs of chains, namely β3-β4 and β4-β5, are β-hairpins. The β5 and β6 elements are also consecutive elements of the polypeptide molecule, but instead of creating a turn, they adopt an extended conformation with both β-strands interacting with β4. Therefore, for topological purposes, β5 and β6 could be considered as one β-chain with an internal loop. The fold is completed by additional elements decorating the central module. These include a parallel β-sheet (strands β1 and β7) as well as helices α1 and α2. The N-terminus of the molecule protrudes from the otherwise globular molecule and adopts the conformation of a 310 helix, designated as η1.
Similarity to other proteins
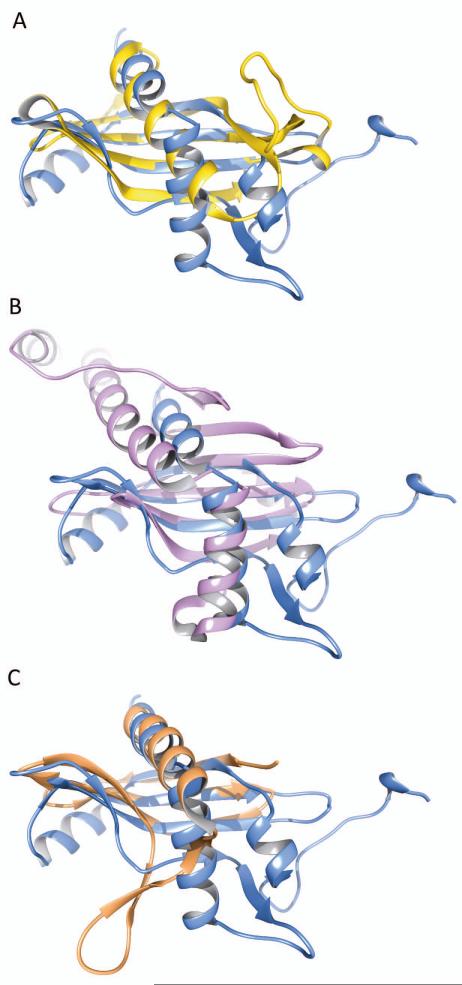
A search for structural homologs using the Dali server [29] does not reveal any significant hits; all 129 non-redundant hits have Z-scores lower than 4.0. Nevertheless, as the central structural motif (a β-sheet gripped around an α-helix) is not without a precedent, some curious matches were identified. For example, the retrieved entries of comparable size to BcRsaM include papain (Z-score 2.6, rmsd 2.8 Å, 55 aligned residues, PDB 3IMA), cystatins (cystatin 2, Z-score 2.4, rmsd 2.7 Å, 57 aligned residues, PDB 3L0R) and Der f 7 allergen (Z-score 3.2, rmsd 4.2 Å, 90 aligned residues, PDB 3UV1). Possibly the closest structural analogy can be observed with protein p22 from Trypanosoma brucei (Z-score 3.6, rmsd 3.5 Å, 79 aligned residues, PDB 3JV1). Another interesting distant relative is a SpoVG protein (Z-score 3.5, rmsd 3.2 Å, 69 aligned residues, PDB 2I9X). As shown in Fig. 2, the overall architecture of the protein core clearly resembles those mentioned above, which enables us to classify it as an (α/β)-roll, but its topology is very different. Therefore, we conclude that BcRsaM has a novel protein fold. Moreover, it does not appear to possess any known DNA-binding motifs. Interestingly, the family of SpoVG, has been shown recently to interact with DNA specifically [30]. The detailed comparison between SpoVG and BcRsaM reveals that our protein does not contain the key β-hairpin. That element in SpoVG bears conserved, positively charged residues crucial for DNA binding. In addition, the SpoVG dimer does not resemble any of the assemblies observed for BcRsaM (see below).
Fig. 2.
Superposition of the BcRsaM monomer with human cystatin C (A, PDB entry 3NX0), with T. brucei p22 (B, PDB entry 3JV1) and with SpoVG (C, PDB entry 2I9X).
Oligomeric state
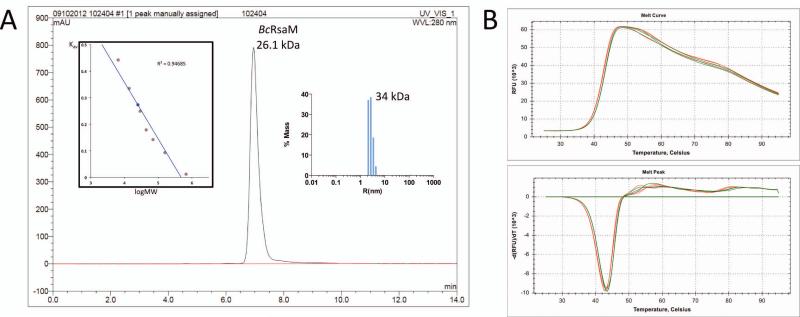
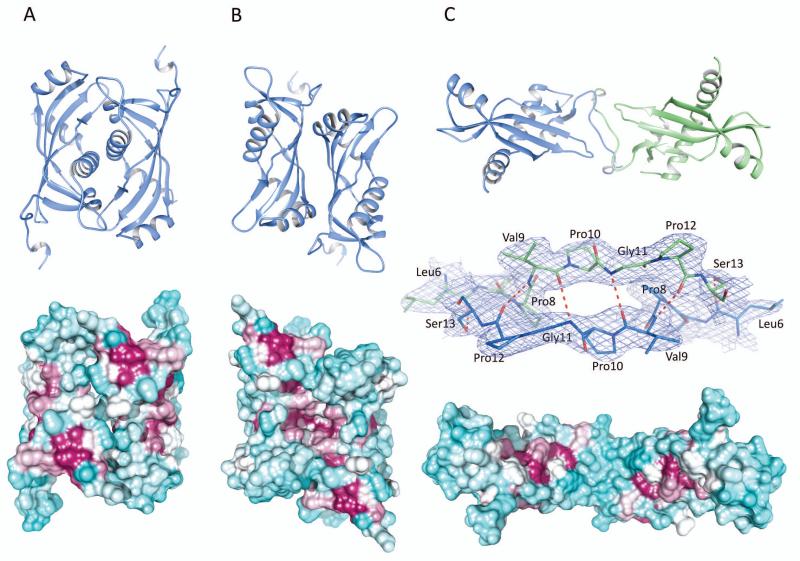
Size exclusion chromatography as well as DLS experiments indicated that the BcRsaM protein is a dimer in solution (26.1 kDa and 34.0 kDa for the two methods with the same protein concentration, respectively, Fig. 3). With a two-fold non-crystallographic and high crystal symmetry, the protein molecules contact with one another through several dimer-like interfaces. As calculated by the PISA server [31], four of them (designated as D1(A), D2(A), D2(B), D3(AB)) involve a buried area greater than 1,000 Å2 (per dimer, Fig. 4) and are considered as potential quaternary structures. The D1(A) assembly (Fig. 4A) can be described as a face-to-face dimer. It is created by molecule A and its crystallographic mate A’ (x-y, -y, -z) with hydrophobic helices α3 being the main interacting elements. Four hydrogen bonds link additional protein fragments as Glu22 (α1) of each subunit binds to Tyr78 (β4) and Gly90 (β5) of the opposite subunit. The dimer generates 1,490 Å2 of buried area, which is ~10% of the total surface area. However, the predicted ΔGdiss is low (1.3 kcal/mol) and the two molecules do not contact with each other through the entire interface area, i.e., two channels are present between the protein chains. Moreover, monomers B do not form an identical arrangement. Although an analogous pattern could be identified, with two B molecules having similar orientations as within the D1(A) assembly, translational components keep the monomers separated (only 53 Å2 of interface area per monomer). These observations suggest that dimer D1(A) is most likely a crystallographic artifact.
Fig. 3.
BcRsaM characterization. A). Molecular weight determination of BcRsaM using size exclusion chromatography and dynamic light scattering. The absorbance at 280 nm is plotted in absorbance units (AU) versus retention time in min. Left insert is a plot of Kav coefficient versus logarithm of molecular weight (red circles correspond to standard proteins: (1) aprotinin (6.5 kDa), (2) ribonuclease A (13.7 kDa), (3) carbonic anhydrase (29 kDa), (4) ovalbumin (44 kDa), (5) conalbumin (75 kDa), (6) aldolase (158 kDa), and (7) thyroglobulin (669 kDa); blue circle corresponds to BcRsaM (26.1 kDa). A clear peak is observed for the BcRsaM dimer, no other species were detected. Right panel shows DLS results representing particle size distribution for BcRsaM. B). DSF data for BcRsaM alone (green) and in the presence of OHL (red). The upper panel is a melting curve and the lower panel is its first derivative.
Fig. 4.
Potential dimers of BcRsaM. In the cartoon models (top), chains are colored according to crystallographic monomers involved in the dimer, with molecule A (or its crystallographic copy) shown in blue and molecule B shown in green. The molecular surfaces (bottom) are colored according to sequence conservation with maroon indicating conserved residues and cyan showing non-conserved ones. Sequence conservation calculation was done in Chimera [61] based on 120 sequences identified in three iterations of PSI-BLAST at NCBI and aligned in the Muscle program [59]. The molecules are oriented to present their more conserved side. For panel C, the middle picture presents the dimer interface shown in 2DFo-mFc electron density map contoured at 1σ level. A). Face-to-face dimer (D1(A)). B). Back-to-back dimer (D2(A)). C). Extended dimer (D3(AB)).
Dimer D2(A) (Fig. 4B) consists of molecule A and its crystallographic copy A” generated through -y, -x, -z-1/6 operation. The A-A” assembly generates 1,840 Å2 (13% of the total surface area) of buried area. An equivalent dimer D2(B) created by monomers B provides a comparable buried area (1,820 Å2) suggesting that the two D2 oligomers represent the same protein state. Nevertheless, both D2 variants have low ΔGdiss (1.7 kcal/mol for A). In fact, predicted ΔGdiss for the B-molecule-based dimer is even lower than for the A-A” dimer. D2 associations are created by back-to-back interaction of the β-sheets. It appears to be mostly hydrophobic as no direct protein-protein hydrogen bonds could be found.
The above dimers have rather globular shapes but a more elongated particle, D3(AB) (Fig. 4C), could also be identified. It includes two polypeptide chains present in the asymmetric unit. Although the buried area of this arrangement is low (1,040 Å2) and its predicted ΔGdiss has the lowest value from all possible assemblies (−0.9 kcal/mol), which indicates an unstable arrangement. The molecules contact with each other through their N-terminal, proline-rich regions (L6HPVPGPS13). Although the interacting fragments do not form typical β-strands that could be recognized by secondary structure analyzing programs, such as DSSP [32], they do generate an intermolecular β-sheet–like association with four main-chain–main-chain hydrogen bonds and two additional side-chain–main-chain interactions (Fig. 4C).
Curiously though, some of the RsaM family members are shorter and do not contain the η1-β1 stretch. On the other hand, there are also homologs with extra elements on the N-terminus (~120 amino acids longer than BcRsaM). The variability of the N-terminus and high sequence conservation of the solvent-exposed face of the β-sheet (see below) suggest that dimers D2 (D2(A)/D2(B)), with two molecules interacting through back-to-back fashion, may represent the biologically relevant oligomeric state of BcRsaM. That is also consistent with the size exclusion chromatography data: the apparent molecular weight of the protein is smaller (26.1 kDa) than the predicted value from the BcRsaM amino acid sequence (31.0 kDa), which indicates a compact, globular shape. The D2 dimer has the largest interface with several conserved residues (Glu59, Asp76, Arg94 and Asn96 patch, see below), it can easily accommodate gene extensions and insertions present in the family, shows good surface electrostatic complementarity and there are a number of hydrophobic residues that are buried on the interface. We believe BcRsaM is not capable of forming a dimer observed in DNA-binding SpoVG.
Sequence conservation
Mapping of the PSI-BLAST-based sequence conservation on the protein structure reveals three conserved regions (Figs. 1D, 4). The most striking one is the core of the molecule occupied by a quartet of strictly preserved tryptophan residues (Trp60, Trp75, Trp77 and Trp125). This hydrophobic core cluster is clearly a unique feature of the RsaM fold. An additional patch of conserved residues is found facing the protein interior, including residues Leu30, Leu116, Thr58 and Ser95. On the protein surface, two regions seem to be maintained across the protein family. One is created by the solvent-facing part of the β-sheet and includes Asp76, a salt bridge between Arg94 – Glu59 and the neighboring Asn96 (part of the D2 dimer interface). The second solvent-exposed conserved fragment is created by the β1-β7 sheet and the loop following the β7 strand, with residues Met98, Gly104, Asp106 and Gly108 being the least variable. The conformation of this protein segment is maintained not only through the main-chain hydrogen bonds within the β-sheet, but also additional interactions, including bifurcated salt bridging of Arg19 (with Asp15 and Asp106) and a side-chain – main-chain hydrogen bond between Asp101 and Gly104 within a β-turn (seen only in chain A; an equivalent fragment of chain B has not been modeled due to poor electron density).
Functional implications
To the best of our knowledge, neither RsaM of B. cenocepacia nor any homologous proteins have been subjected to thorough biochemical analysis. However, one genetic study of BcRsaM and two studies of related proteins have recently been published and provide suggestive evidence that RsaM may play a role in transcriptional regulation. In a study describing a homolog from the B. cenocepacia H111 strain, a transposon insertion between bcam1869 and the adjacent cepR gene was compared to the parent strain using DNA microarrays [28]. This mutation altered the expression of 118 genes, though it was not clear whether the mutation affected bcam1869, or cepR or both. A null mutation in bcam1869 was also constructed using an antibiotic resistance cassette, although this cassette could also influence expression of the flanking cepR and cepI genes. This mutant showed only mild decreases in swarming motility, biofilm formation, protease production, and pathogenicity in C. elegans. Both mutations caused an increase in production of N-acyl-L-homoserine lactones. On the contrary, BcRsaM overexpression caused a decrease in AHL production. Paradoxically, the transposon insertion caused a decrease in transcription of the AHL synthase gene cepI, suggesting that RsaM could act posttranscriptionally. Interestingly, expression of BcRsaM in E. coli strongly blocked the activity of LuxR. This was not due to destruction of exogenous AHL signal molecules, suggesting an interaction between BcRsaM and the LuxR protein or the luxI promoter.
The second study analyzed the RsaM protein from P. fuscovaginae, which is somewhat distantly related to BcRsaM (32% identity). This plant pathogenic bacterium encodes two LuxIR systems, one of which is composed of PfsR and PfsI [22]. The pfsR and pfsI pair flanks the rsaM gene. Random transposon mutants were screened for elevated production of AHL signals, yielding an insertion mutant in the rsaM gene. The defect was only partially complemented by a plasmid copy of rsaM, suggesting that the phenotype could be partly or fully due to cis-acting effects. When the PfsIR system was reconstituted in E. coli, expression of RsaM decreased expression of a pfsI-lacZ fusion (although the data were not provided). The authors concluded that RsaM somehow inhibited the activity or expression of PfsR or PfsI. It is possible that BcRsaM could have similar properties, despite their somewhat weak similarity.
Perhaps the most compelling genetic analysis was obtained using the tofR, tofM, and tofI genes of the plant pathogen B. glumae [33]. TofM resembles BcRsaM (60% identity). Single deletions of tofR, tofM, or tofI reduced production of toxoflavin. A mutant lacking tofR and tofI showed a residual synthesis of toxoflavin, while a tofR, tofI, tofM triple deletion mutant produced no detectable toxoflavin. An unmarked deletion of tofM in an otherwise wild type chromosome also decreased toxoflavin production under some conditions, and this defect was fully complemented using a cloned copy of tofM. These data suggest a level of functional redundancy, in that TofR-AHL complexes can activate production of toxoflavin in the absence of TofM and that TofM can also do so in the absence of TofR-AHL complexes. The authors concluded that TofM is most likely a transcription regulator. The stimulation of transcription by TofM stands in contrast to the inhibitory effects described for BcRsaM and RsaM. Therefore it appears that the functional redundancy is obtained using different mechanisms. TofR regulates by binding to the TofI promoter and TofM regulates TofI by some other unknown mechanism.
Following the above evidence for transcription regulation, we have analyzed BcRsaM interaction with dsDNA. The DNA promoter region in front of the cepI gene in B. cenocepacia carries a semi-palindromic sequence that could potentially serve as a regulator-binding site. However, electrophoretic mobility shift assay (EMSA) experiments with BcRsaM and four different DNA fragments containing the presumed BcRsaM binding site showed no interaction even at 10-fold protein/DNA excess (data not shown). We also did not detect any non-specific binding to dsDNA. It remains possible that this protein could bind to one or more specific DNA sequences located elsewhere, but this is not consistent with a cis acting element. The RsaM electrostatic surface potential is also inconsistent with DNA binding as no major positively charged patches are found needed for interaction with the phosphosugar backbone of the DNA duplex or RNA molecule.
In addition, BcRsaM does not seem to interact with OHL – a cognate signaling ligand for the CepIR system that could influence the regulatory function of the protein. The presence of OHL did not influence RsaM stability or the hydrodynamic properties, suggesting the lack of interaction (Fig. 3). By analogy to other transcription regulators, the ligand molecule might trigger the conformational or oligomerization state change of the protein, switching it from an active to non-active state, or vice versa. Such a significant structural rearrangement should give a clear signal in differential scanning fluorimetry (DSF) and also in dynamic light scattering (DLS) assays. That is not the case for BcRsaM, suggesting that its function is independent of OHL. Moreover, BcRsaM does not contain DNA-binding structural motifs in the monomer (see above) and all reported possible combinations of dimers, so it is very likely that it does not interact with DNA directly. Similar transcription regulators have been described before, for example mannose operon regulator MtlR [34] or TraM – another QS-related regulator [35]. We cannot exclude, however, that BcRsaM influences the QS system at a posttranscriptional and/or posttranslational level.
Conclusion
Previous genetic studies of the RsaM role in CepIR-based QS systems strongly suggested that the protein is a cis-acting transcription regulator. To further investigate this hypothesis, we have examined BcRsaM for interaction with the relevant ligand molecule, OHL, and the cepI promoter, however no OHL and/or DNA binding was observed. Based on SEC, DLS and crystallographic data, BcRsaM appears to form a dimeric assembly. The protein monomer reveals a novel fold, which lacks known DNA binding motifs. Also none of the possible dimeric arrangements, including the most likely D2, bears recognizable DNA-interacting elements. Moreover, no other enzyme active sites signatures or RNA-binding motifs have been detected [36] and the protein does not appear to contain cavities that could accommodate ligand molecules, like OHL. Hence, we propose that the action of RsaM might result from interactions with other components of the transcription or translation machinery rather than from direct association with the DNA promoter, which appears unlikely given the structural features and preliminary biochemical characterization.
Materials and Methods
Cloning to vector pMCSG73 and in vitro TVMV cleavage of NusA-BcRsaM fusion
The full length bcam1869 gene from B. cenocepacia J2315 was amplified from the genomic DNA with KOD DNA polymerase (Novagen, WI, USA) using conditions and reagents provided by the manufacturers and cloned into vector pMCSG73 according to the ligation-independent procedure [37, 38]. Protein targets expressed from vector pMCSG73 are produced as C-terminal fusions to E. coli transcriptional factor NusA in the following form: NusA-(TVMV recognition site)-His6-Strep tag-(TEV recognition site)-TARGET (NusA-ETVRFQ/S-HHHHHH-WSHPQFEK-ENLYFQ/SNA-TARGET) (the underlined sequence shows the TEV recognition site included in the PCR primer, “/” indicates the actual cleavage site). The fusion is subsequently cleaved in vitro using TVMV protease by disrupting protein target expressing cells in the presence of TVMV-producing cells. The amount of TVMV overexpressed in 1 L of growth media is sufficient to cleave the NusA-target fusion from 10 L of target producing bacteria. The cleavage process typically occur within 2 h after the start of sonication.
Protein expression and purification
A bacterial culture of the E. coli BL21-Gold(DE3) strain carrying pMCSG73-bcam1869 was grown in 1 L of enriched M9 medium [39] at 37°C, shaking at 200 rpm until it reached an OD600 of 1.0. Inhibitory amino acids (25 mg each of L-valine, L-isoleucine, L-leucine, L-lysine, L-threonine, L-phenylalanine) and 70 or 90 mg of selenomethionine (SeMet) (Orion Enterprises, IL, USA) were added to the culture, which was then cooled to 4°C for 60 min. Protein expression was induced by 0.5 mM isopropyl-β-D-thiogalactoside. The cells were incubated overnight at 18°C, then harvested and resuspended in lysis buffer (500 mM NaCl, 5% glycerol, 50 mM HEPES pH 8.0, 20 mM imidazole, and 10 mM β-mercaptoethanol). To remove NusA, 3 ml of TVMV cell suspension (OD600 ~ 70) were added to the target cells, which were then disrupted by sonication. Insoluble cellular material was removed by centrifugation. SeMet-labeled protein was purified using Ni-NTA affinity chromatography and the ÄKTAxpress system (GE Healthcare Biosciences, PA, USA) as described previously [40, 41]. This was followed by cleavage of the His6 tag using recombinant His7-tagged TEV protease and an additional step of subtractive Ni-NTA affinity chromatography to remove the protease, affinity tag, and any uncut protein. Native BcRsaM was also produced using the same approach. The cells were grown in the presence or absence of 10 μM OHL using a mixture of M9 and LB media and both proteins were purified using the procedure described above. The pure proteins were concentrated using an Amicon Ultra-15 concentrator (Millipore, MA, USA) in buffer A (20 mM HEPES pH 8.0, 250 mM NaCl, and 2 mM dithiothreitol (DTT)). Protein concentrations were determined from the absorbance at 280 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific, MA, USA).
Size exclusion chromatography
The molecular weight of BcRsaM in solution was determined by size exclusion chromatography using a Dionex HPLC equipped with an AS temperature-controlled autosampler housing two 96-well plate sample racks, a GP50 Gradient Pump, a PDA-100 Photodiode Array Detector (Thermo Scientific, CA, USA), and a SRT SEC-150 column from Sepax Technologies (DE, USA). The column was equilibrated with buffer A and calibrated with aprotinin (6.5 kDa), ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), conalbumin (75 kDa), aldolase (158 kDa), and thyroglobulin (669 kDa). 30 μl of BcRsaM at 5 mg/ml in buffer A was loaded onto the column. The separation was carried out at room temperature at a flow rate of 1 ml/min and absorbance at 280 nm was monitored. The calibration curve of Kav versus log molecular weight was prepared using the equation Kav = (Ve–Vo/)/(Vt–Vo), where Ve = elution volume for the protein, Vo = column void volume, and Vt = total bed volume.
Differential scanning fluorimetry
A DSF assay was performed using a Bio-Rad CFX96 real-time thermal cycler (CA, USA) to determine the thermal stability of RsaM alone and with OHL. The following samples were prepared in 40 μl reactions containing 5× SYPRO Orange (Invitrogen S-6650): (1) 20 μM RsaM control, (2) 20 μM RsaM control containing 0.05% ethyl acetate, and (3) 20 μM RsaM with 50 μM OHL (prepared in ethyl acetate). Each sample was performed in triplicate. Samples were held at 25°C for 1 min, then increased by 0.5°C at 30 s intervals. A first derivative plot (−d(RFU)/dT) was used to determine the melt peaks.
Dynamic light scattering
DLS was used to determine the molecular weight distribution of BcRsaM at 23°C. RsaM was prepared in degassed crystallization buffer at 5 mg/ml. 20 μl RsaM was loaded onto a 384-well clear bottom plate (Corning 3540). The plate was centrifuged at 1000 rpm for 2 min before loading into a DynaPro Plate Reader instrument (Wyatt Technology Corporation, CA, USA). DLS was also used to determine the thermal stability of RsaM in the absence and presence of OHL. The following samples were prepared in 20 μl reactions: (1) RsaM (native) control and (2) RsaM with 50 μM OHL. The samples, at 5 mg/ml, were centrifuged at 13,000 rpm for 5 min. Each sample was loaded onto a 384-well clear bottom plate and overlaid with 7 μl mineral oil. The plate was centrifuged at 1000 rpm for 2 min before loading into a plate reader. Measurements were taken at 1°C intervals between 25°C and 55°C. Melting curves were determined.
Electrophoretic mobility shift assay (EMSA)
EMSA was used to measure protein-nucleic acid interactions [42]. The assays were conducted using four oligonucleotides: a 145 bp fragment containing the cepI promoter containing an imperfect palindrome (underlined) (1) 5′GGTTTTCAATCCCGTTGATCAAGAAACCGTTACCACGTCCCGAATGGCGTCTTTACGCCGTCACCCTGTAAGAGTTACCAGTTACAGGCTCCTCGTGCCGCGCGCTGTAATGCACGCATACAAAAGCACAGATCCGAGGACATCC3′. This fragment was generated by PCR using two primers (5′GGATGTCCTCGGATCTGTGCTTTT3′ and 5′GGTTTTCAATCCCGTTGATCAAGAAACC3′). Three synthetic DNA hairpins containing possible binding sites for BcRsaM were also used ((2) 5′ATGGCGTCTTTACGCCGTCCCCCACGGCGTAAAGACGCCAT3′, (3) 5′ACCCTGTAAGAGTTACCAGTTACAGGCTCCCCCAGCCTGTAACTGGTAACTCTTACAGGGAT3′ and (4) 5′GTTACAGGCTCCTCGTGCCGCGCGCTGTAATCCCCCATTACAGCGCGCGGCACGAGGAGCCTGTAAC3′). Prior to the assay, DNA hairpins were heated to 90°C for 1 min and then allowed to cool to 24°C. Two BcRsaM protein samples were used in EMSA, one produced in the absence of OHL and one produced in the presence of OHL. The samples were prepared in 10 μl reactions in which the DNA amount was constant at 40 ng per reaction. The protein/DNA ratio varied from 1:1, 1:5, and 1:10 and samples were size-fractionated by gel electrophoresis using 6% TBE DNA retardation native gels (Invitrogen EC63655BOX). The reaction for oligo (1) contained 2 μl of 5× binding buffer (Invitrogen E33075): 750 mM KCl, 0.5 mM DTT, 0.5 mM EDTA, and 50 mM Tris pH 7.4. The reactions for oligos (2) through (4) contained 1 μl of 10× binding buffer: 100 mM Tris, 10 mM EDTA, 1 M KCl, 1 mM DTT, and 50% glycerol. The samples were incubated at 37°C for 30 min, then cooled on ice. Prior to loading samples onto the gels, 2 μl of EMSA gel-loading solution (Invitrogen E33075) was added to each sample, then centrifuged. The entire 12 μl volume of each sample was loaded onto the gels. The gels were stained for DNA using SYBR Green (Invitrogen E33075) for 20 min in the dark, then washed twice with 150 ml H2O for 10 s before visualizing. The gels were then stained for protein using SYPRO Ruby (Invitrogen E33075) for 3 h in the dark, then destained with a solution containing 10% methanol and 7% acetic acid for 1 h before visualizing. Protein and DNA alone controls were run as well. As a positive DNA-binding control two DNA binding proteins, LuxT [43] and OccR [44] were used with their known respective DNA binding sites.
Crystallization
Both native and SeMet-labeled BcRsaM were screened for crystallization conditions using a Mosquito liquid dispenser (TTP LabTech, Melbourn, UK) and sitting drop vapor diffusion technique in 96-well CrystalQuick plates (Greiner Bio-one, NC, USA). The protein was set up at 16°C using the MCSG 1–4 screens from Microlytic Inc. (MA, USA). For each condition, 0.4 μl of protein and 0.4 μl of crystallization formulation were mixed and then equilibrated against 140 μl of the reservoir solution. The protein concentration varied depending on the preparation: native protein was set up at 54 mg/ml, SeMet derivatives were set up at 47 mg/ml and 52 mg/ml for the protein expressed in the presence of 90 mg/L and 70 mg/L SeMet in the culture, respectively. The crystals appeared under a number of conditions. For structure solution and refinement, four crystal variants were ultimately used. Two of them were obtained for the protein grown in the presence of SeMet in the culture, designated as crystals X1 (90 mg/L SeMet, 0.2 M calcium acetate, 0.1 M imidazole:HCl pH 8.0, 20% PEG1000) and crystals X4 (70 mg/L SeMet, 0.2 M calcium acetate, 0.1 M HEPES:NaOH pH 7.5, 10% PEG8000). The other two crystals were for the native protein: crystals X2 (0.14 M lithium citrate, 19% PEG3350) and crystals X3 (0.27 M lithium citrate, 25% PEG3350).
Data collection
Crystals X1 were briefly soaked in a cryoprotectant solution containing 15% ethylene glycol in mother liquor. Crystals X2 were soaked in a solution containing 0.25 M lithium citrate, 30% PEG3350 and 0.7 M KI for 40 s. Crystals X3 were soaked in 1 mM K2PtCl4 (in 0.25 M lithium citrate, 30% PEG3350) for 10 min and for another 10 min in 10 mM K2PtCl4. After soaking, all of the derivatized crystals were cryoprotected in a 5% glycerol solution (in 0.25 M lithium citrate, 30% PEG3350) and flash-cooled in liquid nitrogen. Crystals X4 were cryoprotected in reservoir buffer supplemented with 10% glycerol. All X-ray diffraction experiments were performed at the Structural Biology Center 19-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Datasets were collected at 100 K. Diffraction images were processed with the HKL3000 program suite [45]. The resulting sca files for crystals X1, X2 and X3 were directly fed into the autoSHARP program [46]. Structure factor intensities for crystals X4 were converted to amplitudes in Ctruncate [47] from the CCP4 package [48]. Crystals X1, X2 and X3 belong to crystal form 1 (P6222) while crystals X4 represent crystal form 2 (P6122), with a unit cell roughly twice as large as in form X1 due to parameter c doubling. Consequently, while form 1 contains one protein molecule in the asymmetric unit, form 2 accommodates two chains.
Structure solution
The structure was solved by the Multiple Isomorphous Replacement with Anomalous Scattering (MIRAS) approach as implemented in autoSHARP [46] followed by Molecular Replacement (MR) using Phaser [49]. Specifically, in the autoSHARP protocol, crystals X1 were treated as native while crystals X2 and X3 as heavy atom derivatives. The procedure included initial data manipulation (converting intensities to amplitudes in Truncate [47], scaling (SCALEIT [50], identification of the heavy atom sites (SHELXD [51], phasing (SHARP [52] density modification (SOLMON [53] and automatic model building (ARP/wARP [54]. Five iodide and two platinum (II) sites were located and the results of phase determination are provided in Table 1. The automatic model building traced only 58 residues out of 147 and no side chains were assigned. This largely incomplete initial structure was fed into Buccaneer [55] giving a model with 112 residues, in which side chains for 81 residues were autotraced. Despite significant improvement, the model failed to refine against the crystal X1 reflection file and several regions of the electron density map were ambiguous, preventing further manual model building. The unsuccessful refinement could not be attributed to an incorrectly assigned space group and/or twinning, but the diffraction image did show unusual features manifesting in diffuse scattering, which suggests the possibility of order-disorder effects.
Table 1. Data collection and refinement statistics.
| Data collection | ||||
|---|---|---|---|---|
| Crystal | X1 (Native 1) | X2 (KI) | X3 (K2PtCl4) | X4 (Native 2) |
| Space group | P6222 | P6222 | P6222 | P6122 |
| Cell dimensions [Å] | a= 66.6 c=103.4 | a= 66.4 c=102.6 | a= 66.7 c=104.6 | a= 69.1 c=216.8 |
| Temperature [K] | 100 | 100 | 100 | 100 |
| Radiation source | APS, 19-ID | APS, 19-ID | APS, 19-ID | APS, 19-ID |
| Wavelength [Å] | 0.9791 | 1.5498 | 1.0719 | 0.9793 |
| Resolution [Å]a | 30.00 – 2.00 (2.03 – 2.00) |
50.00 – 1.87 (1.90 – 1.87) |
50.00 – 2.75 (2.80 – 2.75) |
50.00 – 2.30 (2.35 – 2.30) |
| Unique reflections | 9,654 (466) | 10,358 (143) | 3,970 (194) | 14,517 (925) |
| Rmergeb | 0.049 (0.812) | 0.044 (above 1) | 0.043 (above 1) | 0.059 (0.847) |
| <I>/<σI> | 42.4 (3.5) | 49.8 (0.23) | 61.3 (0.85) | 42.0 (3.8) |
| Completeness [%] | 92.2 (100) | 88.5 (25.2) | 99.8 (100) | 99.8 (100) |
| Redundancy | 10.7 (11.0) | 11.4 (2.5) | 19.1 (16.3) | 13.5 (14.2) |
| Phasing (for resolution range 33.31 – 1.99 Å) | ||||
| Phasing power (acentric/centric) |
1.33/1.14 | 0.65/0.69 | ||
| Phasing power (anomalous) |
1.483 | 1.143 | ||
| FOM (acentric/centric) | 0.36/0.38 | |||
| Refinement | ||||
| Resolution [Å] | 20.20 -2.30 | |||
| Reflections work/test set |
13688/725 | |||
| Rfree / Rworkc | 0.215/0.257 | |||
| No. of atoms protein/water |
1967/31 | |||
| Average B factor [Å2] protein/water |
65.3/51.5 | |||
| Rms deviations from ideal | ||||
| bond lengths [Å] | 0.011 | |||
| bond angles [°] | 1.07 | |||
| Ramachandran statistics of φ/ψ angles [%] | ||||
| most favored | 98.5 | |||
| outliers | 0 | |||
| Molprobity score | 1.18 | |||
| Clashscore | 0.77 | |||
Values in parentheses correspond to the highest resolution shel .
Rmerge = ΣhΣj|Ihj–<Ih>|/ΣhΣjIhj, where Ihj is the intensity of observation j of reflection h.
R = Σh|Fo|-|Fc|/Σh|Fo| for all reflections, where Fo and Fc are observed and calculated structure factors, respectively. Rfree is calculated analogously for the test reflections, randomly selected and excluded from the refinement.
Therefore, the partial MIRAS model was used as a template for molecular replacement phasing of crystal form X4. MR solution was used to complete and refine the structure. Manual model rebuilding against electron density maps was performed in Coot [56], while crystallographic refinement was carried out in Buster [57]. The protocol included optimization of TLS parameters with seven groups per protein molecule. In the final structure, for chain A residues Thr2 – Asp138 have been modeled while chain B contains residues Ser3 – Ile100 and Ala105 – Asp138. The remaining residues are not well-defined in the electron density maps and have not been built. In addition to the protein molecules, 31 water molecules have been identified. The quality of the final protein model was verified by the Molprobity server [58]. The refinement results are summarized in Table 1. The BcRsaM atomic coordinates were deposited to the PDB under entry 4O2H.
ACKNOWLEDGMENTS
The authors would like to thank Drs. Zbyszek Dauter, Zbyszek Otwinowski and Tom Terwilliger for help with diffraction data analysis and members of the Midwest Center for Structural Genomics and Structural Biology Center for their support. This research has been funded in part by a grant from the National Institutes of Health GM094585 (AJ), and by the U.S. Department of Energy, Office of Biological and Environmental Research, under Contract DE-AC02-06CH11357.
Abbreviations
- AHL
N-acyl-L-homoserine lactone
- Bcc
Burkholderia cepacia complex
- EMSA
Electrophoretic Mobility Shift Assay
- OHL
N-octanoyl-L-homoserine
- QS
quorum sensing
- rmsd
root mean square deviation
Footnotes
Author Contribution Statement
KM performed crystallographic work and structural analysis; GC did expression, purification, crystallization and assays; SC and RJ did cloning; GB and AJ designed EMSA; KM, SW and AJ wrote the paper.
The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Databases: The atomic coordinates and structure factors have been deposited in the Protein Data Bank under entry 4O2H.
References
- 1.Vial L, Chapalain A, Groleau MC, Deziel E. The various lifestyles of the Burkholderia cepacia complex species: a tribute to adaptation. Environ Microbiol. 2011;13:1–12. doi: 10.1111/j.1462-2920.2010.02343.x. [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan LA, Mahenthiralingam E. Biotechnological potential within the genus Burkholderia. Lett Appl Microbiol. 2005;41:8–11. doi: 10.1111/j.1472-765X.2005.01758.x. [DOI] [PubMed] [Google Scholar]
- 3.Lipuma JJ. Update on the Burkholderia cepacia complex. Curr Opin Pulm Med. 2005;11:528–533. doi: 10.1097/01.mcp.0000181475.85187.ed. [DOI] [PubMed] [Google Scholar]
- 4.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int J Syst Evol Microbiol. 2009;59:102–111. doi: 10.1099/ijs.0.001123-0. [DOI] [PubMed] [Google Scholar]
- 5.Vanlaere E, Lipuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int J Syst Evol Microbiol. 2008;58:1580–1590. doi: 10.1099/ijs.0.65634-0. [DOI] [PubMed] [Google Scholar]
- 6.Peeters C, Zlosnik JE, Spilker T, Hird TJ, LiPuma JJ, Vandamme P. Burkholderia pseudomultivorans sp. nov., a novel Burkholderia cepacia complex species from human respiratory samples and the rhizosphere. Syst Appl Microbiol. 2013;36:483–489. doi: 10.1016/j.syapm.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Reik R, Spilker T, Lipuma JJ. Distribution of Burkholderia cepacia complex species among isolates recovered from persons with or without cystic fibrosis. J Clin Microbiol. 2005;43:2926–2928. doi: 10.1128/JCM.43.6.2926-2928.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isles A, Maclusky I, Corey M, Gold R, Prober C, Fleming P, Levison H. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J Pediatr. 1984;104:206–210. doi: 10.1016/s0022-3476(84)80993-2. [DOI] [PubMed] [Google Scholar]
- 9.McClean S, Callaghan M. Burkholderia cepacia complex: epithelial cellpathogen confrontations and potential for therapeutic intervention. J Med Microbiol. 2009;58:1–12. doi: 10.1099/jmm.0.47788-0. [DOI] [PubMed] [Google Scholar]
- 10.Subramoni S, Sokol PA. Quorum sensing systems influence Burkholderia cenocepacia virulence. Future Microbiol. 2012;7:1373–1387. doi: 10.2217/fmb.12.118. [DOI] [PubMed] [Google Scholar]
- 11.Minagawa S, Inami H, Kato T, Sawada S, Yasuki T, Miyairi S, Horikawa M, Okuda J, Gotoh N. RND type efflux pump system MexAB-OprM of pseudomonas aeruginosa selects bacterial languages, 3-oxo-acyl-homoserine lactones, for cell-to-cell communication. BMC Microbiol. 2012:12. doi: 10.1186/1471-2180-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan HB, Greenberg EP. Diffusion of Autoinducer Is Involved in Regulation of the Vibrio-Fischeri Luminescence System. J Bacteriol. 1985;163:1210–1214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohler T, van Delden C, Curty LK, Hamzehpour MM, Pechere JC. Overexpression of the MexEF-OprN multidrug efflux system affects cell-to-cell signaling in Pseudomonas aeruginosa. J Bacteriol. 2001;183:5213–5222. doi: 10.1128/JB.183.18.5213-5222.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsai CS, Winans SC. LuxR-type quorum-sensing regulators that are detached from common scents. Mol Microbiol. 2010;77:1072–1082. doi: 10.1111/j.1365-2958.2010.07279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutter E, Lewenza S, Dennis JJ, Visser MB, Sokol PA. Distribution of quorumsensing genes in the Burkholderia cepacia complex. Infect Immun. 2001;69:4661–4666. doi: 10.1128/IAI.69.7.4661-4666.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewenza S, Conway B, Greenberg EP, Sokol PA. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J Bacteriol. 1999;181:748–756. doi: 10.1128/jb.181.3.748-756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotschlich A, Huber B, Geisenberger O, Togl A, Steidle A, Riedel K, Hill P, Tummler B, Vandamme P, Middleton B, et al. Synthesis of multiple N-acylhomoserine lactones is wide-spread among the members of the Burkholderia cepacia complex. Syst Appl Microbiol. 2001;24:1–14. doi: 10.1078/0723-2020-00013. [DOI] [PubMed] [Google Scholar]
- 18.Malott RJ, Baldwin A, Mahenthiralingam E, Sokol PA. Characterization of the cciIR quorum-sensing system in Burkholderia cenocepacia. Infect Immun. 2005;73:4982–4992. doi: 10.1128/IAI.73.8.4982-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malott RJ, O’Grady EP, Toller J, Inhulsen S, Eberl L, Sokol PA. A Burkholderia cenocepacia orphan LuxR homolog is involved in quorum-sensing regulation. J Bacteriol. 2009;191:2447–2460. doi: 10.1128/JB.01746-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan GT, Wei Y, Winans SC. A LuxR-type repressor of Burkholderia cenocepacia inhibits transcription via antiactivation and is inactivated by its cognate acylhomoserine lactone. Mol Microbiol. 2013;87:94–111. doi: 10.1111/mmi.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gelencser Z, Choudhary KS, Coutinho BG, Hudaiberdiev S, Galbats B, Venturi V, Pongor S. Classifying the topology of AHL-driven quorum sensing circuits in proteobacterial genomes. Sensors (Basel) 2012;12:5432–5444. doi: 10.3390/s120505432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mattiuzzo M, Bertani I, Ferluga S, Cabrio L, Bigirimana J, Guarnaccia C, Pongor S, Maraite H, Venturi V. The plant pathogen Pseudomonas fuscovaginae contains two conserved quorum sensing systems involved in virulence and negatively regulated by RsaL and the novel regulator RsaM. Environ Microbiol. 2011;13:145–162. doi: 10.1111/j.1462-2920.2010.02316.x. [DOI] [PubMed] [Google Scholar]
- 23.O’Grady EP, Viteri DF, Sokol PA. A unique regulator contributes to quorum sensing and virulence in Burkholderia cenocepacia. PLoS One. 2012;7:e37611. doi: 10.1371/journal.pone.0037611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weingart CL, White CE, Liu S, Chai Y, Cho H, Tsai CS, Wei Y, Delay NR, Gronquist MR, Eberhard A, et al. Direct binding of the quorum sensing regulator CepR of Burkholderia cenocepacia to two target promoters in vitro. Mol Microbiol. 2005;57:452–467. doi: 10.1111/j.1365-2958.2005.04656.x. [DOI] [PubMed] [Google Scholar]
- 25.Wei Y, Ryan GT, Flores-Mireles AL, Costa ED, Schneider DJ, Winans SC. Saturation mutagenesis of a CepR binding site as a means to identify new quorumregulated promoters in Burkholderia cenocepacia. Mol Microbiol. 2011;79:616–632. doi: 10.1111/j.1365-2958.2010.07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choudhary KS, Hudaiberdiev S, Gelencser Z, Goncalves Coutinho B, Venturi V, Pongor S. The Organization of the Quorum Sensing luxI/R Family Genes in Burkholderia. Int J Mol Sci. 2013;14:13727–13747. doi: 10.3390/ijms140713727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venturi V, Rampioni G, Pongor S, Leoni L. The virtue of temperance: built-in negative regulators of quorum sensing in Pseudomonas. Mol Microbiol. 2011;82:1060–1070. doi: 10.1111/j.1365-2958.2011.07890.x. [DOI] [PubMed] [Google Scholar]
- 28.Inhülsen S. Ph.D. Dissertation. University of Zurich, Faculty of Science; 2011. Investigations on the Quorum sensing Circuitry in Burkholderia cenocepacia H111. [Google Scholar]
- 29.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One. 2013;8:e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 32.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Chen R, Barphagha IK, Karki HS, Ham JH. Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS One. 2012;7:e52150. doi: 10.1371/journal.pone.0052150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan K, Clancy S, Borovilos M, Zhou M, Horer S, Moy S, Volkart LL, Sassoon J, Baumann U, Joachimiak A. The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria. J Biol Chem. 2009;284:36670–36679. doi: 10.1074/jbc.M109.062679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G, Jeffrey PD, Fuqua C, Shi Y, Chen L. Structural basis for antiactivation in bacterial quorum sensing. Proc Natl Acad Sci USA. 2007;104:16474–16479. doi: 10.1073/pnas.0704843104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskowski RA, Watson JD, Thornton JM. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 2005;33:W89–W93. doi: 10.1093/nar/gki414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LICPCR) Nucleic Acids Res. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eschenfeldt WH, Stols L, Millard CS, Joachimiak A, Mark ID. A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol. 2009;498:105–115. doi: 10.1007/978-1-59745-196-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stols L, Millard CS, Dementieva I, Donnelly MI. Production of selenomethionine-labeled proteins in two-liter plastic bottles for structure determination. J Struct Funct Genomics. 2004;5:95–102. doi: 10.1023/B:JSFG.0000029196.87615.6e. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, et al. High-throughput protein purification and quality assessment for crystallization. Methods. 2011;55:12–28. doi: 10.1016/j.ymeth.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim Y, Dementieva I, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y, Ye Z, Joachimiak G, Videau P, Young J, Hurd K, Callahan SM, Gornicki P, Zhao J, Haselkorn R, et al. Structures of complexes comprised of Fischerella transcription factor HetR with Anabaena DNA targets. Proc Natl Acad Sci USA. 2013;110:E1716–1723. doi: 10.1073/pnas.1305971110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin YH, Miyamoto C, Meighen EA. Purification and characterization of a luxO promoter binding protein LuxT from Vibrio harveyi. Protein Expr Purif. 2000;20:87–94. doi: 10.1006/prep.2000.1285. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Winans SC. The sixty nucleotide OccR operator contains a subsite essential and sufficient for OccR binding and a second subsite required for ligandresponsive DNA bending. J Mol Biol. 1995;253:691–702. doi: 10.1006/jmbi.1995.0583. [DOI] [PubMed] [Google Scholar]
- 45.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 46.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 47.French S, Wilson K. Treatment of Negative Intensity Observations. Acta Crystallogr, Sect A: Found Crystallogr. 1978;34:517–525. [Google Scholar]
- 48.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 49.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Howell PL, Smith GD. Identification of Heavy-Atom Derivatives by Normal Probability Methods. J Appl Cryst. 1992;25:81–86. [Google Scholar]
- 51.Sheldrick GM. A short history of SHELX. Acta Crystallogr, Sect A: Found Crystallogr. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 52.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr, Sect D: Biol Crystallogr. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 53.Abrahams JP, Leslie AG. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr, Sect D: Biol Crystallogr. 1996;52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 54.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr, Sect D: Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 56.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr, Sect D: Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 57.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, et al. BUSTER version 2.10.0. Global Phasing Ltd; Cambridge, United Kingdom: 2011. [Google Scholar]
- 58.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr, Sect D: Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: Extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]