Abstract
Multienzyme complexes catalyze important metabolic reactions in many organisms, but little is known about the complexes involved in biological methane production (methanogenesis). A crosslinking-mass spectrometry (XL-MS) strategy was employed to identify proteins associated with coenzyme M-coenzyme B heterodisulfide reductase (Hdr), an essential enzyme in all methane-producing archaea (methanogens). In Methanosarcina acetivorans, Hdr forms a multienzyme complex with acetyl-CoA decarbonylase synthase (ACDS), and F420-dependent methylene-H4MPT reductase (Mer). ACDS is essential for production of acetyl-CoA during growth on methanol, or for methanogenesis from acetate, whereas Mer is essential for methanogenesis from all substrates. Existence of a Hdr:ACDS:Mer complex is consistent with growth phenotypes of ACDS and Mer mutant strains in which the complex samples the redox status of electron carriers and directs carbon flux to acetyl-CoA or methanogenesis. We propose the Hdr:ACDS:Mer complex comprises a special class of multienzyme redox complex which functions as a “biological router” that physically links methanogenesis and acetyl-CoA biosynthesis pathways.
Introduction
Multienzyme complexes catalyze important reactions in central metabolic processes such as photosynthesis, respiration, and amino acid synthesis. We wanted to determine whether multienzyme complexes are also involved in the central metabolic process of biological methane production (methanogenesis) in methane-producing archaea (methanogens). Methanogens are obligately anaerobic archaea that derive all their energy for growth by reducing carbon sources such as acetate, formate, CO2, methanol, methylamines and methyl-sulfides to methane gas. Metabolic engineering of methanogens is an attractive prospect for increasing the yield and rate of renewable methane production from biomass in anaerobic digesters. However, successful metabolic engineering requires not only an in-depth understanding of methanogen physiology, but also a knowledge of which reactions are physically linked by multienzyme complexes. A detailed, three-dimensional spatial model of methanogenesis proteins would be useful in these efforts.
Metabolic reactions that are linked by multienzyme complexes have clear advantages over reactions that are catalyzed by individual, unlinked enzymes [1]. Complexes channel substrates to prevent diffusion of intermediates into bulk cytoplasm, effectively increasing the relative local concentration of reactants in subsequent pathway steps, speeding the overall rate of production of the final product, and preventing diffusion of toxic intermediates that can damage cell constituents. Complexes can also provide a means of co-regulating pathway enzymes or ensuring proper enzyme dosage (Figure 1). Methanogens obtain up to 1 mole ATP per mole substrate consumed and live near the thermodynamic lower limit of life [2]. Substrate channeling via multienzyme complexes would provide a kinetic advantage by ensuring maximal efficiency for converting substrate to ATP generation. We used in vivo crosslinking, tandem affinity purification, and peptide mass spectrometry (XL-MS) to look for complex formation among methanogenesis enzymes. XL-MS is a reliable technique for identifying protein:protein interactions by identifying crosslinked partners which elute together after affinity column purification. A recent effort in Saccharomyces cerevisiae has successfully demonstrated the ability to use XL-MS to reproduce 30 years of protein:protein interaction data and to predict new interactions which were subsequently verified genetically [3]. Though commonly applied to the study of cell signaling networks, we surmised that XL-MS is a valuable technique for identifying protein:protein interactions between methanogenesis enzymes and electron transfer proteins in the methanogen, Methanosarcina acetivorans.
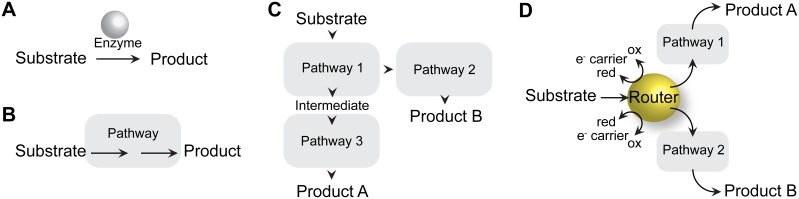
Figure 1. Organization of cellular metabolism.
Metabolic reactions in a cell can be catalyzed by A, individual enzymes, or B, multienzyme complexes that channel substrates and/or sequester intermediates in a pathway. Pathways in the cell can be connected in series, C, or in parallel by D, metabolic “routers” that channel electrons and substrates to either of two metabolic pathways.
The majority of cultivated methanogen strains are restricted to using formate or CO2 as the sole carbon source, and these methanogens use the hydrogenotrophic methanogenic pathway, which relies on reducing equivalents from hydrogen gas to reduce formate or CO2 to methane (Figure 2) [4]. Methanogens which solely utilize the hydrogenotrophic pathway have electron transport systems that are different from the electron transport systems of generalist methanogen species like Methanosarcina acetivorans. The generalist organism M. acetivorans is capable of using the methylotrophic (methanol, methylamines, methylsulfides), carboxidotrophic (CO), and the acetoclastic pathways, but cannot use the hydrogenotrophic or methyl respiration pathways due to the lack of expression of suitable hydrogenases [5], [6], [7], [8].
Figure 2. Comparison of methanogenesis pathways.
A, Hydrogenotrophic methanogenesis in Methanococcus maripaludis. B, Methylotrophic methanogenesis in Methanosarcina acetivorans. Green ovals: energy-conserving reactions. Red ovals: energy-consuming reaction. Please see text for abbreviations.
We wanted to identify proteins that form complexes with coenzyme M-coenzyme B (CoM-S-S-CoB) heterodisulfide reductase (HdrED) in M. acetivorans. HdrED is essential for methylotrophic and aceticlastic growth and is likely to participate in protein:protein interactions with other enzymes of the methanogenesis pathway in Methanosarcina. Previous reports showed that CO oxidation can be coupled to CoM-S-S-CoB reduction in cell extracts in Methanosarcina barkeri MS. The system was then reconstituted using pure Hdr and CO dehydrogenase (a subcomplex of ACDS enzyme) components from Methanosarcina thermophila [9], [10]. The CO:Hdr activity in both reports required the addition of ferredoxin and membranes. XL-MS would seem to be a suitable technique to address whether the CO:Hdr complex occurs in vivo. In this work we have identified proteins that co-purify with the HdrD1 subunit, which contains the CoM-S-S-CoB reductase active site. We report that the proteins with the highest confidence for interacting with HdrD1 are the β subunit of acetyl-CoA decarbonylase/synthase (ACDS), and methylene-tetrahydromethanopterin reductase (Mer).
Results
Strep-tagged HdrD1 protein forms high molecular-weight complexes in vivo
The HdrD1 protein was chosen for XL-MS experiments because it contains the CoM-S-S-CoB heterodisulfide reductase active site and the gene has been shown to be essential for growth on trimethylamine, methanol, methanol + acetate, and acetate [11]. Plasmids expressing N- or C-terminal strep-tagged HdrD1 protein (pNB636 and pNB637, respectively) were recombined onto the M. acetivorans chromosome (Table 1). A StrepTagII peptide affinity tag (WSHPQFEK) was chosen instead of a 6XHistidine tag because His tags have the potential to interfere with assembly of metal clusters. The covalent crosslinker, dimethylsuberimidate (DMS), was added to cells before protein purification to stabilize high molecular weight complexes. DMS is an 11 Å amide crosslinker that crosses cell membranes and is unaffected by residual sulfide present in methanogen cell preparations. N-terminally tagged HdrD1 protein (strepHdrD1) was stably expressed as judged by Western blot (Figure 3a). strepHdrD1 appeared to form two high-molecular weight complexes, observed as bands of approximately 75 kDa and 150 kDa in addition to a band corresponding to strepHdrD1 monomer (45 kDa). C-terminally tagged HdrD1strep protein was not stably expressed and was therefore not used for further experiments (Figure 3a). Instability of C-terminally tagged HdrD1strep protein suggests the 8-amino acid strep tag interferes with correct protein folding.
Table 1. Plasmids and strains used in this study.
| Plasmids and E. coli strains | ||||
| NB # | Genotype | Purpose | Plasmid name | Reference |
| pJK026A derivatives | [18], [36] | |||
| 139 | PmcrBstrephdrD1 | Constitutive production of strepHdrD1protein in Methanosarcina | pNB636 | This study |
| 140 | PmcrBhdrD1strep | Constitutive production of HdrD1strepprotein in Methanosarcina | pNB637 | This study |
| 145 | PmcrBstrephdrD2 | Constitutive production of strepHdrD2protein in Methanosarcina | pNB661 | This study |
| 146 | PmcrBhdrD2strep | Constitutive production of HdrD2strepprotein in Methanosarcina | pNB662 | This study |
| Methanosarcina acetivorans C2A strains | ||||
| NB # | Genotype | Purpose | Reference | |
| 95 | Δhpt::PmcrBtetR/φC31int/attP | Parental strain | same as WWM74 [18] | |
| 75 | Δhpt::PmcrBtetR/φC31int/att pJK026A | control for protein overexpression andnon-specific binding to streptavidinagarose resin | This study | |
| 79 | Δhpt::PmcrBtetR/φC31int/att pNB636 | strephdrD1 constitutive overexpressionfrom hpt locus under PmcrB promoter | This study | |
| 80 | Δhpt::PmcrBtetR/φC31int/att pNB637 | hdrD1strep constitutive overexpressionfrom hpt locus under PmcrB promoter | This study | |
| 41 | Δhpt::PmcrBtetR/φC31int/att pNB661 | strephdrD2 constitutive overexpressionfrom hpt locus under PmcrB promoter | This study | |
| 42 | Δhpt::PmcrBtetR/φC31int/att pNB662 | hdrD2strep constitutive overexpressionfrom hpt locus under PmcrB promoter | This study | |
Figure 3. XL-MS identification of a multienzyme complex in Methanosarcina.
A, Detection of HdrD complexes. 2 µg crosslinked cell lysates from controls or strains expressing strep-tagged HdrD1 protein were analyzed by Western blot. Arrows indicate the position of strep-tagged HdrD1 monomer and crosslinked high molecular weight (HMW) complexes. * = degraded HdrD1strep protein. B, HdrD co-purified proteins detected by mass spectrometry. Node sizes, line opacity and line widths are proportional to the average peptide hit score of the protein detected in biological duplicates. Dotted lines denote an average score below 100, solid lines denote an average score of 100 and above. Image created with Cytoscape [36]. C, Putative model of the Hdr:ACDS:Mer complex. During methylotrophic growth, both ACDS and Mer use methyl-H4MPT as a substrate. Black dotted lines = electron flow between active sites. HdrE (blue) or HdrD (red), Mer is a tetramer (orange), and ACDS is composed of 5 subunits in a (α2ε2)4β8(γδ)8 configuration (green) [13], [16], [37].
HdrD1 interacts with an ACDS:Mer complex
In order to identify constituents of the crosslinked strepHdrD1 complex, we used affinity purification and peptide mass spectrometry. Peptide masses from biological duplicate samples were compared to predicted mass database of M. acetivorans C2A to identify proteins contained in the eluate (Tables S1–S4 in File S1). Protein samples from mock co-purifications conducted with cells expressing β-glucoronidase were used as a control to screen for nonspecific binding to the resin. Sixteen proteins with significant scores were detected from duplicate control samples (Figure S1). After subtracting these nonspecific proteins from the list of proteins that co-purified with strepHdrD1, 29 proteins with significant scores (>100) remained. Of these 29 proteins, the highest score was for HdrD1, as would be expected (Table 2). The second-highest corresponded to the CdhC subunit of acetyl-CoA decarbonylase synthase (ACDS enzyme). CdhC protein is the β subunit of ACDS enzyme and houses a NiFeS “A site” responsible for cleaving acetyl-CoA during growth on acetate or for creating acetyl-CoA from CO2 and CH3-H4MPT during growth on methanol [12], [13], [14]. Mer was also detected in the co-purified samples (Table 2). During growth on methanol, Mer catalyzes the F420-dependent oxidation of CH3-H4MPT to CH2-H4MPT in the oxidative branch of the methylotrophic methanogenesis pathway [15], [16]. The chaperones DnaK and Hsp20 also co-purified with strepHdrD1, suggesting that overexpression may have taxed the protein folding machinery of the cell, an unsurprising result considering that strepHdrD1 expression is driven by the PmcrB promoter, which has the highest expression level in methanogens [17], [18], [19]. The remaining 23 proteins had scores less than 100, suggesting that they are minor constituents of an HdrD1 complex (Figure 3b).
Table 2. HdrD1 protein:protein interactions detected by Mass Spectrometrya.
| Protein gi# | Gene MA# | Function | Average scoreb |
| 20089573 | MA0688 | HdrD1, heterodisulfide reductase, subunit D | 432 |
| 20089889 | MA1014 | CdhC, acetyl-CoA decarbonylase/synthase complex subunit beta | 383 |
| 20092658 | MA3862 | CdhC, acetyl-CoA decarbonylase/synthase complex subunit beta | 383 |
| 20090337 | MA1478 | molecular chaperone DnaK | 220 |
| 20092530 | MA3733 | Mer, methylenetetrahydromethanopterin reductase | 153 |
| 20093358 | MA4574 | hsp20/alpha crystallin family protein | 151 |
Proteins also detected in the control samples have been omitted.
Proteins were identified in duplicate biological samples.
While ACDS, Mer, and molecular chaperones are proteins one would expect are present in high abundance, not all high-abundance proteins co-purified with strepHdrD1. For instance, Methyl-CoM reductase, Mcr, the protein of highest abundance in methanogen cells, was not detected in strep HdrD1 eluates, and none of the other methanogenesis proteins were detected. The highest peptide hits in the control samples were elongation factor EF-2, MtaC1 (methanol-5-hydroxybenzimidazolylcobamide co-methyltransferase, isozyme 1), glutamate-ammonia ligase, and Hsp60. The high peptide hit scores of ACDS and Mer in biological replicates indicates that the co-purification procedure and control screen was sufficiently stringent.
We anticipated that HdrE should be identified. HdrE is an integral membrane b-type cytochrome that delivers electrons to HdrD1. HdrE was identified by purification of CoM-S-S-CoB reductase activity from the membrane fraction of Methanosarcina barkeri. Therefore HdrED interaction was expected to be robust during purification. Batch resin binding with whole cell lysate was used to improve the probability of capturing membrane protein partners, and the main HMW complex band visible in the Western blot corresponds to the expected size of the HdrE:strepHdrD1 crosslinked species (∼75 kDa), suggesting that the HdrE:strepHdrD1 complex should have been detected. It is possible that despite utilization of crosslinker and batch binding of whole lysate to the streptactin resin, HdrE may not have been detectable by mass spectrometry due to problems with complete digestion of membrane proteins or the solubility of hydrophobic peptides. Therefore it is possible that HdrE, along with other highly hydrophobic proteins, may have been missed.
HdrD2 does not form high-molecular-weight complexes
M. acetivorans has a close homolog of HdrD1, HdrD2, which is encoded by gene MA0526. HdrD2 is 31% identical, 47% similar to HdrD1 by primary amino acid sequence. Unlike HdrD1, expression of HdrD2 is not essential for growth [11]. However, we hypothesized that HdrD1 and HdrD2 may share overlapping cellular functions because of their similar sequences. When strep-tagged HdrD2 is expressed in M. acetivorans, we could not detect a high-molecular-weight complex after crosslinking (data not shown). We also did not detect interacting proteins with a MASCOT score above 100 (Table S5 in File S1). These results indicate that although HdrD1 and HdrD2 share significant amino acid sequence similarity, they do not crosslink with the same proteins, and likely have non-overlapping physiological functions.
Discussion
We propose that the Hdr:ACDS:Mer complex is a multienzyme “router” that directs substrates and electrons through either the acetyl-CoA or methanogenesis pathways by connecting the CoM-S-S-CoB, acetyl-CoA, and CH3-H4MPT metabolic nodes (Figure 3c). Despite the importance of several multienzyme complexes in biology (tryptophan synthase, pyruvate carboxylase, polyketide synthases, etc.) it is unusual that acetyl-CoA, a major node involved in carbon fixation, is physically linked with the electron transport system in Methanosarcina by the Hdr:ACDS:Mer multienzyme complex [20], [21], [22].
These studies suggest reduction of CoM-S-S-CoB and oxidation of CH3-H4MPT is physically linked to acetyl-CoA in M. acetivorans by a HdrD:ACDS:Mer complex. The HdrD:ACDS:Mer complex we identified, though detected in cells grown on methanol, likely exists in cells grown on acetate because all three enzymes are essential for both the methylotrophic and aceticlastic methanogenesis pathways (Figure 4). Our findings are consistent and complementary to previous reports of CO:Hdr activity in acetate-grown cell extracts from M. barkeri and in a reconstituted system using purified components from acetate-grown M. thermophila [9], [10]. During methylotrophic growth, ACDS is the enzyme responsible for acetyl-CoA synthesis from CO2, CH3-H4MPT and reduced ferredoxin. During acetoclastic growth, ACDS functions in the opposite direction to cleave acetyl-CoA with the production of CO2, reduced ferredoxin, and CH3-H4MPT, which is reduced to methane [14], [23]. The CdhC β subunit of ACDS houses the NiFe “A site” and catalyzes acetyl-CoA formation from enzyme-bound CO (CO2 reduced by ferredoxin) and enzyme-bound methyl-corrin (derived from CH3-H4MPT) [24]. HdrED, ACDS, and Mer are all essential for methylotrophic and acetoclastic growth, and therefore the HdrD:ACDS:Mer complex likely participates in both methanogenesis pathways.
Figure 4. Enzymes used by M. acetivorans.
Current pathway models for growth of M. acetivorans on A, methanol + acetate or B, acetate as carbon and energy sources. Enzymes in red are essential despite no defined purpose in the pathway. Green ovals = energy conserving step. Red ovals = energy-consuming step. Please see text for abbreviations.
Redox potentials suggest it is possible that electrons can directly flow from CdhC (−540 mV midpoint potential of the A site of Clostridia thermoaceticum ACDS enzyme) to HdrD1 (−142 mV) during acetoclastic growth [25], [26]. However, direct reduction of CoM-S-S-CoB via ACDS:HdrD would bypass Rnf, the proposed sodium-pumping ferredoxin:methanophenazine oxidoreductase, thus preventing formation of a transmembrane ion potential that is necessary for ATP synthesis. During methylotrophic growth ACDS accepts electrons from ferredoxin, and it is possible that HdrD could also compete for electrons from ferredoxin due to its close proximity to CdhC. Such an arrangement would shift flux away from acetyl-CoA biosynthesis towards reduction of CoM-S-S-CoB, albeit at the expense of the ion transmembrane gradient. Genetic and biochemical evidence demonstrates that electrons from ferredoxin are more likely to be oxidized by Rnf and HdrABC. Both HdrABC and Rnf are thought to account for most, but not all, ferredoxin oxidation during methylotrophic methanogenesis (Buan, Kulkarni, Guss, and Metcalf, unpublished data). If HdrD can directly accept electrons from CdhC, or if it competes with CdhC for electrons from ferredoxin, this is expected to be a low-flux pathway that the cell may use to maintain redox balance between ferredoxin, coenzyme A, and CoM-S-S-CoB pools.
If ACDS and Mer conformations are altered as a result of forming a complex in vivo, disruption of the ACDS:Mer interaction is expected to have a negative effect on biosynthesis and oxidative methylotrophic pathways. A recent report by Matschiavelli et al. supports the idea that ACDS and Mer are linked in vivo. The authors showed that deletion of both copies of ACDS results in an increase in the doubling time of M. acetivorans by 14 hours during growth on methanol + acetate [14]. The authors hypothesize that the rate of acetyl-CoA production from acetate via Ack and Pta are limiting in the MCD21 (Δcdh2 Δcdh1 double mutant) and MCD213 (Δcdh2 Δcdh1 ΔcdhA3 triple mutant) strains. However, deletion of ACDS should not show an effect during growth on methanol + acetate because in wild-type cells, methanol is converted to CO2 and methane, while acetate is activated to acetyl-CoA by Ack and Pta, bypassing the need for ACDS. We propose an alternate hypothesis: that flux through the oxidative branch of the methylotrophic pathway is affected due to disruption of the ACDS:Mer complex and a resulting conformational effect on Mer decreases catalytic efficiency of conversion of methyl-H4MPT to methylene-H4MPT.
Reports of CO:Hdr activity in extracts and a reconstituted system, combined with the observation of unexpected methylotrophic growth phenotypes resulting from ACDS mutations, and of methyl oxidation mutations demonstrating a need for acetate supplementation, supports our hypothesis that the HdrD:ACDS:Mer complex we observed has a physiological role in methanogens. Direct interaction between ACDS and Mer may explain why some methyl-H4MPT is oxidized to CO2 during growth on acetate [27], and why attempts to delete the oxidative genes in M. acetivorans have been unsuccessful. Previous reports suggest the oxidative branch is essential during acetoclastic growth due to the need to generate reduced F420 for biosynthetic reactions. In the closely related organism, M. barkeri, a Δmer mutant is viable but can only grow by the methyl respiration pathway, which M. acetivorans cannot use, or mixotrophically on methanol + acetate, albeit very slowly by an unknown Mer/Mtr bypass pathway [28]. Wild-type M. barkeri does not grow mixotrophically on methanol + acetate, but uses methanol for methylotrophic methanogenesis, and acetate for biosynthesis. Therefore, in both M. acetivorans and M. barkeri a Δmer mutant cannot use the acetoclastic methanogenesis pathway despite the fact that M. barkeri can use hydrogen as an energy source and M. acetivorans cannot.
Physical linkage of ACDS and Mer in M. acetivorans has intriguing implications as to how carbon flux through the oxidative branch of methanogenesis and biosynthesis pathways in this organism is controlled. ACDS and Mer both use CH3-H4MPT as substrate, and physically linking these two enzymes means their active sites are in direct competition for substrate. Therefore, as CH3-H4MPT is produced by Mtr, whether or not it is funneled through the oxidative branch of the methylotrophic methanogenesis pathway is dependent on the rate at which it enters the Mer active site. If Mer is not in a favorable conformation to accept substrate, methyl-H4MPT will be available for ACDS to convert into acetyl-CoA for biosynthesis. Furthermore, all the major electron carriers (F420, CoM-S-S-CoB, methanophenazine, ferredoxin) converge on the Hdr:ACDS:Mer complex, and phenotypic behavior of ACDS and Mer mutant strains indicates the Hdr:ACDS:Mer complex acts as an integrated switch that samples the redox status of electron carrier pools. The order of substrate and electron donor/acceptor binding determines whether CH3-H4MPT is fixed as acetyl-CoA by ACDS or is directed to the oxidative branch of the methanogenesis pathway via Mer. By forming a Hdr:ACDS:Mer complex, the cell samples availability of substrates and electron carriers in a minimal spatial location with no need for diffusion of metabolites across cytoplasm. Our data suggests the CH3-H4MPT metabolite is channeled to one of two metabolic fates (acetyl-CoA production or the oxidative branch of methylotrophic methanogenesis) by a single Hdr:ACDS:Mer protein complex, in contrast to enzyme channeling models that propose an “assembly-line” arrangement of enzyme functions [29], [30].
The 3-dimensional spatial organization of metabolism in methanogens may have evolved as a result of the thermodynamic pressure methanogens face. Methanogens obtain very little ATP/mol substrate consumed (approximately 0.5 ATP/acetate or 1 ATP/MeOH), with only acetogens and syntrophs known to survive under even less thermodynamically favorable conditions. The ability to thrive on so little energy could very well result from exquisitely tight control of substrate and electron channeling that is not necessary in, for instance, a facultative aerobic bacteria like E. coli which obtains more energy per mole substrate. Perhaps a fitting analogy would be to describe E. coli as a mechanical machine with metabolic “units” that can be interchanged, whereas Methanosarcina is a solid-state computer, with a hard-wired multienzyme “biological router” that controls flux through acetyl-CoA as well as through methanogenesis (Figure 1). If multienzyme redox routers exist in other organisms, one would predict they may be found in organisms that also live near the thermodynamic limit of life.
Materials and Methods
Growth of E. coli
E. coli strains were grown in LB medium [31] with the appropriate antibiotics or additions in the following concentrations: rhamnose 5 mM, chloramphenicol 35 or 5 µg/ml.
Growth of M. acetivorans
Methanosarcina strains were grown under strictly anaerobic conditions in HS mineral salts medium [32]. For growth on solid medium, cells were plated on HS medium containing 1.4% agar (w/v) with the appropriate carbon source and additions as previously described [33]. All strains were inoculated into 100 mL of high salt media with a methanol carbon source into 250 mL bottles. The cultures were incubated at 35°C in a Thermo Scientific MaxQ 6000 Incubated/Refrigerated Stackable Shaker until exponential phase. The following anaerobic additions were added when appropriate: MeOH (125 mM), acetate (120 mM), TMA (50 mM), Puromycin (2 µg/ml).
Strain construction
Genetic methods for M. acetivorans are well-defined [34]. Expression of tagged proteins is achieved by creating oligos to amplify the genes of interest and cloning the resultant PCR products into the pJK026A shuttle plasmid at the NdeI and BamHI (or HindIII) restriction sites (Table S6 in File S1). The oligos are designed to fuse the strep-tagII peptide (which has been codon optimized for expression in M. acetivorans) to the 5′ or 3′ end of the gene coding sequence [35]. The resulting plasmid (Table 1) is transformed into M. acetivorans to create the strains listed in Table 1. Puromycin-resistant colonies are single-colony purified and screened by PCR to ensure the expression plasmid has integrated at the hpt locus via ΦC31 integrase [18]. Expression of the tagged protein is driven by the constitutive PmcrB promoter on pJK026A.
Strep-tagged affinity co-purification
Protein purification was performed at room temperature under anaerobic conditions. 100 mL cultures were transferred to 15 mL conical tubes and centrifuged at 1228×g in a Thermo Scientific IEC Medilite Microcentrifuge for five minutes. The cell pellet was resuspended in 1 mL of 50 mM NaH2PO4, 0.4 M NaCl, pH 7.2 buffer and transferred into a microfuge tube. 30 mg of DMS cross-linking agent (Dimethyl suberimidate·2 HCl, ThermoFisher Pierce, USA), was added to the remaining cells and mixed for 1 hour. The sample was added to 4 mL of 50 mM TrisCl pH 8 (lysis/quenching buffer). 10 µL of DNase and 50 µL of Halt Protease Inhibitor Cocktail 100X (ThermoFisher Pierce, USA) were added and the sample was incubated for 5 minutes. 100 µL of Streptavidin Agarose Resin (Qiagen, USA) was added, and the sample put on ice for 1 hour with occasional gentle mixing. The sample was placed into 2 mL centrifuge columns and centrifuged for 5 minutes at 1228×g. The resin was washed four times with 2 mL 50 mM NaH2PO4, pH 7.2. Strep-tagged protein was eluted twice with 200 µL biotin eluting buffer (50 mM NaH2PO4, 300 mM NaCl, 0.05% tween 20, pH 8.0, 10 mM biotin), and the resin was stripped with two washes of 200 µL of 8 M guanidine hydrochloride.
SDS PAGE and Western blot
Protein concentrations were measured using the Coomassie Plus Bradford Assay (Bio-Rad, USA). Protein samples were mixed with 6X cracking buffer (58 mM Tris pH 6.8, 58 mM SDS, 100 mM dithiothreitol (DTT), 0.68 mM glycerol, 30 µM bromophenol blue) and boiled for 2 minutes. Proteins and Precision Plus Protein Western Standard ladder were separated on precast 4–20% SDS-PAGE gels, and blotted onto ImmunoBlot PVDF membrane (Bio-Rad, USA). Strep-tagged protein was detected with mouse monoclonal anti-StrepTagII antibody (Qiagen, USA) and sheep anti-mouse HRP-linked secondary antibody (GE Healthsciences, USA). HRP signal was detected using the ECL chemiluminescent detection system (ThermoFisher Pierce, USA). Western blot performed with extracts from vector-only control cells did not detect any protein bands.
Mass spectrometry
For in-solution digests, enriched proteins after elution from the beads were subjected to “shot gun” protein analysis by direct in-solution trypsin digestion of eluent. Eluted protein samples were desalted and dialyzed with 100 mM ammonium bicarbonate using a Millipore Centrifugal Filter Unit (Millipore, USA). Samples were reduced with DTT (7.5 mg), and alkylated with iodoacetamide (0.72 mg) in the dark. Proteins were digested with approximately a 1∶50 trypsin:protein ratio sequencing-grade trypsin (Roche) dissolved in 100 mM ammonium bicarbonate at at 37°C overnight. Tryptic peptides were desalted and concentrated using PepClean C-18 spin columns according to manufacturer’s instructions (Thermo Fisher Scientific, USA).
For in-gel digests, eluted protein was first separated by SDS PAGE and stained with SimplyBlue Safe Stain (Invitrogen). Bands were excised, and gel slices were destained in 1∶1 100 mM ammonium bicarbonate: acetonitrile, and washed in 100% acetonitrile before drying in a speedvac. Tris(2-carboxyethyl)phosphine (TCEP, 100 µl) was added to reduce the protein, and the sample was incubated at 56°C for 45 minutes. Samples were alkylated with iodoacetamide (0.72 mg) and gel slices were washed in 100 mM ammonium bicarbonate. Gel slices were washed twice with 100% acetonitrile and dried in a speed vac. Trypsin was added and gel slices were allowed to swell at 4°C. Gel slices were incubated in 50 mM ammonium bicarbonate at 37°C overnight, and eluted tryptic peptides were desalted as above. Biological replicates of the digested peptide samples were submitted to the University of Nebraska-Lincoln Redox Metabolomics and Proteomics Core Facility.
One dimensional LC-MS/MS was performed with an ultimate 3000 Dionex MDLC system (Dionex Corporation, USA) integrated with a nanospray source and LCQ Fleet Ion Trap mass spectrometer (ThermoFisher Scientific, USA). LC-MS/MS included an on-line sample pre-concentration and desalting using a monolithic C18 trap column (Pep Map, 300 µm I.D×5 mm, 100 Å, 5 µm, Dionex, USA). Desalted peptides were eluted and separated on a C18 Pico Frit analytical column (75 µm I.D×15 cm, 3 µm, 100 Å, New Objective, USA) by applying an acetonitrile (ACN) gradient (ACN plus 0.1% formic acid, 90 minute gradient at a flow rate of 300 µl/min) and were introduced into the mass spectrometer using the nano spray source. The LCQ Fleet mass spectrometer was operated with the following optimized parameters: nano spray voltage, 2.0 kV; heated capillary temperature, 200°C; full scan m/z range, 400–2,000). The mass spectrometer was operated in data dependent mode with 4 MS/MS spectra for every full scan, 5 microscans averaged for full scans and MS/MS scans, a 3 m/z isolation width for MS/MS isolations, and 35% collision energy for collision-induced dissociation.
The MS/MS spectra were searched against M. acetivorans proteome database using MASCOT (Version 2.2 Matrix Science, London, UK). Database search criteria were as follows: enzyme: trypsin, missed cleavages: 2; mass: monoisotropic; fixed modification: carbamidomethyl (C); variable modification: oxidation (M); peptide tolerance: 1.5 Da; MS/MS fragment ion tolerance: 1 Da. Protein identifications were accepted with a statistically significant MASCOT protein score that corresponds to an error probability of p<0.05. Datasets from duplicate vector only control mock purifications were used as a screen. Raw datasets can be found in Tables S2-S6 in File S1. Protein hits were required to be identified (score >0) in at least two independent purifications for inclusion in Table 2.
MASCOT results were loaded into a MySQL database as a list of identified proteins (nodes) and potential interactions with HdrD for each purification sample. Queries compared samples with the control to identify interactions in both independent purification samples. These results were then visualized using Cytoscape [36].
Supporting Information
Analysis of XL-MS results. Peptide hits from duplicate biological replicates after crosslinking and strep-tag affinity purification were compared. A, control protein samples. B, samples from cells overexpressing strepHdrD1 protein. blue: hits found only in one control sample, yellow: hits found in both control samples, orange: hits found in one strepHdrD1 sample, red: hits found in both strepHdrD1 samples. Data was visualized using Cytoscape.
(TIF)
Supporting tables. Table S1, XL-MS data for control sample 1. Table S2, XL-MS data for control sample 2. Table S3, XL-MS data for strepHdrD1 sample 1. Table S4, XL-MS data for strepHdrD1 sample 2. Table S5, XL-MS data for HdrD2strep sample. Table S6, Oligos used for strain construction.
(XLSX)
Funding Statement
This material is based upon work supported by the National Science Foundation under Grant No. IOS-1449525 to NRB; by the National Institute of General Medical Sciences of the National Institutes of Health under award numbers F32 GM078796 to NRB, P20 RR-17675, and P30GM103335; a Pepsi UCARE Fellowship to DJL; by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Award Number DE-FG02-02ER15296 to WWM; and by the Nebraska Tobacco Settlement Biomedical Research Development Funds. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation, the National Institutes of Health, the Department of Energy, or Pepsi. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Agapakis CM, Boyle PM, Silver PA (2012) Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat Chem Biol 8: 527–535. [DOI] [PubMed] [Google Scholar]
- 2. Schlegel K, Welte C, Deppenmeier U, Muller V (2012) Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J 279: 4444–4452. [DOI] [PubMed] [Google Scholar]
- 3. Herzog F, Kahraman A, Boehringer D, Mak R, Bracher A, et al. (2012) Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337: 1348–1352. [DOI] [PubMed] [Google Scholar]
- 4. Lie TJ, Costa KC, Lupa B, Korpole S, Whitman WB, et al. (2012) Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc Natl Acad Sci U S A 109: 15473–15478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deppenmeier U, Lienard T, Gottschalk G (1999) Novel reactions involved in energy conservation by methanogenic archaea. FEBS Lett 457: 291–297. [DOI] [PubMed] [Google Scholar]
- 6. Wang M, Tomb JF, Ferry JG (2011) Electron transport in acetate-grown Methanosarcina acetivorans. . BMC Microbiol 11: 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rother M, Metcalf WW (2004) Anaerobic growth of Methanosarcina acetivorans C2A on carbon monoxide: an unusual way of life for a methanogenic archaeon. Proc Natl Acad Sci U S A 101: 16929–16934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oelgeschlager E, Rother M (2009) In vivo role of three fused corrinoid/methyl transfer proteins in Methanosarcina acetivorans . Mol Microbiol 72: 1260–1272. [DOI] [PubMed] [Google Scholar]
- 9. Fischer R, Thauer RK (1990) Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS). FEBS Lett 269: 368–372. [DOI] [PubMed] [Google Scholar]
- 10. Peer CW, Painter MH, Rasche ME, Ferry JG (1994) Characterization of a CO: heterodisulfide oxidoreductase system from acetate-grown Methanosarcina thermophila . J Bacteriol 176: 6974–6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buan NR, Metcalf WW (2010) Methanogenesis by Methanosarcina acetivorans involves two structurally and functionally distinct classes of heterodisulfide reductase. Mol Microbiol 75: 843–853. [DOI] [PubMed] [Google Scholar]
- 12. Funk T, Gu W, Friedrich S, Wang H, Gencic S, et al. (2004) Chemically distinct Ni sites in the A-cluster in subunit beta of the acetyl-CoA decarbonylase/synthase complex from Methanosarcina thermophila: Ni L-edge absorption and X-ray magnetic circular dichroism analyses. J Am Chem Soc 126: 88–95. [DOI] [PubMed] [Google Scholar]
- 13. Gong W, Hao B, Wei Z, Ferguson DJ Jr, Tallant T, et al. (2008) Structure of the alpha2epsilon2 Ni-dependent CO dehydrogenase component of the Methanosarcina barkeri acetyl-CoA decarbonylase/synthase complex. Proc Natl Acad Sci U S A 105: 9558–9563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matschiavelli N, Oelgeschlager E, Cocchiararo B, Finke J, Rother M (2012) Function and Regulation of Isoforms of Carbon Monoxide Dehydrogenase/Acetyl Coenzyme A Synthase in Methanosarcina acetivorans . J Bacteriol 194: 5377–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. te Brommelstroet BW, Geerts WJ, Keltjens JT, van der Drift C, Vogels GD (1991) Purification and properties of 5,10-methylenetetrahydromethanopterin dehydrogenase and 5,10-methylenetetrahydromethanopterin reductase, two coenzyme F420-dependent enzymes, from Methanosarcina barkeri . Biochim Biophys Acta 1079: 293–302. [DOI] [PubMed] [Google Scholar]
- 16. Aufhammer SW, Warkentin E, Ermler U, Hagemeier CH, Thauer RK, et al. (2005) Crystal structure of methylenetetrahydromethanopterin reductase (Mer) in complex with coenzyme F420: Architecture of the F420/FMN binding site of enzymes within the nonprolyl cis-peptide containing bacterial luciferase family. Protein Sci 14: 1840–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Metcalf WW, Zhang JK, Apolinario E, Sowers KR, Wolfe RS (1997) A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc Natl Acad Sci U S A 94: 2626–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guss AM, Rother M, Zhang JK, Kulkarni G, Metcalf WW (2008) New methods for tightly regulated gene expression and highly efficient chromosomal integration of cloned genes for Methanosarcina species. Archaea 2: 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baneyx F, Mujacic M (2004) Recombinant protein folding and misfolding in Escherichia coli. . Nat Biotechnol 22: 1399–1408. [DOI] [PubMed] [Google Scholar]
- 20. Yu LP, Xiang S, Lasso G, Gil D, Valle M, et al. (2009) A symmetrical tetramer for S. aureus pyruvate carboxylase in complex with coenzyme A. Structure. 17: 823–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aparicio JF, Fouces R, Mendes MV, Olivera N, Martin JF (2000) A complex multienzyme system encoded by five polyketide synthase genes is involved in the biosynthesis of the 26-membered polyene macrolide pimaricin in Streptomyces natalensis . Chem Biol 7: 895–905. [DOI] [PubMed] [Google Scholar]
- 22. Miles EW (2013) The tryptophan synthase alpha2beta2 complex: a model for substrate channeling, allosteric communication, and pyridoxal phosphate catalysis. J Biol Chem 288: 10084–10091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krzycki JA, Lehman LJ, Zeikus JG (1985) Acetate catabolism by Methanosarcina barkeri: evidence for involvement of carbon monoxide dehydrogenase, methyl coenzyme M, and methylreductase. J Bacteriol 163: 1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gencic S, Grahame DA (2008) Two separate one-electron steps in the reductive activation of the A cluster in subunit beta of the ACDS complex in Methanosarcina thermophila . Biochemistry 47: 5544–5555. [DOI] [PubMed] [Google Scholar]
- 25. Gorst CM, Ragsdale SW (1991) Characterization of the NiFeCO complex of carbon monoxide dehydrogenase as a catalytically competent intermediate in the pathway of acetyl-coenzyme A synthesis. J Biol Chem 266: 20687–20693. [PubMed] [Google Scholar]
- 26. Heiden S, Hedderich R, Setzke E, Thauer RK (1994) Purification of a 2-Subunit Cytochrome-B-Containing Heterodisulfide Reductase from Methanol-Grown Methanosarcina barkeri . European Journal of Biochemistry 221: 855–861. [DOI] [PubMed] [Google Scholar]
- 27. Krzycki JA, Wolkin RH, Zeikus JG (1982) Comparison of unitrophic and mixotrophic substrate metabolism by acetate-adapted strain of Methanosarcina barkeri . J Bacteriol 149: 247–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Welander PV, Metcalf WW (2008) Mutagenesis of the C1 oxidation pathway in Methanosarcina barkeri: new insights into the Mtr/Mer bypass pathway. J Bacteriol 190: 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. de Kok A, Hengeveld AF, Martin A, Westphal AH (1998) The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim Biophys Acta 1385: 353–366. [DOI] [PubMed] [Google Scholar]
- 30. Iverson TM, Luna-Chavez C, Cecchini G, Rees DC (1999) Structure of the Escherichia coli fumarate reductase respiratory complex. Science 284: 1961–1966. [DOI] [PubMed] [Google Scholar]
- 31. Uetake H, Luria SE, Burrous JW (1958) Conversion of somatic antigens in Salmonella by phage infection leading to lysis or lysogeny. Virology 5: 68–91. [DOI] [PubMed] [Google Scholar]
- 32. Metcalf WW, Zhang JK, Shi X, Wolfe RS (1996) Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J Bacteriol 178: 5797–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Metcalf WW, Zhang JK, Wolfe RS (1998) An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl Environ Microbiol 64: 768–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Buan NR, Kulkarni G, Metcalf WW (2011) Genetic methods for Methanosarcina species. In: Rosenzweig, A and S Ragsdale, eds Methods in Methane Metabolism (Methods in Enzymology) 494: 23–42. [DOI] [PubMed] [Google Scholar]
- 35. Korndorfer IP, Skerra A (2002) Improved affinity of engineered streptavidin for the Strep-tag II peptide is due to a fixed open conformation of the lid-like loop at the binding site. Protein Sci 11: 883–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, et al. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kung Y, Ando N, Doukov TI, Blasiak LC, Bender G, et al. (2012) Visualizing molecular juggling within a B12-dependent methyltransferase complex. Nature 484: 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of XL-MS results. Peptide hits from duplicate biological replicates after crosslinking and strep-tag affinity purification were compared. A, control protein samples. B, samples from cells overexpressing strepHdrD1 protein. blue: hits found only in one control sample, yellow: hits found in both control samples, orange: hits found in one strepHdrD1 sample, red: hits found in both strepHdrD1 samples. Data was visualized using Cytoscape.
(TIF)
Supporting tables. Table S1, XL-MS data for control sample 1. Table S2, XL-MS data for control sample 2. Table S3, XL-MS data for strepHdrD1 sample 1. Table S4, XL-MS data for strepHdrD1 sample 2. Table S5, XL-MS data for HdrD2strep sample. Table S6, Oligos used for strain construction.
(XLSX)