Abstract
Colorectal cancer rates in Latin American countries are less than half of those observed in the United States. Latin Americans are the resultant of generations of an admixture of Native American, European, and African individuals. The potential role of genetic admixture in colorectal carcinogenesis has not been examined. We evaluate the association of genetic ancestry with colorectal neoplasms in 190 adenocarcinomas, 113 sporadic adenomas and 243 age- and sex-matched controls enrolled in a multicentric case–control study in Colombia. Individual ancestral genetic fractions were estimated using the STRUCTURE software, based on allele frequencies and assuming three distinct population origins. We used the Illumina Cancer Panel to genotype 1,421 sparse single-nucleotide polymorphisms (SNPs), and Northern and Western European ancestry, LWJ and Han Chinese in Beijing, China populations from the HapMap project as references. A total of 678 autosomal SNPs overlapped with the HapMap data set SNPs and were used for ancestry estimations. African mean ancestry fraction was higher in adenomas (0.13, 95% confidence interval (95% CI)=0.11–0.15) and cancer cases (0.14, 95% CI=0.12–0.16) compared with controls (0.11, 95% CI=0.10–0.12). Conditional logistic regression analysis, controlling for known risk factors, showed a positive association of African ancestry per 10% increase with both colorectal adenoma (odds ratio (OR)=1.12, 95% CI=0.97–1.30) and adenocarcinoma (OR=1.19, 95% CI=1.05–1.35). In conclusion, increased African ancestry (or variants linked to it) contributes to the increased susceptibility of colorectal cancer in admixed Latin American population.
Keywords: colorectal neoplasm, pedigree, polymorphism, genetic, Latin America, Colombia
Introduction
Colorectal cancer incidence in Latin America has steadily increased during the last decades;1 however, these rates are less than half of those observed in African Americans and Caucasian Americans in North America.2 Colorectal cancer incidence rates largely vary across continents, showing the highest rates in those countries with mainly Caucasian populations.1 Such differences, and even the recent rising trends observed in developing countries, have been attributed to the high and increasing prevalence of risk factors associated with a ‘westernized' lifestyle, such as obesity and physical inactivity.3 The reasons behind the higher colorectal cancer risk among African Americans compared with non-Hispanic Whites (Caucasians) in the United States are still not clear. Several studies propose health disparities as the main reason behind these differences.4, 5 However, prospective evidence showed that such factors explain roughly 27% of the excess risk in African Americans relative to Whites,6 suggesting that variation in genetic susceptibility across populations may have an important role.7, 8
The observed increase in colorectal cancer incidence has possibly been accompanied, if not led, by an increase in the incidence of colorectal adenomas.9, 10 Adenomas are the main precursor lesions to most sporadic colorectal cancers and develop through the complex interactions of environmental and genetic risk factors.11 Although recent reports have suggested that the risk of colorectal adenoma may be influenced by racial differences12, 13 in admixed populations, these findings are based on reported ethnicity rather than measured genetic ancestry. Self-reported measures of ethnicity in admixed populations are notoriously inaccurate regarding the individual ancestry.14 This is especially important in populations of Latin America where the admixture between people of at least three continents (Africa, Europe and America) has been widespread since the seventeenth century.
In this study we used a sparse set of single-nucleotide polymorphisms (SNPs) to evaluate the association of genetic ancestry with the risk of colorectal adenomas and adenocarcinomas in the Colombian population controlling for well-known colorectal cancer and adenoma risk factors.
Material and methods
Study population and enrollment
Cases and controls were randomly extracted from a larger multicenter case–control study aimed at identifying the environmental and genetic risk factors of colorectal cancer in Colombia. After ethical approval from the Ethics Board of The National Cancer Institute of Colombia, we recruited incident cases (diagnosed at enrollment) of colorectal adenoma and adenocarcinomas at major colonoscopy medical centers in six of the largest Colombian cities (Barranquilla, Bogota, Bucaramanga, Cali, Cartagena and Santa Marta) from January 2008 to February 2011. Colombia has not yet established a colorectal cancer screening program; therefore, most of the colonoscopy examinations were medically indicated. Cases were originally diagnosed after a complete and satisfactory colonoscopy examination, but only pathologically confirmed cases were finally enrolled in the study. Eligible cases were Colombians, residents in the city of enrollment, aged between 30 and 75 years at the time of colonoscopy, willing and mentally capable to participate, and did not have any personal history of colorectal cancer, ulcerative colitis or Crohn's disease. Controls were approached at the waiting room of primary care units, nearby or in the same hospital where the cases were recruited among individuals attending for medical conditions different from gastrointestinal discomfort and willing to participate; they were unrelated to cases and had no personal history of cancer or colorectal adenomas. Controls were matched by sex and age group (±5 years) to the cases.
Participants gave written informed consent, donated a blood sample and answered a full epidemiological survey, a food frequency questionnaire designed for the study15 and the short version of the IPAQ16 (International Physic Activity Questionnaire), looking into the best-known risk factors for colorectal adenoma and adenocarcinoma.17 Buffy coats were kept in portable liquid nitrogen containers until transferred in dry ice to the National Cancer Institute Facilities in Bogotá for final storage at −80 °C. Questionnaires were processed centrally. We used Teleform, version 5.2 software package (Cardiff Software, Inc., Highland Park, IL, USA) to increase the efficiency of data management and reduce typing error. By the end of the recruitment phase, we enrolled 506 controls, 322 adenocarcinomas and 239 colorectal adenomas. Because of funding constraints, we restricted our genetic analyses to a random subset sample of 264 controls, 206 adenocarcinomas and 126 adenomas. Adenomas included into the analysis were large (≥1 cm) without severe dysplasia, and with histopathological diagnosis of tubular, tubulovillous or villous adenoma (<20%, 20–80% and ≥80% of villous component, respectively).
SNP genotyping
DNA was extracted from buffy coat samples, using the QIAamp DNA Blood Mini KIT (Qiagen, Hilden, Germany), as recommended by the manufacturer and eluted in 100 μl of Nuclease-free Water (Ambion, Carlsbad, CA, USA). Two hundred and fifty nanograms (250 ng) of DNA were resuspended in 5 μl of TE Buffer, denatured and bound to paramagnetic beads for high-throughput genotyping using the protocols described for the highly multiplexed GoldenGate assay18 (Illumina Inc., San Diego, CA, USA). Briefly, two allele-specific oligonucleotide (ASO) probes, linked to universal primer sequences (labeled with either Cy3 or Cy5 for each allele), along with one locus-specific oligonucleotide (LSO) probe, also linked to a universal primer and an address sequence, are hybridized to the DNA. Extension of the ASO and ligation to the LSO is carried out, and the product is amplified by PCR. The amplified product was hybridized to the chips containing sequences complementary to each unique address sequence and the alleles were determined by the scanner according to the fluorescent emitted (Cy3, Cy5 or both). Microarray analysis was done at the LSUHSC/LCRC Genomic Facility in the Stanley S Scott Cancer Center at Louisiana State University Health Sciences Center, New Orleans, LA, USA. The SNP panel used for this study (Illumina Cancer Panel) consist of 1421 thoroughly screened and validated SNP loci, covering all chromosomes and tagging 408 genes chosen from the National Cancer Institute's Cancer Genome Anatomy Project SNP500 Cancer Database.19 According to the manufacturer, the mean minor allele frequency (MAF) across all the SNPs in the genotyping panel was 0.25, 0.22 and 0.21 for Caucasians, Han Chinese/Japanese and Yoruba Africans, respectively.
Quality control
We followed a standard quality control (QC) protocol for case–control genetic association studies20 using the PLINK software.21 SNPs were excluded from the analysis if they departed from Hardy–Weinberg equilibrium (P<0.01), there was a significant difference between missing genotype rates among cases and controls (P<0.01), the SNP overall call rate was <0.95 or the MAF was <0.04. Participants with call rates ≤0.95, or with heterozygosity rates >3 SD from the sample mean, were also excluded. In addition, we excluded one individual of each pair featuring an identity by descendant value >0.375 from the analysis, avoiding duplicated, related or contaminated samples. Gender could not be reliably estimated from the limited number of SNPs available on the X chromosome (N=13), and we relayed on our recorded gender. Eighteen percent of the controls (n=21), 10% of the adenomas (n=13), 7.7% of adenocarcinomas (n=16) and 15.8% of the SNPs (n=225) did not pass through the QC, leaving 238 controls, 113 adenomas and 190 adenocarcinomas for the analysis.
Inference of ancestry proportions
We used the Bayesian clustering algorithm STRUCTURE version 2.2 (Pritchard et al22) under an admixture model to estimate the proportions of European, African and Amerindian ancestry in each of our samples. We used a flat prior before running a burn-in period of 5000 iterations and kept 1 in 5000 iterations. Under the admixture model, the genotype information of each individual is modeled assuming that they inherited a fraction of their genome from ancestors originating from one of the kth populations of origin. We included a set of overlapping SNPs (N=804) genotyped in three reference ancestral populations (k=3) from the HapMap3 project:23 Utah residents with Northern and Western European ancestry, Luhya in Webuye, Kenya and Han Chinese in Beijing, China. The well-established similarities of the allele frequencies between the latter population and Amerindians24 made it a useful alternative to discriminate the Amerindian from African and European ancestry in the study sample. Six hundred and seventy-eight autosomal SNPs remained for analysis after pruning for linkage disequilibrium, excluding one of each pair with R2>0.5, in a windows size of 50 SNPs and a window shift of 5 SNPs.
To verify the admixture estimations using the selected set of SNPs, we also estimated the ancestry fractions of individuals from two admixed populations included in the HapMap3 database: Mexican ancestry from Los Angeles (MEX) and African ancestry in Southwest USA (ASW). Finally, we calculated the informativeness for assignment measure (In) proposed by Rosenberg et al25 to estimate the ancestral information that each SNP included provides.
Statistical analysis
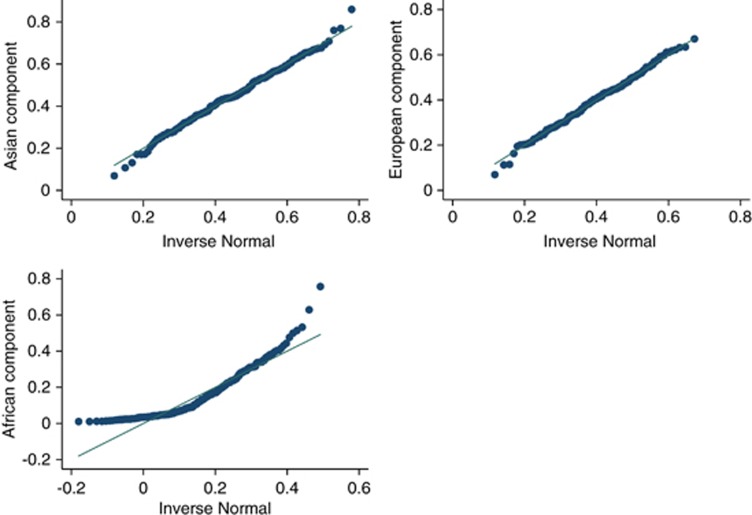
We compared the mean ancestry fractions between cases (adenomas and carcinomas) and controls using one way ANOVA tests. To evaluate the association of genetic ancestry with adenoma and cancer separately, we used binary conditional logistic regression models controlling for potential confounding factors. As the three ancestry fractions are dependant on each other, they cannot be handled as independent variables at once. To overcome this limitation, without leaving out from the analysis any of the ancestry fractions, we include into the model two parameters: first, the arithmetic difference between European and Amerindian ancestry fractions (main genetic substitution in Latin American populations) and, second, the estimated African ancestry fraction. The latter was modeled log transformed, as it was positively skewed in the study population (Figure 1). These two parameters were fitted alternatively as raw continuous (to measure the linear trend) and as categorical variable to measure the variation in risk per 10% increase of African ancestry (from 0.01 to ≥0.30) and European ancestry substitution increase (from ≤−0.30 to ≥0.30). We avoid the pairwise comparison of the resulting ancestry fraction categories in the logistic regression analysis, but we report their distribution for descriptive purposes.26 The multivariate analysis, which had city of enrollment as conditional variable, included the following: gender, age, attained education level (none, elementary school, secondary school, technical studies (ie, college), university or higher), family history of colorectal cancer in first-degree relatives, history of alcohol intake (no intake, <12.50 and ≥12.50 g/day), cigarette smoking (<0.5, 0.5–0.9 and ≥1 packs/year ), red meat consumption (< 2, 2–4 and ≥5 servings per week), physical activity (<10, 10–19, ≥20 h/week), non-steroid anti-inflammatory drugs (at least one per week during the last 6 months, yes or no), dietary fiber and total energy intake (quartiles regarding the distribution among controls).
Figure 1.
Quantile-normal plot per ancestry fraction in the study population.
Results
Cases were slightly older than controls. Although adenomas where positively associated with higher attained education (P=0.02), adenocancinomas showed the opposite, being positively associated with lower educational level instead (P<0.01). Cancer cases also showed an inverse association with BMI at diagnosis, probably due to reverse causation (Table 1). No other differences were observed among the risk factors included into the analysis.
Table 1. Main demographic characteristics and risk factors in the study population.
| Controls | Adenoma | Adenocarcinoma | ||||||
|---|---|---|---|---|---|---|---|---|
| (n=243) | (n=113) | (n=190) | ||||||
| Risk factor | n | % | n | % | P-values | n | % | P-values |
| Sex | ||||||||
| Female | 141 | 58.02 | 54 | 47.79 | 90 | 47.4 | ||
| Male | 102 | 41.98 | 59 | 52.21 | 0.13 | 100 | 52.6 | 0.14 |
| Age | ||||||||
| Mean years, (SD) | 54.2 (0.76) | 57.4 (0.94) | 0.01 | 57.9 (12.3) | 0.01 | |||
| Education attained | ||||||||
| None | 6 | 2.5 | 2 | 1.8 | 18 | 9.5 | ||
| Primary school | 91 | 37.4 | 33 | 29.2 | 75 | 39.5 | ||
| Secondary | 89 | 36.6 | 37 | 32.7 | 61 | 32.1 | ||
| College | 28 | 11.5 | 11 | 9.7 | 11 | 5.8 | ||
| University | 29 | 11.9 | 30 | 26.5 | 0.02 | 25 | 13.2 | <0.01 |
| Colorectal cancer family history | 113 | |||||||
| No | 231 | 95.0 | 104 | 92.0 | 176 | 92.6 | ||
| Yes | 13 | 5.0 | 9 | 8.0 | 0.26 | 14 | 7.4 | 0.29 |
| Alcohol intake (g/day) | ||||||||
| No intake | 135 | 55.6 | 72 | 63.7 | 107 | 56.3 | ||
| <12.5 | 70 | 28.8 | 22 | 19.5 | 49 | 25.8 | ||
| 12.5 | 38 | 15.6 | 18 | 15.9 | 0.17 | 33 | 17.4 | 0.75 |
| Smoking (packs/year) | ||||||||
| Non smoker | 146 | 60.1 | 71 | 62.8 | 112 | 58.9 | ||
| <1 | 51 | 21.0 | 16 | 14.2 | 32 | 16.8 | ||
| 1 | 46 | 18.9 | 26 | 23.0 | 0.35 | 45 | 23.7 | 0.27 |
| Red meat (servings/week) | ||||||||
| <2 | 42 | 17.3 | 20 | 18.6 | 35 | 18.4 | ||
| 2–5 | 121 | 49.8 | 51 | 44.2 | 80 | 42.1 | ||
| 5 | 78 | 32.1 | 34 | 30.1 | 0.83 | 62 | 32.6 | 0.59 |
| Energy intake (quartile) | ||||||||
| Q1 | 60 | 24.7 | 30 | 26.5 | 29 | 15.3 | ||
| Q2 | 61 | 25.1 | 29 | 25.7 | 41 | 21.6 | ||
| Q3 | 61 | 25.1 | 19 | 16.8 | 50 | 26.3 | ||
| Q4 | 60 | 24.7 | 25 | 22.1 | 0.53 | 56 | 29.5 | 0.13 |
| BMI | ||||||||
| <20 | 6 | 2.5 | 5 | 4.4 | 35 | 18.4 | ||
| 20–24 | 85 | 35.0 | 36 | 31.9 | 82 | 43.2 | ||
| 25–29 | 111 | 45.7 | 47 | 41.6 | 47 | 24.7 | ||
| 30 | 45 | 18.5 | 23 | 20.4 | 24 | 12.6 | <0.01 | |
| NSAID | ||||||||
| No | 184 | 75.7 | 87 | 77.0 | 148 | 77.9 | ||
| Yes | 59 | 24.3 | 26 | 23.0 | 0.84 | 42 | 22.1 | 0.6 |
| Dietary fiber (quartile) | ||||||||
| Q1 | 61 | 25.1 | 24 | 21.2 | 44 | 23.2 | ||
| Q2 | 61 | 25.1 | 23 | 20.4 | 39 | 20.5 | ||
| Q3 | 61 | 25.1 | 25 | 22.1 | 48 | 25.3 | ||
| Q4 | 59 | 24.3 | 33 | 29.2 | 0.63 | 46 | 24.2 | 0.88 |
| Physical activity (IPAQ categories) | ||||||||
| Low | 102 | 42.0 | 58 | 51.3 | 77 | 40.5 | ||
| Moderate | 72 | 29.6 | 27 | 23.9 | 49 | 25.8 | ||
| High | 69 | 28.4 | 28 | 24.8 | 0.17 | 64 | 33.7 | 0.6 |
| City of enrollment | ||||||||
| Bucaramanga | 69 | 28.4 | 35 | 31.0 | 40 | 21.1 | ||
| Bogota | 30 | 12.3 | 10 | 8.8 | 23 | 12.1 | ||
| Cartagena | 33 | 13.6 | 12 | 10.6 | 26 | 13.7 | ||
| Cali | 33 | 13.6 | 10 | 8.8 | 33 | 17.4 | ||
| SantaMarta | 26 | 10.7 | 20 | 17.7 | 27 | 14.2 | ||
| Barranquilla | 52 | 21.4 | 26 | 23.0 | 0.51 | 41 | 21.6 | 0.56 |
| Anatomic location | ||||||||
| Colon NOS | 8 | 7.1 | 8 | 4.2 | ||||
| Right colon | 40 | 35.4 | 48 | 25.3 | ||||
| Left colon | 43 | 38.1 | 42 | 22.1 | ||||
| Rectum | 22 | 19.5 | 92 | 48.4 | ||||
Abbreviations: BMI, body mass index; IPAC, International Physic Activity Questionnaire; NOS, no otherwise specified; NSAID, nonsteroidal anti-inflammatory drug.
Among adenomas cases, European (mean=0.44, 95% confidence interval (95% CI)=0.42–0.46) and African (mean=0.13, 95% CI=0.11–0.15) ancestry proportion were higher compared with controls (European: mean=0.39; 95% CI=0.38–0.41; African: mean=0.11, 95% CI=0.10–0.12), whereas the Amerindian mean proportion behave just inversely proportional to the European. In contrast, cancer cases showed a higher mean proportion of African ancestry compared with controls (0.14 vs 0.11 P<0.01; Table 2). When comparing the categorical distribution of African ancestry and European ancestry substitution (European minus Amerindian ancestry fractions) in the study population, we observed similar results: an association of African ancestry with both adenomas (P=0.07) and cancer (P=0.02), whereas the European genetic substitution only was associated with adenoma (P=0.001) but not with cancer cases (P=0.95; Table 3). Ancestry fractions estimated for the MEX and ASW population were very similar to those reported in the literature27 despite the low In values featured by the SNP included in our analysis (max=0.34, mean=0.03, SD=0.04), reassuring the reliability of the ancestry estimations.
Table 2. Mean ancestry fraction and 95% CIs in controls, adenomas, adenocarcinomas and admixed population (MEX and ASW) included into the analysis.
| Ancestry fraction | |||
|---|---|---|---|
| European | Amerindiana | African | |
|
Group/population |
Mean (95% CI) |
Mean (95% CI) |
Meanb
(95% CI) |
| Controls (n=238) | 0.39 (0.38–0.41) | 0.45 (0.43–0.46) | 0.11 (0.10–0.12) |
| Adenoma (n=115) | 0.44*** (0.42–0.46) | 0.39*** (0.37–0.41) | 0.13* (0.11–0.15) |
| Cancer (n=190) | 0.38 (0.37–0.40) | 0.43 (0.41–0.45) | 0.14** (0.12–0.16) |
| MEX (n=77) | 0.48 (0.45–0.51) | 0.48 (0.45–0.51) | 0.04 (0.03–0.05) |
| Ref valuec | 0.5 | 0.45 | 0.05 |
| ASW (n=83) | 0.18 (0.15–0.20) | 0.05 (0.04–0.07) | 0.77 (0.75–0.79) |
| Ref valuec | 0.2 | − | 0.8 |
Abbreviations: ANOVA, analysis of variance; ASW, African ancestry in Southwest USA; CHB, Han Chinese in Beijing, China; CI, confidence interval; MEX, Mexican ancestry from Los Angeles.
P-value for one-way ANOVA test: ***<0.001, **<0.01 and *<0.1*.
Based on CHB reference population.
Back transformated from log.
Seldin et al.27
Table 3. Categorical distribution of African and European ancestry substitutiona in controls, adenomas and colorectal cancer cases included into the analysis.
| Control | Adenomas | Cancer | |
|---|---|---|---|
| n (%) | n (%) | n (%) | |
| African Ancestry | |||
| 0.01–0.09 | 110 (45.27) | 39 (34.51) | 58 (30.53) |
| 0.10–0.19 | 58 (23.87) | 38 (33.63) | 64 (33.68) |
| 0.20–0.29 | 39 (16.05) | 24 (21.24) | 34 (17.89) |
| 0.30–0.39 | 36 (14.81) | 12 (10.62) | 34 (17.89) |
| P-valueb | 0.07 | 0.02 | |
| European ancestry substitutiona | |||
| ≤−0.30 | 55 (22.63) | 12 (10.62) | 46 (24.21) |
| −0.29 to −0.20 | 40 (16.46) | 17 (15.04) | 34 (17.89) |
| −0.19 to −0.10 | 49 (20.16) | 20 (17.7) | 39 (20.53) |
| 0–0.09 | 58 (23.87) | 22 (19.47) | 36 (18.95) |
| 0.10–0.19 | 41 (16.87) | 42 (37.17) | 35 (18.42) |
| 0.20–0.29 | 9 (3.7) | 11 (9.73) | 8 (4.21) |
| ≥0.30 | 11 (4.53) | 16 (14.16) | 10 (5.26) |
| P-valueb | 0.001 | 0.87 | |
European ancestry substitution=European minus Amerindian ancestry fractions.
P-value for χ2-test.
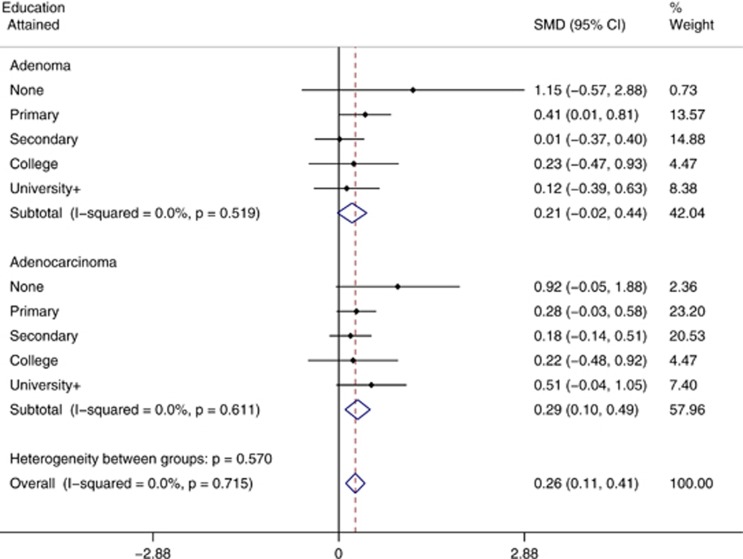
After controlling for confounding, conditional logistic regression analysis results were consistent with the crude ones, showing a positive marginal association of increasing African ancestry with colorectal adenomas (risk variation per 10% increase (odds ratio (OR))=1.122; 95% CI=0.97, 1.30; P for linear trend=0.08) and statistically significant with adenocarcinomas (risk variation per 10% increase (OR)=1.19; 95% CI=1.05, 1.35; P for trend=0.003; Table 4). In contrast, increasing European ancestry was positively associated only to adenoma (risk variation per 10% increase (OR)=1.25; 95% CI=1.08, 1.46; P for trend=0.001) but not to cancer (risk variation per 10% increase (OR)=1.02; 95% CI=0.90–1.16; P for trend=0.75). In addition, adenoma was associated with university or higher education compared with primary school (the most prevalent category; OR=3.81, 95% CI=1.59–9.17), whereas colorectal cancer risk was marginally associated with no education attained (OR=2.5, 95% CI=0.89–7.36). Adjusting by age and sex did not modify the results significantly (Table 4). There was no evidence of heterogeneity in the mean differences of African ancestry when comparing adenomas or adenocarcinomas with controls across education strata (Figure 2). When exploring the ancestry association stratified by distal and proximal colorectal neoplasms, we did not observed any differences from the overall results (results not shown).
Table 4. OR and 95% CIs of age and sex adjusted and fully adjusted conditional regression models for genetic ancestry fractions and known risk factor of colorectal adenoma and adenocarcinoma.
| Adenoma | Adenocarcinoma | |||||
|---|---|---|---|---|---|---|
| Characteristic | OR | 95% CI | P-values | OR | 95% CI | P-values |
| Age- and sex-adjusted model | ||||||
| African ancestry | ||||||
| Risk variation per 10% increase | 1.13 | (0.99, 1.27) | 0.06 | 1.18 | (1.06, 1.31) | 0.001 |
| P for trend | 0.09 | 0.002 | ||||
| European ancestry a | ||||||
| Risk variation per 10% increase | 1.25 | (1.08, 1.46) | 0.001 | 0.98 | (0.88, 1.10) | 0.79 |
| P for trend | 0.0004 | 0.91 | ||||
| Fully adjusted model | ||||||
| African ancestry | Ref | − | Ref | − | ||
| Risk variation per 10% increase | 1.12 | (0.97, 1.30) | 0.15 | 1.19 | (1.05, 1.35) | 0.006 |
| P for trend | 0.08 | 0.003 | ||||
| European ancestry b | ||||||
| Risk variation per 10% increase | 1.25 | (1.08, 1.46) | 0.007 | 1.02 | (0.90, 1.16) | 0.75 |
| P for trend | 0.001 | 0.68 | ||||
| Education attained | ||||||
| None | 0.77 | (0.11, 5.24) | 0.79 | 2.56 | (0.89, 7.36) | 0.08 |
| Primary school | Ref | − | Ref | − | ||
| Secondary | 1.5 | (0.74, 3.06) | 0.26 | 0.77 | (0.45, 1.30) | 0.4 |
| College | 2.11 | (0.78, 5.75) | 0.14 | 0.55 | (0.23, 1.31) | 0.23 |
| University | 3.82 | (1.59, 9.17) | 0.01 | 1.03 | (0.60, 2.52) | 0.95 |
| Colorectal cancer family history c | ||||||
| Yes | 1.73 | (0.60, 5.02) | 0.83 | 1.72 | (0.67, 4.40) | 0.74 |
| Alcohol intake (g/day) c | ||||||
| <12.5 | 0.65 | (0.30, 1.44) | 0.29 | 1.08 | (0.56, 2.07) | 0.82 |
| 12.5 | 0.86 | (0.40, 1.83) | 0.69 | 0.97 | (0.48, 1.94) | 0.92 |
| Smoking (packs/years) | ||||||
| <0.5 | Ref | − | Ref | − | ||
| 0.5–0.9 | 0.65 | (0.30, 1.44) | 0.29 | 1.08 | (0.56, 2.07) | 0.82 |
| 1 | 0.86 | (0.40, 1.83) | 0.69 | 0.97 | (0.48, 1.94) | 0.92 |
| Red meat (times/week) | ||||||
| <2 | Ref | − | Ref | − | ||
| 2–4 | 0.87 | (0.41, 1.84) | 0.71 | 1.34 | (0.67, 2.67) | 0.41 |
| 5 | 0.84 | (0.37, 1.88) | 0.67 | 1.66 | (0.80, 3.42) | 0.17 |
| Energy intake (quartile) | ||||||
| Q1 | Ref | − | Ref | − | ||
| Q2 | 0.81 | (0.35, 1.88) | 0.62 | 0.64 | (0.31, 1.33) | 0.23 |
| Q3 | 1.21 | (0.49, 2.99) | 0.69 | 0.68 | (0.30, 1.52) | 0.34 |
| Q4 | 1.36 | (0.51, 3.65) | 0.54 | 0.44 | (0.18, 1.08) | 0.09 |
| NSAID | ||||||
| Yes | 0.6 | (0.29, 1.21) | 0.15 | 0.93 | (0.52, 1.66) | 0.82 |
| Dietary fiber (quartile) | ||||||
| Q1 | 1 | |||||
| Q2 | 0.81 | (0.35, 1.88) | 0.62 | 0.64 | (0.31, 1.33) | 0.23 |
| Q3 | 1.21 | (0.49, 2.99) | 0.69 | 0.68 | (0.30, 1.52) | 0.34 |
| Q4 | 1.36 | (0.51, 3.65) | 0.54 | 0.44 | (0.18, 1.08) | 0.07 |
| Physical activity (IPAQ categories) | ||||||
| Low | ||||||
| Moderate | 0.67 | (0.34, 1.32) | 0.24 | 0.77 | (0.43, 1.37) | 0.37 |
| High | 0.76 | (0.37, 1.54) | 0.44 | 0.89 | (0.50, 1.60) | 0.7 |
Abbreviations: CI, confidence interval; IPAC, International Physic Activity Questionnaire; NSAID, nonsteroidal anti-inflammatory drug; OR, odds ratio.
European ancestry substitution effect (European−Amerindian ancestry) adjusted only for age, sex and city of enrollment.
European ancestry substitution effect (European−Amerindian ancestry) adjusted for age, sex, city of enrollment and all other factors listed.
Absence of the exposure considered as reference value.
Figure 2.
Forest plot for the standardized mean differences (SMD) of African ancestry fraction (log scale) in adenomas and adenocarcinomas stratified by education level attained.
Discussion
To the best of our knowledge, this is the first report on the association of genetic ancestry and sporadic colorectal adenomas and adenocarcinomas in an admixed population. Our findings add evidence to the hypothesis that genetic ancestry influences cancer risk in Latino populations. A similar positive association has been reported previously between European ancestry and breast cancer in the Mexican population.28 Genetic ancestry fractions estimations have recently drawn the attention in clinical practice as they have been proposed as a genome-wide biomarker useful to evaluate relapse in children undergoing therapy for acute lymphoblastic leukemia.29
The association of African ancestry not only with adenocarcinoma but also marginally with adenoma supports our hypothesis of the role of genetic ancestry in early stages of colorectal carcinogenesis and may rely on differences in allele frequencies in polymorphism related to colorectal cancer risk.30 We found that this association was not confounded by well-known risk factors including education attained. Education level included into the analysis, was chosen as proxy of socioeconomic status (SES), because it is attained early in life and does not change greatly after the third decade of life. SES has previously been associated independently to colorectal cancer worldwide and to genetic ancestry in Latin America. The nature of the association between SES and colorectal cancer risk is discrepant across continents.31 Whilst studies in Europe, East Asia and Australia, in general, have found a positive association, in the United States and Canada the association observed is inverse.31, 32 This discrepancy is not fully understood but it is partially explained by differences in screening coverage31 and the way environmental factors (mediators) are interrelated with SES (determinants).32, 33 Our results are contrary to a previous study reporting a positive association of colorectal cancer and SES in Colombia.34 Here we found an inverse association of education level with adenocarcinoma, but also a positive association on higher education level with adenoma (Table 4). A previous report has also shown the positive association on European ancestry with higher education among Latinos.35 Thus, the interplay of wealth, European ancestry and education among the Latino population, could partially explain the positive association of European ancestry and adenoma risk found in this analysis.
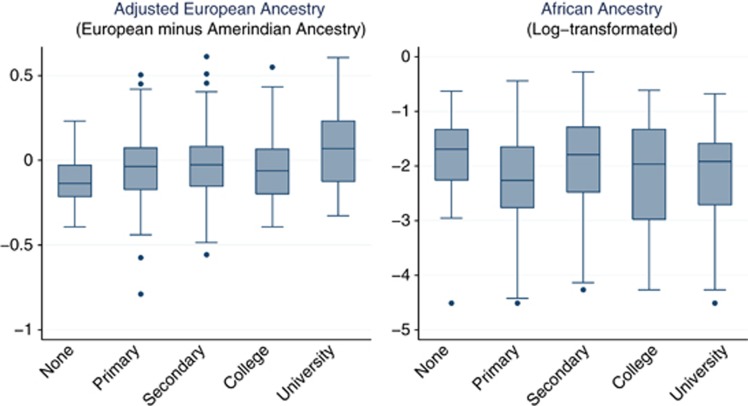
In contrast, the positive association of African ancestry to both adenoma and adenocarcinoma reported here is hard to explain due to differences in SES given that in our study, first, African Ancestry was not associated with education level (P for trend=0.76; Figure 3) and, second, adenomas and adenocarcinomas showed opposite association to education level. It is worth mentioning that Afro-Colombians are an ethnic minority with large disparities compared with the overall population. In our study population, the African ancestry was not high (interquartile range=0.6–0.24) as African Americans (80%) and its likely that within this range most of the individuals did not identify themselves as being of African descent.
Figure 3.
European and African ancestry fraction box plot per education level attained in the study population.
Our study features several strengths. It includes both preneoplasic and neoplasic lesions confirmed by histopathology, allowing us to evaluate the association of ancestry in the early events of colorectal carcinogenesis; cases and controls were sampled from the same population (all controls were recruited at general practice consult and cases were mostly referred by general physicians to colonoscopy), assuring a better control for selection bias. Our ancestry estimations are reliable, as those estimated for the ASW and MEX individuals were similar to those published in the literature.27 The differences in the anatomic distribution of adenomatous polyps and cancer showing a shift to the left for cancer cases was observed as previously described,36 and the similar age range in adenomas and adenocarcinomas does not suggest a selection bias.37
There are some limitations of our study that should be considered when interpreting the results. It is likely that educational levels do not reflect, nor control, the entire variability of the socioeconomic status; thus, residual confounding may exist. Nevertheless, here we assessed and included into the analysis the most relevant nutritional and lifestyle factors that may mediate the association of SES with colorectal adenoma and adenocarcinoma. There could be a differential access to colonoscopy, where wealthy people may have better access to these procedures. However, the chance of underrepresentation of individuals with a higher level of education in the control group is not likely, as recent census data38 in Colombia showed that only 9% of the population have university or a higher education degree, a similar value observed in our control sample (11%). Our sample size provides limited statistical power to detect small effects, especially regarding the observed effect in the association between African ancestry and adenomas. Nevertheless, we reduced multiple hypothesis testing to the minimum. In addition, the point estimates observed showed narrow CIs and the crude and adjusted results were consistent. If African ancestry is actually associated with colorectal cancer, the association with adenoma (its main precursor lesion) could also be expected. However, the use of a common set of controls could also explain such association and therefore the results should be interpreted with caution. As an observational study, residual confounding cannot be ruled out. Replication of these results is warranted to validate our results.
The positive association of African genetic ancestry with adenoma and colorectal cancer is consistent with a recent publication reporting that colorectal cancer risk is likely to be mediated through genetic susceptibility to adenomas.39 Early detection of adenomas is a key issue in colorectal cancer control. Newly published evidence supports that detecting adenomas and removing them not only decreases the incidence but also the mortality of colorectal cancer.40 There is now promising evidence showing that genetic markers could discriminate population at increased risk of colorectal cancer. It has been shown, for example, that adding information of SNPs associated with colorectal cancer to family history increases the absolute risk estimation of having the disease at population level.41
Further research should address how these SNPs, discovered mainly in European Caucasian population, influence the genetic association here reported. Admixture mapping42 could be the next step to further explore the mechanism behind this association. There is no admixture mapping analysis published so far on colorectal cancer despite the large number of GWAS study on this cancer. A variant in chromosome 8q24 initially described by admixture mapping for prostate cancer in African Americans also showed a positive association with colorectal cancer risk30 and, recently, a case–control study reported an association of one 8q24 loci variant (rs380284) and adenoma risk in Caucasians.39
In conclusion, we report for the first time that African ancestry (or variants linked to it) contributes to the susceptibility of colorectal cancer in admixed Latin American population. Our results are promising as they may help get insights of colorectal carcinogenesis and even more to find biomarkers useful to stratify colorectal cancer risk in the Latino populations, where colorectal mortality rates are increasing, although not high enough to recommend mass screening programs.43
Acknowledgments
We are most grateful to the participants in the study. We thank all the in-field work team (nurses, gastroenterologists and pathologists) in all cities, who worked in enrolling eligible individuals, and all the administrative support staff at the National Cancer Institute, who collaborated with this study. We also thank all healthcare institutions that participated in this study: Bogota: Hospital San Rafael, Hospital Universitario Mederi, Clinica Colombia; Barranquilla: Clinica General de Norte, UGASEND; Bucaramanga: Hospital Universitario de Santander, (Cali), Fundación Oftalmológica de Santander; Cali: Hospital Univeristario del Valle, Fundación del Valle de Lili, Centro Medico Imbanaco; Cartagena: Hospital San Juan de Dios, Hospital Universitario del Caribe, Hospital Naval; Santa Marta: Laboratorio de patologia CITOPAT, Hospital Universitario Fernando Troconis, Unidad Endoscopica GASTROMAG. Finally, we thank the UICC for the ASCBI fellowship grant, which allow us work on the analysis of data for this manuscript at the Roslin Institute. This study was funded by the National Cancer Institute of Colombia (internal grant number 41030310-24); Colombian National Research Council, Colciencias (grant number 210145921617); Union International Cancer Control (UICC; ACSBI Fellowship 2011 grant (to GH)); NIGMS – P20GM103501; Louisiana Cancer Research Consortium; AT acknowledges support from Cancer Research UK grant C12229/A13154 and the Biotechnology and Biological Sciences Research Council through the Roslin Institute Strategic Programme Grant.
The authors declare no conflict of interest.
References
- Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59:366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- Rim SH, Seeff L, Ahmed F, King JB, Coughlin SS. Colorectal cancer incidence in the United States, 1999-2004. Cancer. 2009;115:1967–1976. doi: 10.1002/cncr.24216. [DOI] [PubMed] [Google Scholar]
- Eheman C, Henley SJ, Ballard-Barbash R, et al. Annual report to the nation on the status of cancer, 1975-2008, featuring cancers associated with excess weight and lack of sufficient physical activity. Cancer. 2012;118:2338–2366. doi: 10.1002/cncr.27514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal S, Bhupinderjit A, Bhutani MS, et al. Colorectal cancer in African Americans Am J Gastroenterol 2005100515–523.discussion 514. [DOI] [PubMed] [Google Scholar]
- Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, van Ballegooijen M, Zauber AG, Jemal A. Contribution of screening and survival differences to racial disparities in colorectal cancer rates. Cancer Epidemiol Biomarkers Prev. 2012;21:728–736. doi: 10.1158/1055-9965.EPI-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollberding NJ, Nomura AM, Wilkens LR, Henderson BE, Kolonel LN. Racial/ethnic differences in colorectal cancer risk: the multiethnic cohort study. Int J Cancer. 2011;129:1899–1906. doi: 10.1002/ijc.25822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovov B, Araujo-Perez F, Sigel CS, et al. Differential gene expression between African American and European American colorectal cancer patients. PLoS One. e30168. 2012;7 doi: 10.1371/journal.pone.0030168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou J, Carbonero F, Zoetendal EG, et al. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Weng YR, Jiang B, et al. Epidemiological study of colorectal adenoma and cancer in symptomatic patients in China between 1990 and 2009. J Dig Dis. 2011;12:371–378. doi: 10.1111/j.1751-2980.2011.00531.x. [DOI] [PubMed] [Google Scholar]
- Wilson LS, Lightwood J. Model of estimated rates of colorectal cancer from polyp growth by year of surveillance. J Med Screen. 2001;8:187–196. doi: 10.1136/jms.8.4.187. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Fitzpatrick FA. Convergence of hormones, inflammation, and energy-related factors: a novel pathway of cancer etiology. Cancer Prev Res (Phila) 2009;2:922–930. doi: 10.1158/1940-6207.CAPR-08-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffeld SM, Loffeld RJ. Colorectal cancer and adenomas are rare in individuals of Turkish descent living in the Zaanstreek region in the Netherlands. J Cancer Res Clin Oncol. 2010;136:1439–1443. doi: 10.1007/s00432-010-0799-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace K, Ahnen D, Burke C, et al. Race and risk of large bowel polyps in younger and older patients. Cancer Epidemiol Biomarkers Prev. 2012;21:566. [Google Scholar]
- Sucheston LE, Bensen JT, Xu Z, et al. Genetic ancestry, self-reported race and ethnicity in African Americans and European Americans in the PCaP cohort. PLoS One. 2012;7:e30950. doi: 10.1371/journal.pone.0030950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herran OF, Ardila MF, Rojas MP, Hernandez GA. [Design of dietary questionnaires to study the relationships between diet and cancer prevalence in Colombia] Biomedica. 2010;30:116–125. [PubMed] [Google Scholar]
- Craig CL, Marshall AL, Sjostrom M, et al. International physical activity questionnaire: 12-country reliability and validity. Med Sci Sports Exerc. 2003;35:1381–1395. doi: 10.1249/01.MSS.0000078924.61453.FB. [DOI] [PubMed] [Google Scholar]
- Terry MB, Neugut AI, Bostick RM, et al. Risk factors for advanced colorectal adenomas. Cancer Epidemiol Biomarkers Prev. 2002;11:622–629. [PubMed] [Google Scholar]
- Cunningham JM, Sellers TA, Schildkraut JM, et al. Performance of amplified DNA in an Illumina GoldenGate BeadArray assay. Cancer Epidemiol Biomarkers Prev. 2008;17:1781–1789. doi: 10.1158/1055-9965.EPI-07-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer BR, Yeager M, Burdett L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Zhang W, Cox NJ, Dolan ME. FstSNP-HapMap3: a database of SNPs with high population differentiation for HapMap3. Bioinformation. 2008;3:139–141. doi: 10.6026/97320630003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins-Schramm HE, Chima B, Morii T, et al. Mexican American ancestry-informative markers: examination of population structure and marker characteristics in European Americans, Mexican Americans, Amerindians and Asians. Hum Genet. 2004;114:263–271. doi: 10.1007/s00439-003-1058-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Li LM, Ward R, Pritchard JK. Informativeness of genetic markers for inference of ancestry. Am J Hum Genet. 2003;73:1402–1422. doi: 10.1086/380416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers AJ, Bennette C. Against Quantiles: categorization of continuous variables in epidemiologic research, and its discontents. BMC Med Res Methodol. 2012;12:21. doi: 10.1186/1471-2288-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seldin MF, Pasaniuc B, Price AL. New approaches to disease mapping in admixed populations. Nat Rev Genet. 2011;12:523–528. doi: 10.1038/nrg3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejerman L, Romieu I, John EM, et al. European ancestry is positively associated with breast cancer risk in Mexican women. Cancer Epidemiol Biomarkers Prev. 2010;19:1074–1082. doi: 10.1158/1055-9965.EPI-09-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cheng C, Devidas M, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43:237–241. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson N, Yu F, et al. A genomewide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarts MJ, Lemmens VE, Louwman MW, Kunst AE, Coebergh JW. Socioeconomic status and changing inequalities in colorectal cancer? A review of the associations with risk, treatment and outcome. Eur J Cancer. 2012;46:2681–2695. doi: 10.1016/j.ejca.2010.04.026. [DOI] [PubMed] [Google Scholar]
- Doubeni CA, Laiyemo AO, Major JM, et al. Socioeconomic status and the risk of colorectal cancer. Cancer. 2012;118:3636–3644. doi: 10.1002/cncr.26677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leufkens AM, Van Duijnhoven FJ, Boshuizen HC, et al. Educational level and risk of colorectal cancer in EPIC with specific reference to tumor location. Int J Cancer. 2012;130:622–630. doi: 10.1002/ijc.26030. [DOI] [PubMed] [Google Scholar]
- Haenszel W, Correa P, Cuello C. Social class differences among patients with large-bowel cancer in Cali, Colombia. J Natl Cancer Inst. 1975;54:1031–1035. [PubMed] [Google Scholar]
- Florez JC, Price AL, Campbell D, et al. Strong association of socioeconomic status with genetic ancestry in Latinos: implications for admixture studies of type 2 diabetes. Diabetologia. 2009;52:1528–1536. doi: 10.1007/s00125-009-1412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenoglio L, Castagna E, Comino A, et al. A shift from distal to proximal neoplasia in the colon: a decade of polyps and CRC in Italy. BMC Gastroenterol. 2010;10:139. doi: 10.1186/1471-230X-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Mori M, Yoshizumi T, Sugimachi K. Cancer and adenomatous polyp distribution in the colorectum. Am J Gastroenterol. 1999;94:191–193. doi: 10.1111/j.1572-0241.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- Ministerio de Educacion, Colombia http://www.mineducacion.gov.co/cvn/1665/article-99519.html , 2006
- Carvajal-Carmona LG, Zauber AG, Jones AM, et al. Much of the genetic risk of colorectal cancer is likely to be mediated through susceptibility to adenomas. Gastroenterology. 2013;144:53–55. doi: 10.1053/j.gastro.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauber AG, Winawer SJ, O'Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366:687–696. doi: 10.1056/NEJMoa1100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop MG, Tenesa A, Farrington SM, et al. Cumulative impact of common genetic variants and other risk factors on colorectal cancer risk in 420.103 individuals. Gut. 2012;62:871–881. doi: 10.1136/gutjnl-2011-300537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ. A twist on admixture mapping. Nat Genet. 2011;43:178–179. doi: 10.1038/ng0311-178. [DOI] [PubMed] [Google Scholar]
- Lambert R, Sauvaget C, Sankaranarayanan R. Mass screening for colorectal cancer is not justified in most developing countries. Int J Cancer. 2009;125:253–256. doi: 10.1002/ijc.24371. [DOI] [PubMed] [Google Scholar]