Abstract
Fragile X syndrome is a common inherited form of intellectual disability and autism spectrum disorder. Most patients exhibit a massive CGG-repeat expansion mutation in the FMR1 gene that silences the locus. In over two decades since the discovery of FMR1, only a single missense mutation (p.(Ile304Asn)) has been reported as causing fragile X syndrome. Here we describe a 16-year-old male presenting with fragile X syndrome but without the repeat expansion mutation. Rather, we find a missense mutation, c.797G>A, that replaces glycine 266 with glutamic acid (p.(Gly266Glu)). The Gly266Glu FMR protein abolished many functional properties of the protein. This patient highlights the diagnostic utility of FMR1 sequencing.
Keywords: fragile X syndrome, mutation, missense, FMR1 sequencing
Introduction
Fragile X syndrome (FXS) is an X-linked disorder presenting in males and, less frequently, in females with developmental delay. It is characterized by intellectual disability, speech and language delay, and a characteristic physical appearance of a long narrow face with prominent ears and jaw and macroorchidism in males.1 Most patients suffer from severe social anxiety and hyperarousal with 60% of patients meeting diagnostic criteria for some form of autism spectrum disorder (ASD). Indeed, FXS is the most common single gene cause of ASD.2
FXS was one of the first examples of a trinucleotide repeat expansion disorder with the discovery of its causal gene, FMR1.3 This gene harbors a CGG-repeat in its 5′ untranslated region. In normal individuals, repeat length is polymorphic, with 29–30 repeats being the most common allele.4 In patients with FXS, there is a large expansion of the repeat sequence to over 200 repeats, referred to as the full mutation. Once the repeat length reaches 200, an epigenetic event is triggered that results in methylation of the entire FMR1 gene and silences transcription.1
The absence of FMR1 transcription leads to the loss of the encoded protein, FMRP. FMRP is a selective RNA-binding protein that regulates translation of its target mRNAs at the synapse in an activity-dependent manner.5 Precise translation of these messages modulates synaptic strength and in the absence of FMRP, synaptic strength is defective with excess internalization of AMPA receptors from the synaptic membrane.6 This mimics excessive group 1 metabotropic glutamate receptor signaling, which is an insight that has initiated several FXS clinical trials with therapeutic approaches directly targeting this pathway.7, 8, 9
FXS is almost exclusively diagnosed by molecular testing for CGG-repeat expansion, even though the causal link to FXS is not directly because of repeat expansion but rather because of the loss of the encoded FMR protein. This CGG-repeat test is among the most frequently ordered genetic tests and is standard of care for any child not meeting developmental milestones. Consequently, the actual diagnostic yield is only 1–2%.10 Whereas it is clear that CGG-repeat expansion is the most frequent cause of FXS, more conventional mutations, particularly FMR1 deletions, have also been reported in FXS.11 Deletions are typically found as an anomaly of the CGG-repeat test or by microarray analysis, and sequencing of FMR1 is not frequently carried out. This lack of clinical FMR1 sequencing is not surprising as it was assumed that sequencing would not uncover a significant number of mutations, and therefore negatively affected insurance coverage of diagnostic sequencing. The lack of FMR1 sequence testing has led to a marked deficit of conventional mutations, particularly missense mutations, even though the full mutation is a null allele like many conventional mutations. Indeed, in over two decades since the discovery of FMR1 only a single missense mutation, isoleucine 304 to asparagine (p.(Ile304Asn)), has been reported.12 Here we report a second missense mutation in FMR1, glycine 266 to glutamic acid (p.(Gly266Glu)), leading to FXS. This highlights the clinical utility of FMR1 sequencing, particularly at a time when gene sequencing is now more affordable and targeted therapeutics for FXS are being developed.
Materials and methods
Patient
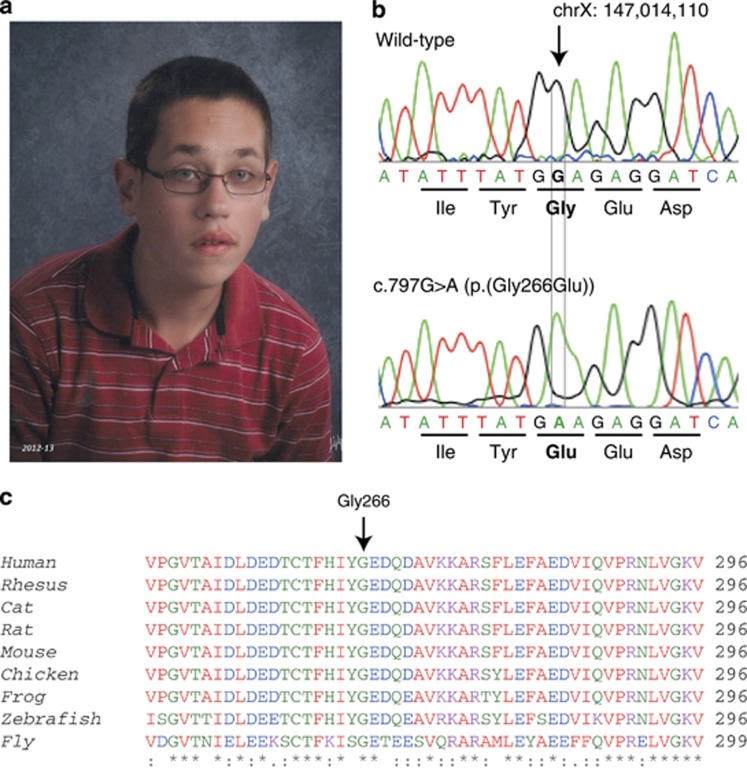
The patient was delivered vaginally at 41 weeks' gestation without complication and an occipital frontal circumference at 50th percentile. No abnormalities were noted except a bifid uvula. However, by 6 months developmental delay was noticed with walking achieved at 24 months. At the age of 13, the patient's gross motor skills, visual-motor problem solving, speech and language skills, and general conceptual abilities were all around the 3- to 4-year level. The patient achieved urinary continence at the age of 15. Examination at the age of 16 revealed a severely intellectually impaired male. The patient's height was 172.5 cm (45th percentile), weight 52.3 kg (17th percentile), and occipital frontal circumference 56 cm (66th percentile). Ear length was 7.5 cm (>95th percentile). The patient was diagnosed with an autistic spectrum disorder based upon observed or reported difficulty with eye contact, little interaction with peers, little social and emotional reciprocity, no indications of make-believe play, and difficulty with transitions and rituals in activities of daily living. Multiple dysmorphic features were noted (Figure 1a) including a tall forehead, long face, large ears, hypermobility of the elbows and small joints of the hands, flat feet, several café-au-lait macules, and macroorchidism. In adolescence, the patient began having disruptive outbursts with some aggression, poor attention span and hyperactivity, and was diagnosed with attention deficit with hyperactivity disorder (ADHD) that responded favorably to treatment with methylphenidate. There is no history of seizures. There is no other family history of developmental disorder in the proband's parents, three brothers and two sisters, or two maternal uncles.
Figure 1.
Identification of a patient with a novel FMR1 missense mutation. (a) Patient's characteristic facial features that are consistent with Fragile X Syndrome, including tall forehead, elongated face, and large ears. (b) DNA chromatogram of the wild-type and patient alleles showing the single nucleotide substitution (NM_002024.5:c.797G>A) that replaces the glycine at residue 266 with glutamic acid (p.(Gly266Glu)). (c) ClustalW alignment across multiple species of FMRP amino acids 247–296. FMRP at residue 266 is highly conserved from human through Drosophila.
The patient was negative for FXS testing of CGG-repeat length (23 repeats). Karyotype analysis showed normal 46, XY. Array comparative genomic hybridization studies showed no copy number variants, metabolic studies were all within normal limits, and brain MRI was normal. Full sequencing of the patient's FMR1 gene revealed a guanine to adenine transition at position chrX:g.147014110G>A (NM_002024.5:c.797G>A) leading to a missense mutation at amino acid 266, converting the highly conserved glycine residue to glutamic acid (Figures 1b and c). The patient's mother is unaffected but is found to carry the Gly266Glu mutation, and all three of his unaffected brothers do not carry the mutation. This variant has been submitted to the FMR1 variant database (http://databases.lovd.nl/shared/genes/FMR1).
Mutation analysis
Full-length FMR1, with and without the G266E mutation, was cloned into a lentiviral vector. Subsequent lentivirus was used to transduce primary neuron cultures dissected from Fmr1 KO mice at embryonic day 16.5 or immortalized mouse embryonic fibroblasts (MEFs) from Fmr1 knockout mice. Analysis of AMPA receptor trafficking, polyribosome profiling, and mRNA binding were carried out as previously described (see Supplementary Materials). This study was approved by the Emory University Internal Review Board.
Results
In order to determine whether the c.797G>A (p.(Gly266Glu)) mutation is pathological, we tested the mutation using various established functions of FMRP in vitro. One of the penultimate consequences of FMRP loss is exaggerated AMPA receptor internalization.5 Cultured mouse hippocampal neurons derived either from wild-type (WT) or Fmr1 knockout (KO) mice have been shown to exhibit marked differences in AMPA receptor trafficking, with the KO neurons showing significantly greater internalization than WT neurons.13 We infected Fmr1 KO neurons with either WT or G266E-FMRP lentivirus and measured AMPA receptor internalization. We found that G266E-FMRP was unable to rescue the exaggerated AMPA receptor internalization in KO neurons, indicating that this mutant protein leads to impaired synaptic function (Figure 2a).
Figure 2.
Functional analysis of mutant G266E-FMRP. (a) Constitutive AMPA receptor assay showing that G266E-FMRP is unable to rescue exaggerated AMPA receptor internalization in KO neurons. Hippocampal neurons from Fmr1 KO mice were cultured for 18 days, infected with either WT or G266E-FMRP, and the percentage of internalized to total AMPA receptors was calculated from individual dendrites. G266E-FMRP-infected neurons were statistically different from WT-FMRP-infected neurons (one-way ANOVA; n=30; F=609.92, P<0.001, Tukey post hoc analysis: ***P<0.001 for all pairwise comparisons except WT versus KO+WT-FMRP P=0.42). As the variance in uninfected KO neurons was so low, G266E-infected neurons were still statistically different from KO neurons even though the mutant protein clearly does not rescue AMPA receptor internalization like WT-FMRP. Data are represented as boxplot with whiskers from minimum to maximum. (b) Polyribosome assay showing that G266E-FMRP does not associate with polyribosome fractions. The top graph is a representative A254 absorbance profile from Fmr1 KO MEF cells infected with either WT or G266E-FMRP, and the monosome (80S) and polyribosome peaks are indicated. Below is the distribution of FMRP by western blot analysis for each fraction corresponding to the same region of the linear sucrose gradient above. S6 ribosomal protein is also shown to verify sample loading in each well. These are representative blots from n=3 experiments. (c) RNA co-immunoprecipitation showing that G266E-FMRP does not bind three well-validated FMRP targets using qPCR analysis of the relative mRNA enrichment of Map1B, PSD95, and CamKII mRNAs after FMRP immunoprecipitation. Cortical neurons from Fmr1 KO mice were cultured for 10 days, infected with GFP, WT-FMRP, or G266E-FMRP lentivirus, and then processed for FMRP-RNA co-immunoprecipitation. The relative mRNA level for each primer set was normalized to each sample's β-actin mRNA and also relative FMRP expression level as determined by western blot densitometry. When mRNA enrichment (IP:input) for WT-FMRP is set to equal 1.0, G266E-FMRP mRNA enrichment drops by twofold to the same levels as GFP-infected neurons (paired Student's t-test; n=4; t=16.92, ***P<0.001 for Map1B; t=12.544, **P=0.001 for PSD95; t=12.919, **P=0.001 for CamKII). Data are represented as mean±SD.
Impaired synaptic function by Gly266Glu suggests that the ability of FMRP to regulate protein synthesis may be defective. A canonical property of this protein in translation is its association with polyribosomes.14 To determine whether G266E-FMRP is able to associate with polyribosomes, we observed the distribution of FMRP in sucrose gradients of lysates from Fmr1 KO MEFs that were infected with either WT or G266E-FMRP. WT-FMRP showed robust FMRP staining in polyribosome fractions, whereas G266E-FMRP was virtually absent in these fractions (Figure 2b). Thus, mutant G266E-FMRP is defective in its ability to associate with polyribosomes.
FMRP associates with polyribosomes, in part, because of its selective binding of mRNA,15 and a number of specific targets have been extensively validated.1 In order to determine whether G266E-FMRP is able to bind mRNA, we analyzed the relative mRNA levels of three well-characterized FMRP targets in co-immunoprecipitation experiments from WT or G266E-FMRP-infected Fmr1 KO cortical neurons. We found that the relative mRNA enrichment for Map1B, PSD95, and CamKII mRNAs in G266E-FMRP pull-down experiments was similar to the background levels from GFP-infected negative controls (Figure 2c). These values were significantly lower than WT-FMRP pull-down experiments, indicating that G266E-FMRP is unable to bind known FMRP mRNA targets.
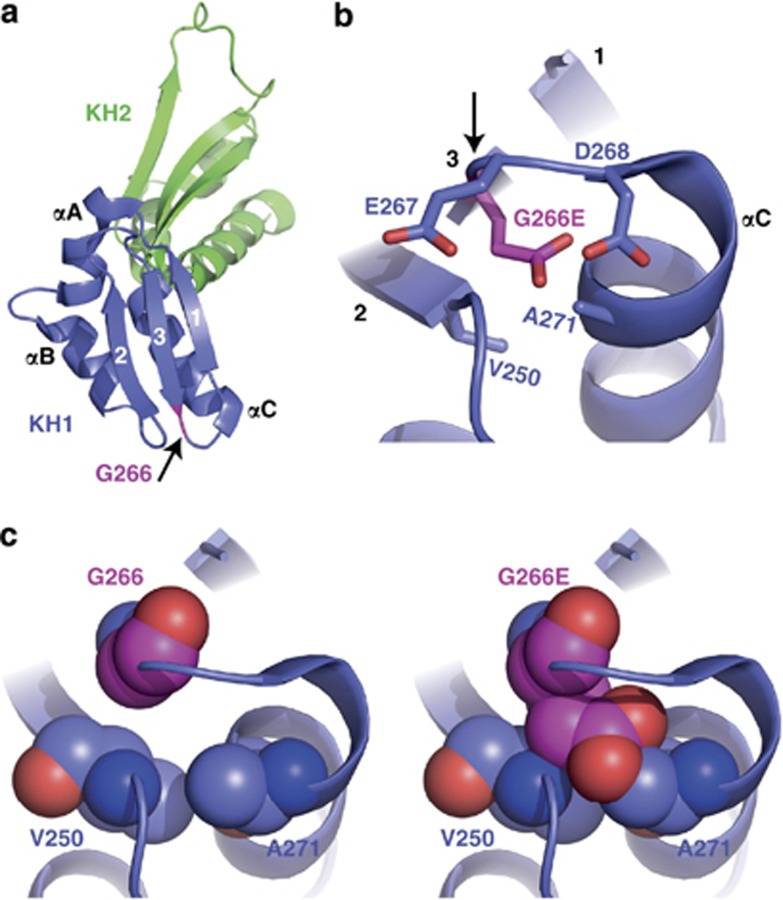
The Gly266Glu mutation resides within the well-conserved KH1 RNA-binding domain of FMRP. High-resolution crystal structure of FMRP's KH1–KH2 domain is available (Protein Data Bank code 2QND)16 and it indicates that Gly266 is located at the carboxyl end of β-strand 3, which is part of the β-sheet consisting of antiparallel strands 2, 3, and 1 (Figure 3a). Modeling the glutamic acid substitution at residue 266 introduces a large and negatively charged side chain that clashes into neighboring A271 of the following α-helix C and V250 of β-strand 3 (Figure 3c). In addition, glutamic acid at 266 would be subjected to repulsion forces from the surrounding negatively charged residues, E267 and D268 (Figure 3b). These data suggest that Gly266Glu mutation will cause significant disruptive structural change consistent with its loss-of-function in the above functional assays.
Figure 3.
Structural analysis of mutant G266E-FMRP. (a) Ribbon representation of the FMRP KH1 (blue) and KH2 (green) domains from Protein Data Bank code 2QND. The three β-strands (1, 2, and 3) and three α-helices (αA, αB, and αC) of KH1 are labeled, and the position of Gly266 is highlighted in pink as indicated by the arrow. (b) Stick representation of residues 266–268 showing the close proximity of three negatively charged amino acids when residue 266 is converted from glycine (neutral) to glutamic acid (negative). Arrow points to residue 266. (c) Sphere representations of glycine (left) or glutamic acid (right) at residue 266. There is high probability for glutamic acid to crash into residues V250 and A271 due to the space constraints predicted by this structural model.
Discussion
We have identified a novel missense mutation in FMR1 that leads to FXS in the absence of CGG-repeat expansion. This marks only the second time a missense mutation has been reported to cause FXS since the identification of the FMR1 gene over 20 years ago. This single nucleotide substitution, c.797G>A (p.(Gly266Glu)), was discovered in a male patient with characteristic FXS phenotype.
Several lines of a priori evidence indicate this is a pathologic change. Glycine 266 is highly conserved from human through Drosophila and the glutamic acid substitution is judged as damaging by the prediction algorithms SIFT, Polyphen-2, and Provean (0, 1, and −7.53, respectively). Moreover, Gly266, located in the KH1 RNA-binding domain of FMRP, is found in the invariant β-stand 3 of KH domains in general. Indeed, Gly266 is found to be conserved at position 60 in 12 out of 15 KH domains from unrelated RNA-binding proteins17 and a missense mutation of this residue in the Drosophila Bicaudal-C protein (Gly295Arg) creates a loss-of-function mutation.18 Structural analysis of the previously determined FMRP KH1–KH2 domain structure16 also suggests that Gly266Glu is damaging. The sharp turn between β-strand 3 and α-helix C where residue 266 is located requires a relatively small and flexible amino acid, such as glycine. Exchanging this glycine for glutamic acid will almost certainly interfere with normal KH folding due to the substantial steric disturbance that is created by the much larger and negatively charged side chain.
Consistent with a loss-of-function mutation, we have demonstrated that the Gly266Glu missense produces a functional null protein that is unable to perform many key FMRP functions including mRNA binding, polyribosome association, and mGluR-mediated AMPA receptor trafficking. We conclude that the loss of these FMRP functions due to Gly266Glu mutation is the basis for the FXS phenotype in this patient.
This report of a Gly266Glu mutation joins the report of the only other pathogenic missense mutation known of FMRP, the Ile304Asn mutation (c.911T>A) reported in a patient in 1993.12 One might ask why so few missense mutations have been identified in FMR1? This gene does not appear any less mutable than other typical X-linked genes. A recent study sequencing FMR1 in 963 developmentally delayed males observed five silent (synonymous) sites and two replacement (nonsynonymous) sites,19 which compares favorably with X-linked genome-wide averages of ∼3.7 silent sites and 2.5 replacement sites.20 However, if one examines other X-linked monogenic causes of intellectual disability and/or developmental delay (ID/DD), the small number of FMR1 missense mutations is striking. For example, there are 143 unique missense mutations of MECP2, leading to Rett syndrome (as of 7/2013; Human Gene Mutation Database). These two genes are of comparable size with FMR1 having 1896 coding bases and MECP2 having 1458 coding bases. The most parsimonious explanation is that the majority of patients presenting with a distinctive Rett syndrome phenotype do, in fact, have MECP2 mutations, whereas for FXS this is not true. Indeed, only ∼1.4% of patients clinically tested for the full mutation are positive.10 Partly, this is due to the nonspecific and variable phenotype of FXS and that testing for FXS is among the primary tests ordered for any child not reaching developmental milestones, therefore a low positive test rate is expected. Yet missense FMR1 mutations must exist in the population, perhaps leading to phenotypes that are not usually brought to medical attention, such as children with learning disabilities or who struggle in regular classrooms but not considered intellectually impaired. FMR1 mutations may also underlie nonspecific ID/DD phenotypes without distinctive features to prompt FMR1 sequencing.
It is clear that mutations of all classes that lead to classic FXS are essentially functional null mutations. The full mutation leads to transcriptional silencing of FMR1 and is the most common cause of FXS with a prevalence of ∼1 in 5000 males.21 Diagnostic testing for FXS is largely limited to full mutation screening. However, a number of conventional mutations have also been demonstrated to lead to FXS.11, 12 Many FMR1 deletions have been reported, with most uncovered through absent or unexpected bands on a Southern blot used to diagnose the full mutation, or by microarray analysis. Nonsense and splice site mutations have also been reported in a limited number of patients.22, 23, 24 However, the overall prevalence of conventional FMR1 mutations among children with ID/DD is unclear, as diagnostic laboratories do not currently perform routine FMR1 sequencing. If one posits that perhaps 1 per 500 males with ID/DD carry a deleterious conventional FMR1 mutation, not an unreasonable estimate compared with other X-linked ID/DD genes,25 then the diagnostic yield for FXS testing could be improved by ∼14%. Identification of conventional FMR1 mutations is not only important for genetic counseling, educational and program planning, but also for treatment strategy as mechanism-targeted therapeutics for FXS are increasingly entering clinical trials. Patients with functional null mutations, such as the Gly266Glu patient described above, would be expected to benefit similarly to patients with the full mutation of FMR1. In an era where sequencing costs are dropping exponentially, FMR1 sequencing should now be incorporated into the standard of care for any child presenting with developmental delay as an adjunct to repeat expansion testing. This will require that affordable, easily accessible, and insurance-covered testing be made available to clinicians.
Acknowledgments
This study was supported by grants from the National Institutes of Health (HD24064 and MH087977 to Dr Warren). We thank the patient and his family for their cooperation. We thank David Cutler for his helpful discussion and the Emory Viral Vector Core for lentivirus production.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: a twenty-year perspective. Ann Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- Wang LW, Berry-Kravis E, Hagerman RJ. Fragile X: leading the way for targeted treatments in autism. Neurotherapeutics. 2010;7:264–274. doi: 10.1016/j.nurt.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Fu YH, Kuhl DP, Pizzuti A, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakar AL, Dolen G, Bear MF. The pathophysiology of fragile X (and what it teaches us about synapses) Ann Rev Neurosci. 2012;35:417–443. doi: 10.1146/annurev-neuro-060909-153138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry-Kravis E, Hessl D, Coffey S, et al. A pilot open label, single dose trial of fenobam in adults with fragile X syndrome. J Med Genet. 2009;46:266–271. doi: 10.1136/jmg.2008.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont S, Curie A, des Portes V, et al. Epigenetic modification of the FMR1 gene in fragile X syndrome is associated with differential response to the mGluR5 antagonist AFQ056. Sci Transl Med. 64ra1. 2011;3 doi: 10.1126/scitranslmed.3001708. [DOI] [PubMed] [Google Scholar]
- Berry-Kravis EM, Hessl D, Rathmell B, et al. Effects of STX209 (arbaclofen) on neurobehavioral function in children and adults with fragile X syndrome: a randomized, controlled, phase 2 trial. Sci Transl Med. 152ra27. 2012;4 doi: 10.1126/scitranslmed.3004214. [DOI] [PubMed] [Google Scholar]
- Strom CM, Crossley B, Redman JB, et al. Molecular testing for Fragile X Syndrome: lessons learned from 119,232 tests performed in a clinical laboratory. Genet Med. 2007;9:46–51. doi: 10.1097/gim.0b013e31802d833c. [DOI] [PubMed] [Google Scholar]
- Coffee B, Ikeda M, Budimirovic DB, Hjelm LN, Kaufmann WE, Warren ST. Mosaic FMR1 deletion causes fragile X syndrome and can lead to molecular misdiagnosis: a case report and review of the literature. Am J Med Genet. 2008;146A:1358–1367. doi: 10.1002/ajmg.a.32261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boulle K, Verkerk AJ, Reyniers E, et al. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- Nakamoto M, Nalavadi V, Epstein MP, Narayanan U, Bassell GJ, Warren ST. Fragile X mental retardation protein deficiency leads to excessive mGluR5-dependent internalization of AMPA receptors. Proc Natl Acad Sci USA. 2007;104:15537–15542. doi: 10.1073/pnas.0707484104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- Valverde R, Pozdnyakova I, Kajander T, Venkatraman J, Regan L. Fragile X mental retardation syndrome: structure of the KH1-KH2 domains of fragile X mental retardation protein. Structure. 2007;15:1090–1098. doi: 10.1016/j.str.2007.06.022. [DOI] [PubMed] [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, et al. Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell. 2000;100:323–332. doi: 10.1016/s0092-8674(00)80668-6. [DOI] [PubMed] [Google Scholar]
- Mahone M, Saffman EE, Lasko PF. Localized Bicaudal-C RNA encodes a protein containing a KH domain, the RNA binding motif of FMR1. EMBO J. 1995;14:2043–2055. doi: 10.1002/j.1460-2075.1995.tb07196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SC, Bray SM, Suhl JA, et al. Identification of novel FMR1 variants by massively parallel sequencing in developmentally delayed males. Am J Med Genet. 2010;152A:2512–2520. doi: 10.1002/ajmg.a.33626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee B, Keith K, Albizua I, et al. Incidence of fragile X syndrome by newborn screening for methylated FMR1 DNA. Am J Hum Genet. 2009;85:503–514. doi: 10.1016/j.ajhg.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronskov K, Brondum-Nielsen K, Dedic A, Hjalgrim H. A nonsense mutation in FMR1 causing fragile X syndrome. Eur J Hum Genet. 2011;19:489–491. doi: 10.1038/ejhg.2010.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugenbeel KA, Peier AM, Carson NL, Chudley AE, Nelson DL. Intragenic loss of function mutations demonstrate the primary role of FMR1 in fragile X syndrome. Nat Genet. 1995;10:483–485. doi: 10.1038/ng0895-483. [DOI] [PubMed] [Google Scholar]
- Wang YC, Lin ML, Lin SJ, Li YC, Li SY. Novel point mutation within intron 10 of FMR-1 gene causing fragile X syndrome. Hum Mut. 1997;10:393–399. doi: 10.1002/(SICI)1098-1004(1997)10:5<393::AID-HUMU10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Tarpey PS, Smith R, Pleasance E, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–543. doi: 10.1038/ng.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.