Abstract
Uniparental disomy (UPD) describes the inheritance of a pair of chromosomes from only one parent. It may occur as isodisomy, heterodisomy or a combination of both and may involve only chromosome segments. UPD can affect each chromosome. The incidence is estimated to be around 1:3500 in live births. Some parts of chromosomes are subject to ‘parent-of-origin imprinting' and the phenotypic effect in UPD syndromes is mainly due to functional imbalance of imprinted genes. Isodisomy can result in mutation homozygosity in autosomal-recessive inherited diseases. UPD causes several well-defined imprinting syndromes associated with intellectual disability (ID). Although knowledge on frequency and size of UPDs in patients with unexplained ID remains largely unknown as no efficient genome-wide screening technique was available for detection of both isodisomic and heterodisomic UPDs. SNP microarrays have been proven to be capable to detect UPDs through Mendelian errors. The correct subclassification of UPD requires child–parent trio experiments. To further elucidate the role of UPD in patients with unexplained ID, we analyzed a total of 322 child–parent trios. We were not able to detect UPDs (isodisomies and heterodisomies) within our cohort spanning whole chromosomes or chromosomal segments. We conclude that UPD is rare in patients with unexplained ID.
Introduction
The frequency of developmental delay and intellectual disability (DD/ID) in the general population is estimated to be 1%1 and the majority of patients lack specific syndromic features.2 Several studies reported causative submicroscopic chromosomal aberrations that were only detectable by chromosomal microarray analysis (CMA)3 and a consensus statement was published that CMA is recommended as first-tier clinical diagnostic test for patients with DD/ID.4 Besides the detection of submicroscopic deletions and duplications, SNP microarrays can be used for detection of uniparental disomy (UPD). UPD is the inheritance of a chromosome pair from one parent.5, 6 Based on different mechanisms, UPD can present as isodisomy (two copies of one parental homolog), heterodisomy (both homologs of one parent) or a combination thereof and can involve complete chromosomes or only chromosomal segments. The mode of inheritance can be maternal or paternal. UPD is generally thought to occur at a rate of 1:3500 live births.7 Clinical relevant UPD phenotypes have been described for several autosomes.8 The pathogenesis of UPD is determined by both epigenetic imprinting (e.g. Silver–Russell syndrome, Beckwith–Wiedemann syndrome, Prader–Willi syndrome or Angelman syndrome) as well as demasking of autosomal-recessive diseases (homozygosity by isodisomy). Mosaicism for trisomy and early gestational effects of partial trisomic rescue are also clinical relevant problems associated with UPD.7
Although the concept of UPD was known for several decades,9 efficient genome-wide screening and correct subclassification were not available until recently. Current techniques for UPD detection (eg, methylation-specific PCR, methylation-specific MLPA and microsatellite analysis) are limited by number of markers and are generally restricted to distinct genomic regions or well-known imprinting syndromes. The usability of SNP microarrays for UPD detection has been proven and requires child–parent trio and special software for correct subclassification of UPDs10, 11, 12 or parental exclusions from microsatellite analysis. UPD detection from trio experiments exceeds standard analysis that infers isodisomy from loss of heterozygosity13 and requires specialized bioinformatic tools that automatically analyze occurrences of inheritance errors (Mendelian errors (MEs)) in large cohorts.14, 15 As no genome-wide screening is carried out routinely, only little is known about occurrence and size of UPDs in patients with unexplained DD/ID.
Materials and methods
We screened 324 parent–patient trios with unexplained syndromic and non-syndromic ID from three different centers of the German Mental retardation Network (MRNET) under the hypothesis that UPD may explain the genetic cause of ID in some patients. Most patients included in the MRNET presented with ID and minor dysmorphic features and/or congenital malformation including pre- and/or postnatal growth disturbance. All patients were initially analyzed for pathogenic copy number variants (CNV) using Mapping 6.0 SNP arrays (Affymetrix, Santa Clara, CA, USA) and seen by a group of experts (pediatrician and/or clinical geneticist with primary focus on disorders with DD/ID and experienced in syndromic disorders). Clinically suspected diseases including recognizable well-known imprinting syndromes were ruled out where appropriate.
Informed consent was obtained from patients and their parents. Genomic DNA was isolated from peripheral blood leukocytes by routine procedures and quality was assessed with the Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). All patients were analyzed using the Genome-Wide Human SNP Array 6.0 (Affymetrix). All processing steps were carried out following the manufacturer's protocols. The Chromosome Analysis suite (v. CytoB-N1.2.2.271, Affymetrix) was used for subsequent quality control and exclusion of CNVs because CNVs may mimic isodisomic stretches. We filtered for quality parameters (median absolute pairwise difference <0.4, call rate >97%) and confirmed the consistency of each trio. UPDtool was used for genome-wide detection of UPDs within the child–parent trios.14 In brief, UPDtool detects MEs and uses them to build coherent stretches. Each stretch is further classified according to inheritance of MEs, frequency of MEs, frequency of homozygous genotypes and frequency of genotypes identical to one parent. Finally, all stretches and their interpretation can be found in a tab-delimited text file. Current limitations of UPDtool include detection of low-level UPD mosaics and segmental UPDs in consanguineous families.
Results
Two positive controls with known combined UPDs were used to adopt UPDtool's standard parameters for smaller UPDs. For example, our second positive control (lower panel part a of Figure 1) contained an isodisomic stretch of 7 Mb at the beginning of chromosome 7 that was initially overseen using standard parameters because it contained <300 MEs. We adjusted the minimum length of ME stretches to 100 SNPs and the minimum number of MEs per stretch to 10. Thus, our results contained more fragmented regions and we decided to analyze our cohort of 324 patient–parent trios as a two-step process: we used standard parameters (stretches with ≥300 MEs, size of sliding window ≥1000 SNPs, min. fraction of MEs ≥1%) for a first screening and applied a set of relaxed parameters for a more detailed analysis (stretches with ≥10 MEs, size of sliding window ≥100 SNPs, min. fraction of MEs ≥1%).
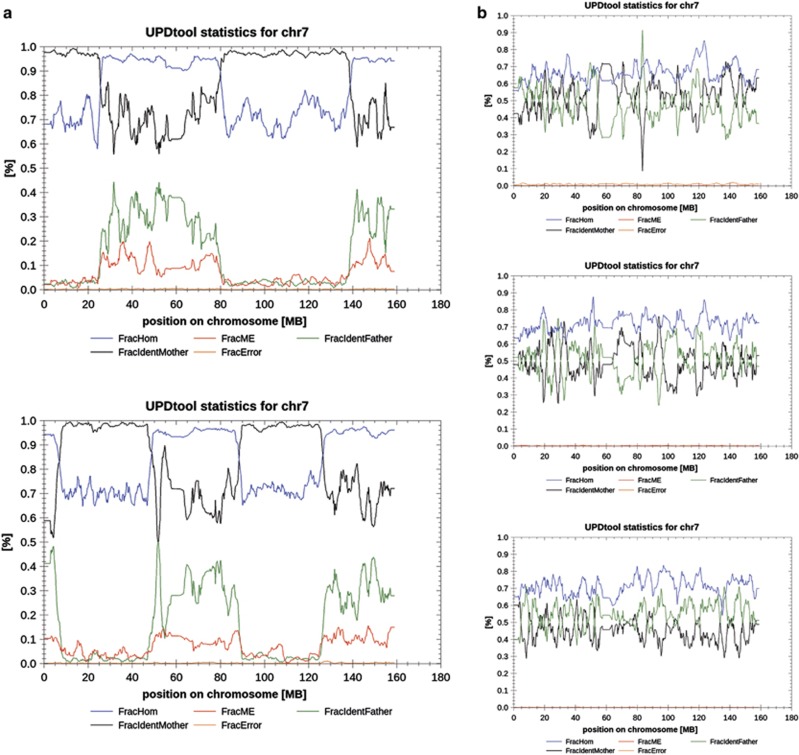
Figure 1.
Exemplary results of UPDtool for chromosome 7 in two positive controls and three selected patients. (a) illustrates two positive controls with combined hetero-iso-disomies from maternal origin on chromosome 7. The upper image shows a sequence of hetero- and isodisomic stretches: heterodisomy from 45 kb to 26 Mb; isodisomy from 26 to 80 Mb; heterodisomy from 80 to 139 Mb; isodisomy from 139 to 159 Mb. The lower image from a different positive control is characterized by a similar allelic pattern of UPD: isodisomy from 40 kb to 7 Mb; heterodisomy from 7.5 to 48 Mb; isodisomy from 48 to 88 Mb; heterodisomy from 88 to 126 Mb; isodisomy from 126 to 159 Mb. The first isodisomic stretch of the second positive control (panel a, lower image) was only detected after adjustment of standard parameters, because this contained <300 MEs. (b) shows exemplary plots for chromosome 7 of selected patients from our cohort. There are no indications for UPD in these images. Legend: FracHom=fraction of genotypes in 1k-window that is homozygous, FracME=fraction of MEs in a 1k-window, FracIdentFather=fraction of genotypes within a 1k-window where both alleles are identical to the fathers' alleles, FracIdentMother=fraction of genotypes within a 1k-window where both alleles are identical to the mothers' alleles, FracError=fraction of errors (other than MEs) within a 1k-window.
A total of 322 trios passed primary quality filters and were eligible for further UPD detection. Within our cohort, we identified 0.893 MEs per 1000 SNPs (MEs/1k SNPs, SD: ±0.644 MEs/1k SNPs) 0.3 MEs per MB (SD: ±0.25), respectively. We identified MEs distributed over all chromosomes at a rate comparable to that in the literature and MEs originated alternating from both parents.
Figure 1 compares the results of our positive controls and three randomly chosen cases from our patient cohort. We were not able to detect any occurrence of UPD within our cohort in both analysis runs. Notably, we did not detect segmental or small (partial) UPDs in our patient cohort. As a proof of principle and increased sensitivity of the adjusted parameters, we were able to detect small inherited and de novo deletions that can mimic isodisomic stretches down to a length of 90 kb (ie, in patient 32 with 21 MEs and length of 102 SNPs). This finding can be considered as a measure for sensitivity representing the resolution of the algorithm used.
Discussion
To our knowledge, this is the largest multicenter study providing information on UPDs (hetero- and isodisomies) in patients with DD/ID and their parents. Our retrospective study demonstrates that detection of UPDs including hetero- and isodisomies can be easily implemented into laboratory workflows using software packages publicly available.14, 15 In summary, we were not able to detect any occurrence of complete or segmental iso- or heterodisomy in our patient cohort both with standard and adjusted filter parameters.
A different approach for SNP array-based genome-wide UPD detection by Papenhausen et al11 focused on detection of regions of isoallelism in a set of over 13 000 index cases.11 This group analyzed long contiguous stretches of homozygosity (LCSH) in patients with DD, autism or congenital anomalies. A threshold for possible UPD correlation was set at 13.5 Mb for isolated LCSH and 15 Mb if two LCSH in a single chromosome were found. Applying these parameters, 96 patients with UPD qualifying LCSH were identified, 46 of those had additional molecular parent–child trio testing and a total of 29 cases of UPD (isodisomies and mixed hetero-/isodisomies) were confirmed, suggesting a frequency of UPD in ID patients of at least 0.002 (29/13.000). This included patients with well-known recognizable recurrent UPD phenotypes such as Angelman syndrome, Prader–Willi syndrome, Silver–Russell syndrome, Beckwith–Wiedemann syndrome, UPD14 or transient neonatal diabetes.11 In contrast, our cohort was clinically prescreened and patients positive for these entities were excluded. However, as our method has the advantage of detecting complete heterodisomies in combination with a higher sensitivity for detection of very small segmental isodisomies, we expected to find 1–2 cases of UPD within our cohort.
In a more recent study, Sasaki et al16 examined the genotypes of 173 healthy trios using the Genome-Wide Human SNP Array 6.0 data. UPD criteria were defined as a minimum of 6 consecutive MEs within segments extending over 200 kb. No whole chromosome UPD was detected, but one case of segmental paternal isodisomy (8.2 Mb) on 17p13.3-13.1. The authors estimated the rate of segmental UPD to be 1 per 3806 chromosome pairs. However, DNA used in this study was derived from lymphoblastoid cell lines and somatic recombination or events during cell culture cannot be excluded.
In summary, estimates on birth prevalence of UPD in the general population or patient cohorts derived from these SNP array studies are of limited value, mainly due to the application of different, not comparable approaches and their limitations in study design. This includes different sample sizes, sample types (genomic DNA from uncultured cells versus cultured cell lines), patient cohorts (unselected versus prescreened) and sensitivity of UPD detection method (LCSH versus trio genotyping).
We conclude that UPD, hetero- and isodisomy as well as segmental non-mosaic UPD, other than recurrent, clinically well-defined UPD phenotypes, is a rare cause of unexplained DD/ID.
Acknowledgments
The Institutes for Human Genetics Erlangen, Heidelberg and Tübingen, Germany, were members of the ‘German Mental Retardation Network' (MRNET), which was funded by the German Federal Ministry of Education and Research (BMBF) as a part of the National Genome Research Network (http://www.ngfn.de/en/retardierung.html, project reference numbers 01GS0816, 01GS08160, 01GS08162, 01GS08168).
The authors declare no conflict of interest.
References
- Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32:419–436. doi: 10.1016/j.ridd.2010.12.018. [DOI] [PubMed] [Google Scholar]
- Moeschler JB, Shevell M,, American Academy of Pediatrics Committee on Genetics: Clinical genetic evaluation of the child with mental retardation or developmental delays. Pediatrics. 2006;117:2304–2316. doi: 10.1542/peds.2006-1006. [DOI] [PubMed] [Google Scholar]
- McMullan DJ, Bonin M, Hehir-Kwa JY, et al. Molecular karyotyping of patients with unexplained mental retardation by SNP arrays: a multicenter study. Hum Mutat. 2009;30:1082–1092. doi: 10.1002/humu.21015. [DOI] [PubMed] [Google Scholar]
- Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazawa K, Ogata T, Ferguson-Smith AC. Uniparental disomy and human disease: an overview. Am J Med Genet C Sem Med Genet. 2010;154C:329–334. doi: 10.1002/ajmg.c.30270. [DOI] [PubMed] [Google Scholar]
- Liehr T. Cytogenetic contribution to uniparental disomy (UPD) Mol Cytogenet. 2010;3:8. doi: 10.1186/1755-8166-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays. 2000;22:452–459. doi: 10.1002/(SICI)1521-1878(200005)22:5<452::AID-BIES7>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Gardner RJM, Sutherland GR, Shaffer LG.Chromosome Abnormalities and Genetic Counseling4th ednNew York, USA: Oxford University Press; 2011 [Google Scholar]
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–143. doi: 10.1002/ajmg.1320060207. [DOI] [PubMed] [Google Scholar]
- Altug-Teber O, Dufke A, Poths S, et al. A rapid microarray based whole genome analysis for detection of uniparental disomy. Hum Mutat. 2005;26:153–159. doi: 10.1002/humu.20198. [DOI] [PubMed] [Google Scholar]
- Papenhausen P, Schwartz S, Risheg H, et al. UPD detection using homozygosity profiling with a SNP genotyping microarray. Am J Med Genet A. 2011;155A:757–768. doi: 10.1002/ajmg.a.33939. [DOI] [PubMed] [Google Scholar]
- Bruce S, Leinonen R, Lindgren CM, et al. Global analysis of uniparental disomy using high density genotyping arrays. J Med Genet. 2005;42:847–851. doi: 10.1136/jmg.2005.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastepe M, Altug-Teber O, Agarwal C, Oberfield SE, Bonin M, Juppner H. Paternal uniparental isodisomy of the entire chromosome 20 as a molecular cause of pseudohypoparathyroidism type Ib (PHP-Ib) Bone. 2011;48:659–662. doi: 10.1016/j.bone.2010.10.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder C, Sturm M, Dufke A, et al. UPDtool: a tool for detection of iso- and heterodisomy in parent-child trios using SNP microarrays. Bioinformatics. 2013;29:1562–1564. doi: 10.1093/bioinformatics/btt174. [DOI] [PubMed] [Google Scholar]
- Ting JC, Roberson ED, Miller ND, et al. Visualization of uniparental inheritance, Mendelian inconsistencies, deletions, and parent of origin effects in single nucleotide polymorphism trio data with SNPtrio. Hum Mutat. 2007;28:1225–1235. doi: 10.1002/humu.20583. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Mishima H, Miura K, Yoshiura K. Uniparental disomy analysis in trios using genome-wide SNP array and whole-genome sequencing data imply segmental uniparental isodisomy in general populations. Gene. 2013;512:267–274. doi: 10.1016/j.gene.2012.10.035. [DOI] [PubMed] [Google Scholar]