Abstract
The constellation of clinico-pathological and laboratory findings including massive hepatomegaly, steatosis, and marked hypertriglyceridemia in infancy is extremely rare. We describe a child who is presented with the above findings, and despite extensive diagnostic testing no cause could be identified. Whole exome sequencing was performed on the patient and parents' DNA. Mutations in GPD1 encoding glycerol-3-phosphate dehydrogenase that catalyzes the reversible redox reaction of dihydroxyacetone phosphate and NADH to glycerol-3-phosphate (G3P) and NAD+ were identified. The proband inherited a GPD1 deletion from the father determined using copy number analysis and a missense change p.(R229Q) from the mother. GPD1 protein was absent in the patient's liver biopsy on western blot. Low normal activity of carnitine palmitoyl transferases, CPT1 and CPT2, was present in the patient's skin fibroblasts, without mutations in genes encoding for these proteins. This is the first report of compound heterozygous mutations in GPD1 associated with a lack of GPD1 protein and reduction in CPT1 and CPT2 activity.
Keywords: GPD1, hypertriglyceridemia, steatohepatitis, hepatomegaly
Introduction
GPD1 (MIM 138420) encodes cytoplasmic NAD-dependent glycerol-3-phosphate dehydrogenase 1 (GPD1), a 349-amino-acid protein of 37.5 kD size that catalyzes the reversible redox reaction of dihydroxyacetone phosphate (DHAP) and NADH to glycerol-3-phosphate (G3P) and NAD+.1, 2 GPD1 together with a mitochondrial enzyme, GPD2 also has an important role in the transport of reducing equivalents from the cytosol to the mitochondria.2 Recently, a homozygous splicing mutation in GPD1, c.361-1G>C, predicted to encode p.Ile119fsX94, was identified in 10 family members from four highly consanguineous Israeli Arab families presenting with early onset hepatomegaly, hepatic steatosis, and hypertriglyceridemia.2
Monogenic hypertriglyceridemia is caused by mutations in lipoprotein lipase (LPL), apolipoprotein C-II (APOC2), glycosyl-phosphatidyl-inositol-anchored HDL-binding protein (GPIHBP1), ApoA-V (APOA5), and lipase maturation factor 1 (LMF1).2, 3, 4 Hepatic steatosis as a monogenic disease in early infancy is extremely rare and has been described with Chanarin–Dorfman syndrome due to abhydrolase domain containing 5 (ABHD5) mutations, and mitochondrial fatty acid oxidation defects including CPT1A, CPT2, and very long chain acyl-CoA dehydrogenase (ACADVL) deficiency.5, 6, 7 These patients often have other features including hypoglycemia, encephalopathy and cardiomyopathy, myopathy, and liver failure. A combination of profound hypertriglyceridemia and severe hepatomegaly in early infancy is extremely rare.
Here we describe a female patient of Caucasian descent with massive hepatomegaly, fatty liver and severe hypertriglyceridemia carrying a compound heterozygous mutation in GPD1 (MIM 138420). This is the first instance of a GPD1 mutation identified outside of the Israeli Arab population. In addition, we show that the GPD1 protein was absent in the patient's liver tissue. There was no compensatory increase in the GPD2 levels in response to GPD1 deficiency. Further, although the activity of carnitine palmitoyltransferases CPT1A and CPT2 was reduced in the patient's fibroblasts, no mutations of these genes were identified, and the amount of CPT1A in her liver tissue sample was not altered.
Patient and methods
Enrollment of patient and sample collection
The patient is a female infant, delivered normally at birth at term gestation and adopted aged three days. Abdominal enlargement was present since birth and a history of frequent spitting up/vomiting prompted an ultrasound at six weeks that ruled out pyloric stenosis. At five months, she was noted to have marked hepatomegaly (liver enlarged 2 cm below the umbilicus) and a small head circumference of 40.5 cm, (5th centile) prompting further evaluation. Liver function tests were abnormal with AST of 4.88 μkat/l (normal 0.034–0.68 μkat/l), ALT 2.67 μkat/l (normal 0.051–0.92 μkat/l), and GGT 6.03 μkat/l (normal 0.13–0.58 μkat/l). Serum total bilirubin was 6.84 μmol/l, within normal limits (reference range: 4–17 μmol/l). The liver ultrasound at five months revealed diffuse enlargement and increased echogenicity (Figures 1b and c) and biopsy done few days later showed diffuse and marked micro- and macrovesicular steatosis (Figure 1a), consistent with non-alcoholic steatohepatitis (NASH) or fatty liver disease. No evidence of cholestasis, cytoplasmic inclusions, significant iron deposition, significant inflammation or fibrosis was apparent. A lipid profile revealed significantly elevated plasma triglycerides (9.48 mmol/l; normal 0.57–1.47) and elevated cholesterol (5.1 mmol/l; normal<4.4 mmol/l). Plasma triglycerides remained high; the last level measured at 1.5 years of age was 6.06 mmol/l. An 8-h fasting study revealed no hypoglycemia or urine ketones. Plasma electrophoresis revealed chylomicronemia consistent with Fredrickson's type 5 hyperlipidemia. Apolipoprotein A1 and B, acid lipase and lysosomal enzyme screening panel for 12 different enzymes were all normal. Several genes were sequenced to identify pathogenic mutations, including G6PC (glycogen storage disease (GSD) type IA), GAA (GSD type II), AGL (GSD type III), SLC37A4 (GSD 1B, 1C, 1D), LPL (LPL deficiency type I), APOC2 (hyperlipidemia), and INSR; all were reported as normal. No significant copy number variation was identified on chromosomal microarray analysis. Urine organic acids, acylglycines, and plasma amino acids and acylcarnitines were normal. The activity of CPT1 and CPT2 enzymes was low normal in cultured skin fibroblasts (CPT1=0.47; normal 0.4–1.58 and CPT2=0.11; normal 0.16–0.57 nmol/min/mg protein). In response, the CPT1A and CPT2 genes were sequenced, both reported as normal. By early infancy, she had failure to thrive (FTT) and was managed with foods high in calories and low in fat with medium chain triglycerides supplementation. Her head circumference increased from 42 cm (1st centile) at 13 months to 46 cm (10th centile) at 30 months of age with improved nutrition. MRI and MR spectroscopy of the brain was performed at 1.5 years of age and was reported as normal. Developmentally, the patient was appropriate for her age.
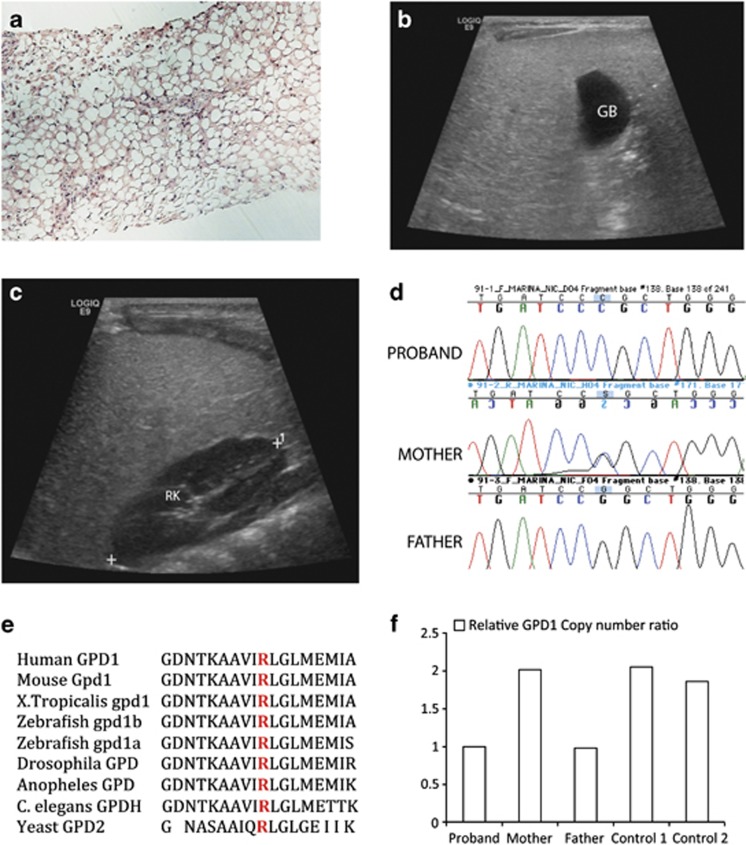
Figure 1.
Radio-pathologic and genetic findings in proband with GPD1 mutation. (a) Liver biopsy: the liver parenchyma shows macro- and microvesicular fatty change on Hematoxylin and Eosin staining (H&E), magnification × 10. (b and c) Ultrasound demonstrates diffuse hyperechogenicity of the liver with a fine granular pattern. In comparison, the gall bladder (GB) (b) and right kidney parenchyma (RK) (c) are hypoechoic. (d) DNA sequence analysis of genomic PCR products illustrates three genotypes for GPD1 c.686G>C seen in the family with proband homozygous, mother heterozygous and father WT for the allele. (e) Altered arginine residue (red) is evolutionarily well conserved in GPD1 and GPD1L proteins in vertebrates and lower organisms including yeast. (f) Relative GPD1 expression levels using copy number assay revealed father and proband to be GPD1 haploinsufficient relative to the mother and two controls.
The proband (0091-01) and her parents (0091-02 and 0091-03) were enrolled in an IRB-approved study by the Gene Discovery Core of The Manton Center for Orphan Disease Research while she was an inpatient at Boston Children's Hospital (BCH). Blood samples were collected and processed for DNA extraction by the Research Connection Biobank Core at BCH. The remaining tissue from the diagnostic liver biopsy was collected for additional molecular analyses.
Sequencing study and analysis
DNA from the proband and both biological parents was sent for whole exome sequencing (WES) to Axeq Technologies, Rockville, MD, USA. Samples were prepared as an Illumina sequencing library and enriched for exomic sequences using the Illumina Exome Enrichment protocol. The captured libraries were sequenced using Illumina HiSeq 2000 Sequencers. The reads were mapped to the human genome assembly UCSC hg19 using Burrows–Wheeler Alignment (version 0.5.8). Single nucleotide polymorphisms (SNPs) and small insertions/deletions were called with SAMtools (version 0.1.7). The variants or SNPs were filtered using NHLBI exome variant server (http://evs.gs.washington.edu/EVS/) and dbSNP Build 137 databases.
PCR was performed to amplify the candidate gene (GPD1), reference sequence used was GPD1; NG_032168.1, NM_005276.3. Variants identified in GPD1 were submitted to the Leiden Open Variant Database (LOVD) http://databases.lovd.nl/shared/genes/GPD1.
The following primers were used: GPD1_1F: AGGAGGGGTCTTTTCTCAC and GPD1_1R: ATCAGGTCAGCAACACCACA, and the resultant amplified DNA was sent for Sanger Sequencing. Results were analyzed using the Sequencer 5.0 software and aligned with the wild-type (WT) gene sequence. Primers were also designed to sequence various SNPs adjacent to GPD1. The SNPs included: rs836180, rs200359712, rs201128732, rs2640533, rs147189770, rs10875996, rs4898546, rs7139363, rs836170, rs836171, rs113783111, rs836177, rs10783334, rs7964522, rs112386170, rs7964698, rs10735822, rs71464996, rs71441320, rs55639096, and rs12229758.
Protein expression study
Western blot was performed to measure the levels of GPD1, GPD2, and CPT1A proteins in the liver tissue samples available from the patient and an age-matched control. The tissue was stored at –80 °C until analysis. Transferred proteins were probed with antibodies against GPD1 (HPA044620, 1:200 dilution, Sigma-Aldrich, St. Louis, MO, USA), GPD2 (Ag11036, 1:2000 dilution, Proteintech, Chicago, IL, USA), CPT1A ((Ag7202, 1:500 dilution, Proteintech, Chicago, IL, USA), and GAPDH (FL-335, 1:1000 dilution, Santa Cruz Biotechnology, Santa Cruz, CA, USA) as control, and visualized using enhanced chemiluminescence. Quantification of protein levels was normalized to GAPDH using the program Quantity One 4.2.1 (Bio-Rad Laboratories, Inc., Hercules, CA, USA) on an Image Station 440 (Kodak DS; Eastman Kodak co., Rochester, NY, USA).
Taqman copy number assay
Taqman copy number assay (Life Technologies, Woburn, MA, USA) was performed on DNA from the proband, both parents and two controls. The probes for both GPD1 (Hs00911535_cn, FAM labeled) and copy number reference gene (4403326, VIC-labeled RNase P (RPPH1)) are commercially available. The assay was repeated twice and was performed in triplicate. Twenty nanogram (1 ul) of DNA was used per well with 12 ul of Taqman Universal PCR master mix (Applied Biosystems, Woburn, MA, USA) and 1 ul of each of the specific probes (target and control). A 7300 Real time PCR system from Applied Biosystems was used to perform the PCR and Sequence Detection software v1.4 for the interpretation of results. The DNA amplification based on the amplification plots was compared and mean cycle threshold (Ct) values were obtained. The fold change in target and reference gene copy number, normalized to control was calculated using relative quantity=2−ΔΔCt.
Results
Proband carried GPD1 mutation
WES was performed on DNA samples from the trio (both biological parents and the proband). The proband carried what appeared to be a homozygous mutation c.686G>C in the GPD1 gene postulated to cause p.(R229P) amino-acid change. Sanger Sequencing confirmed the mutation (Figure 1d), and simultaneously, research group in Ontario using candidate gene analysis independently obtained the same result (personal communication from Robert Hegele). This residue is evolutionarily conserved (Figure 1e) and the mutation was not present in any of the publicly available SNP databases, whereas c.686G>A potentially causing p.(R229Q) change was identified in 1/13 005 chromosomes (frequency <0.0001) in exome variant server database. Further, Polyphen-28 score was 1, consistent with mutation to be probably damaging, and SIFT analysis9 revealed that this change was potentially not tolerated. Interestingly, although the mother was heterozygous for the mutation, the father was apparently homozygous for the WT allele. We hypothesized that the father may carry a deletion at the same locus inherited by the proband, or she may exhibit uniparental disomy for the maternal allele.
GPD1 haploinsufficiency in the proband and father
Taqman copy number assay was performed on the trio and two control DNA samples to evaluate if the father and the proband were haploinsufficient for GPD1. The sequence strand used to design the Taqman probe Hs00911535_cn included the GPD1 c.686 mutation locus (Chr 12:g.50,501,423). Probe 4403326 for RPPH1 was used as reference. A GPD1 deletion was observed in the proband and the father indicated by the presence of a single copy as compared with two copies present in the mother and two controls (Figure 1f). We suspect that the deletion is >1.85 kb in size as at a different GPD1 locus (rs836180, Chr 12:g.50,503,269), the father was apparently homozygous for a SNP while both the proband and the mother were homozygous for the WT allele. To evaluate if the deletion involved genes adjacent (5′ and 3′) to GPD1, we genotyped several known SNPs located within SMARD1 and COX14 genes. Many of these SNPs were informative and there was no evidence of a deletion. On the basis of the location of genomic coordinates, we hypothesize that the deletion is smaller than 28.7 kb. In conclusion, the proband is compound heterozygous for two GPD1 mutations, carrying a deletion from the father and missense change from the mother.
Lack of GPD1 protein in liver tissue sample from the proband
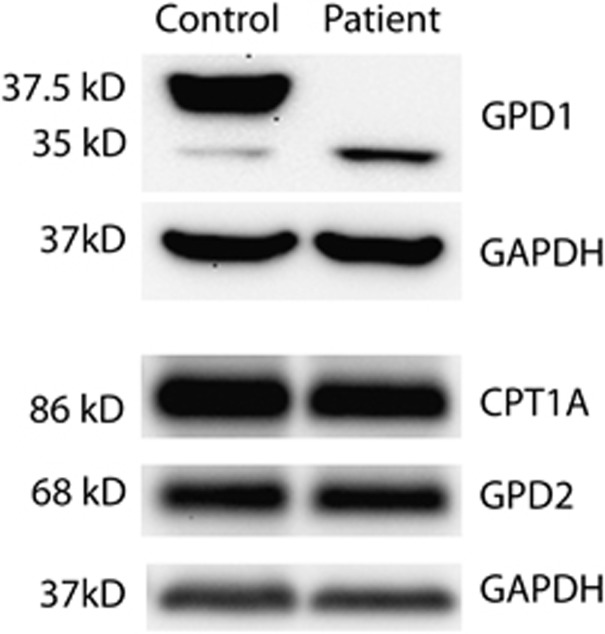
To evaluate the effects of these mutations, the available liver biopsy samples from the proband and an age- and sex-matched control were obtained from the Pathology Department. GPD1 antibody was used to probe for the GPD1 protein. Two bands were identified in the control corresponding to the two GPD1 isoforms of 37.5 kD (isoform 1) and 35 kD molecular weight (isoform 2) (Figure 2). Interestingly, isoform 1 was highly expressed in the control but was absent in the patient. In comparison, isoform 2 was expressed in small amounts in the control, whereas it was slightly increased in the patient. We suspect that for isoform 1, the missense mutation c.686G>A may affect splicing thereby creating an aberrant transcript that undergoes nonsense-mediated decay. Further, the slight increase in isoform 2 noted in the patient may be secondary to the lack of isoform 1.
Figure 2.
Western blot analysis on liver biopsy sample from the proband and age-matched control. GPD1 protein was absent in the proband although similar amounts of GPD2 and CPT1A were present in the proband and control.
The level of GPD2, the mitochondrial isoform, was compared to determine if it was altered in response to GPD1 deficiency. We probed for GPD2 using a specific antibody and there was no evidence of a change in GPD2 levels in the proband.
The activity of both CPT1 and CPT2 enzymes was reduced in cultured skin fibroblasts from the proband but sequencing both the genes did not identify any pathogenic mutation. We assayed the amounts of CPT1 A, the liver isoform of CPT1 using western blot, and did not find any significant difference between the patient and the control.
Discussion
We report for the first time a patient who inherited two heterozygous GPD1 mutations, a deletion and a missense change. Our studies conclusively demonstrate that the two mutations resulted in GPD1 deficiency. The patient presented with FTT, vomiting, marked hepatomegaly, and hypertriglyceridemia since early infancy. The clinical findings were very similar to the 10 individuals from four consanguineous Israeli Arab families carrying a homozygous founder mutation c.361-1G>C in GPD1, who presented with vomiting, FTT, enlarged livers, and fasting hypertriglyceridemia (TG 258-6244 mg/dl).2
GPD1 is a member of the NAD-dependent GPD family. It has a major role in lipid biosynthesis by converting DHAP to G3P, which in turn is dephosphorylated to glycerol followed by acylation with fatty acids to form acylglycerol species. DHAP, an intermediate of glycolysis may not be converted to G3P due to GPD1 deficiency, causing relative excess of DHAP and a reduction in G3P. Indeed, in a mouse model of GPD1 deficiency, DHAP concentrations were significantly higher, and G3P lower when compared with the levels in WT mice in liver, muscle, and kidneys.10 The mechanism of fatty liver in our patient may be due to acylation of excess DHAP. An acyl-DHAP pathway has been previously described where DHAP may first be acylated followed by reduction to 1-acyl-sn-G3P.11, 12 The acyl DHAP pathway is known to have a significant role in glycerolipid synthesis in mouse liver.12
The CPT1 and CPT2 activity were low normal in the patient's fibroblasts, but no mutations were identified in CPT1A and CPT2 genes. Further, there was no reduction in the amounts of CPT1A protein in the liver. We suspect that the reduction in CPT1 and CPT2 activity may be secondary to the buildup of the DHAP and/or neutral fat in the cytoplasm. Malonyl CoA, an intermediate in the pathways of de novo fatty acid biosynthesis and fatty acid elongation, is also known to allosterically inhibit CPT.13
The diagnosis of GPD1 deficiency had significant implications in the management of the patient. Liver transplantation, previously considered for her, was not recommended after the molecular diagnosis. This was partly based on the outcome data in patients carrying GPD1 mutation.2 The oldest patient in the GPD1 splicing mutation-carrying cohort from the Middle East was 23 years old, and asymptomatic. Further, in eight of the ten affected patients, the triglyceride levels declined over time.
In conclusion, GPD1 deficiency represents a defect in the glucose and lipid pathways associated with severe hypertriglyceridemia and NASH. WES provided a diagnosis in this case where extensive clinical testing had failed to find a cause. The molecular diagnosis provided anticipatory information to the family, and guided the clinical management. Although developmentally appropriate at this time, this patient will need close follow-up as lack of GPD1 may interfere with the glycerol phosphate shuttle, and not allow certain cells to derive all their energy from glycolysis, specifically in brain cells that do not completely rely on the malate–aspartate shuttle to deliver cytosolic reducing equivalents to the mitochondrial matrix.
Acknowledgments
We acknowledge the financial support from the Gene Discovery Core of The Manton Center for Orphan Disease Research at Boston Children's Hospital. Sanger sequencing was performed by the Molecular Genetics Core Facility of the IDDRC at Boston Children's Hospital supported by National Institute of Health (NIH) P30HD18655. PBA was supported by K08 AR055072 from the NIH.
The authors declare no conflict of interest.
References
- Menaya J, Gonzalez-Manchon C, Parrilla R, Ayuso MS. Molecular cloning, sequencing and expression of a cDNA encoding a human liver NAD-dependent alpha-glycerol-3-phosphate dehydrogenase. Biochim Biophys Acta. 1995;1262:91–94. doi: 10.1016/0167-4781(95)00069-s. [DOI] [PubMed] [Google Scholar]
- Basel-Vanagaite L, Zevit N, Har Zahav A, et al. Transient infantile hypertriglyceridemia, fatty liver, and hepatic fibrosis caused by mutated GPD1, encoding glycerol-3-phosphate dehydrogenase 1. Am J Hum Genet. 2012;90:49–60. doi: 10.1016/j.ajhg.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterfy M, Ben-Zeev O, Mao HZ, et al. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39:1483–1487. doi: 10.1038/ng.2007.24. [DOI] [PubMed] [Google Scholar]
- Brahm A, Hegele RA. Hypertriglyceridemia. Nutrients. 2013;5:981–1001. doi: 10.3390/nu5030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefont JP, Djouadi F, Prip-Buus C, Gobin S, Munnich A, Bastin J. Carnitine palmitoyltransferases 1 and 2: biochemical, molecular and medical aspects. Mol Aspects Med. 2004;25:495–520. doi: 10.1016/j.mam.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Mathur A, Sims HF, Gopalakrishnan D, et al. Molecular heterogeneity in very-long-chain acyl-CoA dehydrogenase deficiency causing pediatric cardiomyopathy and sudden death. Circulation. 1999;99:1337–1343. doi: 10.1161/01.cir.99.10.1337. [DOI] [PubMed] [Google Scholar]
- Cakir M, Bruno C, Cansu A, Cobanoglu U, Erduran E. Liver cirrhosis in an infant with Chanarin-Dorfman syndrome caused by a novel splice-site mutation in ABHD5. Acta Paediatr. 2010;99:1592–1594. doi: 10.1111/j.1651-2227.2010.01869.x. [DOI] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald MJ, Marshall LK. Mouse lacking NAD+-linked glycerol phosphate dehydrogenase has normal pancreatic beta cell function but abnormal metabolite pattern in skeletal muscle. Arch Biochem Biophys. 2000;384:143–153. doi: 10.1006/abbi.2000.2107. [DOI] [PubMed] [Google Scholar]
- Hajra AK, Bishop JE. Glycerolipid biosynthesis in peroxisomes via the acyl dihydroxyacetone phosphate pathway. Ann N Y Acad Sci. 1982;386:170–182. doi: 10.1111/j.1749-6632.1982.tb21415.x. [DOI] [PubMed] [Google Scholar]
- Agranoff BW, Hajra AK. The acyl dihydroxyacetone phosphate pathway for glycerolipid biosynthesis in mouse liver and Ehrlich ascites tumor cells. Proc Natl Acad Sci USA. 1971;68:411–415. doi: 10.1073/pnas.68.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]