Summary
In Arabidopsis thaliana, the amino acid sequences of membrane-associated acyl-CoA-binding proteins ACBP1 and ACBP2 are highly-conserved. We have previously shown that in developing seeds, ACBP1 accumulates in the cotyledonary cells of embryos and ACBP1 was proposed to be involved in lipid transfer. We show here by immunolocalization using ACBP2-specific antibodies, that ACBP2 is also expressed in the embryos at various stages of seed development in Arabidopsis.
Phenotypic analyses of acbp1 and acbp2 single mutants revealed that knockout of either ACBP1 or ACBP2 alone did not affect their life cycle since both single mutants exhibited normal growth and development similar to wild type. However, the acbp1acbp2 double mutant was embryo lethal and was also defective in callus induction.
On lipid and acyl-CoA profiling analyses, siliques but not leaves of the acbp1 mutant were shown to accumulate galactolipid monogalactosyldiacylglycerol (MGDG) and 18:0-CoA but the levels of most polyunsaturated species of phospholipids such as phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and phosphatidylserine (PS) had declined.
Since recombinant ACBP1 and ACBP2 bind unsaturated PC and acyl-CoA esters in vitro, we propose that ACBP1 and ACBP2 are essential in lipid transfer during early embryogenesis in Arabidopsis.
Keywords: acyl-CoA measurement, acyl-CoA-binding protein, embryo development, lipid metabolism, phosphatidylcholine, phospholipids
Introduction
During embryogenesis, which is an early essential step in the life cycle of higher plants, cell differentiation and division occur and a multicellular organism is produced from one single cell (Yeung & Meinke, 1993; Devic, 2008). In Arabidopsis thaliana, seed development can be divided into two major phases: embryo morphogenesis and maturation. Embryo morphogenesis is initiated by the fusion of the male and female gametes forming a zygote, after which rapid cell division occurs and the embryo pattern is soon established. Subsequently, during embryo maturation, cell expansion and accumulation of reserves follow. Finally, the seed undergoes desiccation and dormancy sets in (Harada, 1997; Fan et al., 2008; Baud & Lepiniec, 2009).
To date, a large number of Arabidopsis genes essential in seed development (EMBs) have been identified using forward- and reverse-genetic approaches (Tzafrir et al., 2004; Meinke et al., 2008). Analyses of embryo development in these mutants have indicated that the majority display aborted phenotypes at or before the globular stage, reinforcing this to be most critical in embryo development (Devic, 2008). Some EMB genes affect lipid transport and metabolism (9 genes), or CoA transport and metabolism (4 genes), implicating the essential role of lipids in plant embryo development (Tzafrir et al., 2003; http://www.seedgenes.org). Studies have revealed that depletion of LPAAT1 causes embryo lethality in Arabidopsis, with embryo development arrested at the heart-torpedo stage (Kim & Huang, 2004). LPAAT1 encodes a plastid-localized lysophosphatidic acid acyltransferase for the acylation of the sn-2 position of lysophosphatidic acid in phosphatidic acid biosynthesis and was deemed essential for heart-torpedo stage establishment in embryo development in Arabidopsis (Kim & Huang, 2004). In contrast, the knockout mutant of Arabidopsis LPAT2 lacking an endoplasmic reticulum (ER)-located lysophosphatidic acid acyltransferase is defective in female, but not male, gametophyte development (Kim et al., 2005). Downregulation of ACCase in Brassica napus by an antisense approach produced wrinkled seeds accompanied by a decline in lipid content (Sellwood et al., 2000). In Arabidopsis, two genes ACC1 and ACC2 encode multifunctional isoforms of ACCase (Yanai et al., 1995) and knockout mutants of ACC1 were embryo lethal (Baud et al., 2003). These results demonstrate that processes affecting plant lipid metabolism, especially those related to very long-chain fatty acid elongation are important during the early stages of seed development.
ACBPs are a family of proteins that facilitate the binding of long-chain acyl-CoA esters at a conserved acyl-CoA-binding domain (Xiao & Chye, 2009). In Arabidopsis, six genes encode ACBPs which are subcellularly localized in different compartments (Chye et al., 1999; Li & Chye, 2003; Chen et al., 2008; Xiao et al., 2008a). Since they bind different acyl-CoA esters with varying affinities, they do not seem to have redundant roles in vivo (Chye, 1998; Chye et al., 2000; Leung et al., 2004; 2006; Gao et al., 2009; Xiao et al., 2009). Besides the presence of the conserved ACBP domain, some ACBPs contain other functional domains, such as ankyrin repeats in ACBP1 and ACBP2, and kelch motifs in ACBP4 and ACBP5, which mediate interaction with protein partners (Li & Chye; 2004; Li et al., 2008; Gao et al., 2009). Three cytosolic ACBPs (ACBP4, ACBP5 and ACBP6) are known to bind phosphatidylcholine in vitro (Chen et al., 2008; Xiao et al., 2009). Immunogold labeling analysis has previously indicated that ACBP1 is localized to the plasma membrane of epidermal cells and in the cotyledonary cells at various stages (heart, torpedo and cotyledon) of embryo development in developing seeds (Chye et al., 1999). It has been proposed that ACBP1 may be involved in lipid transfer originating from the endoplasmic reticulum (ER) to the plasma membrane during lipid metabolism in these seeds (Chye et al., 1999). Consistently, ACBP1-GFP and ACBP2-GFP fusion proteins were localized at the plasma membrane and ER in onion epidermal cells by particle bombardment and results were confirmed using subcellular fractionation followed by western blot analysis (Li & Chye, 2003). The N-terminal transmembrane domains, in both ACBP1 and ACBP2, are essential for their membrane association (Li & Chye, 2003). Given the importance of proteins/enzymes related to lipid metabolism in Arabidopsis seed development (Baud et al., 2003; Rylott et al., 2003), the accumulation of ACBP1 in seeds and embryos suggests possible roles in embryo development or seedling establishment. Thus, it was pertinent to investigate whether ACBP1 and/or ACBP2 affect early development by analysis of single mutants as well as the acbp1acbp2 double mutant.
Materials and methods
Plant materials and growth conditions
The acbp2 T-DNA insertion mutant was identified from a T-DNA insertional library from the Torrey Mesa Research Institute of Syngenta (www.tmri.org). After surface-sterilization and chilling at 4°C for 2 days, seeds of Arabidopsis thaliana wild-type (ecotype Columbia), acbp1 and acbp2 mutants were germinated and grown on MS medium (Murashige & Skoog, 1962) supplemented with 2% sucrose grown under cycles of 8 h dark (21°C) and 16 h light (23°C). Soil-grown plants were also grown under 8 h dark (21°C) and 16 h light (23°C) cycles.
Immunohistochemical localization of ACBP2 using light microscopy
Immunohistochemical localization of ACBP2 using the anti-ACBP2 specific antibodies (Chye et al., 2000; Li & Chye, 2003) was performed as previous described (Chye et al., 1999). Briefly, the Arabidopsis siliques containing developing seeds at various stages of embryos were fixed and embedded in paraffin following the procedure described by Chye et al. (1999). Blocking and antibody reactions were carried out in 1% BSA in phosphate buffered saline (PBS). The sections were incubated for 5 min with PBS and 0.1% saponin (Sigma) before incubation with PBS containing 0.1% saponin, 1% BSA and 2% goat serum for 1 h at room temperature. Sections were incubated with rabbit anti-ACBP2 specific antibodies (1:1000 [v/v]) at 4°C overnight and with the secondary antibody biotinylated alkaline-phosphatase-conjugated goat anti-rabbit antibodies (1:1000 [v/v]; BioRad) at room temperature for 2 h. Levamisole (1 mM; Sigma) was included in the alkaline phosphatase reaction to inhibit endogenous phosphatase activity and this reaction was carried out following the instructions of manufacturer (BioRad).
Identification of the acbp2 mutant
The homozygous acbp2 mutant was isolated by PCR amplification using 2 primer pairs (i) ACBP2 gene-specific forward primer ML251 (5′-ATCGGCGTTGGTTTTTCGTTTTTGAGAAT-3′) with reverse primer ML252 (5′-TTGCCGCCAAAGTCGGTTATTTATTCGTT-3′) and (ii) ML205 (5′-CGTCACCCAGAGGAGTC-3′) with the T-DNA left border primer Oligo113 (O113; 5′-TAGCATCTGAATTTCATAACCAATCTCGATACAC-3′). The PCR products were separated by electrophoresis on 0.8% agarose and DNA was transferred to a nylon membrane (Hybond-N, Amersham). The blot was hybridized overnight at 42 °C to a random-primed 32P-labeled full-length ACBP2 gene probe. The blot was washed in 0.1 × SSC, 0.1% SDS at 65 °C for 10 min. The position of the T-DNA insertion was confirmed by DNA sequence analysis of the resultant PCR products.
Western blot analysis
Total plant protein was extracted (Chye et al., 1999) from mature silique-bearing plants of wild-type Arabidopsis or the acbp2 mutant. Protein concentration was determined using the Bio-Rad Protein Assay Kit following the method of Bradford (1976). Ten μg of total protein was loaded per well in SDS-polyacrylamide gel electrophoresis. The proteins were electrophoretically transferred to Hybond-C membrane (Amersham) from the SDS-PAGE gel using the Trans-Blot cell (Bio-Rad). Affinity-column purified ACBP2-specific antibodies (Chye et al., 2000; Li & Chye, 2003) were used in Western blot analysis. The Amplified Alkaline Phosphatase Goat Anti-rabbit Immuno-blot Assay Kit (BioRad) was used following the instructions of the manufacturer in detection of cross-reacting bands.
Semi-quantitative reverse transcription (RT)-PCR analysis
Total RNA was isolated using TRIzol reagent (Invitrogen, Cat No. 15596-018) from rosettes and siliques of 7-week-old wild-type, acbp1 and acbp2 plants. First-strand synthesis was carried out using the Superscript™ First-strand synthesis system (Invitrogen, Cat No. 12371-019). Gene-specific primers for RT-PCR were used as described previously (Xiao et al., 2008b) and the cycle numbers of amplification with each primer pair was adjusted to be within the linear range.
Screening of the acbp1acbp2 double mutant
The acbp1 (Xiao et al., 2008b) and acbp2 homozygous mutants were crossed and their resultant F2 population was screened for acbp1acbp2 double mutants. F2 seeds were sterilized and grown on kanamycin-containing MS medium. From kanamycin resistant (for acbp2 allele) plants, DNA was extracted and primer combinations ML179/ML209 and ML179/SLB1 (Xiao et al., 2008b), O113/ML206 (5′-TCGGGGTGGGGATGA TGC-3′) and ML206/ML252 (Fig. 3a) were used in PCR to screen for the acbp1 and acbp2 alleles, respectively. Since acbp1acbp2 double mutants were not obtained from >200 F2 plants screened, acbp1ACBP2+/− (i.e., homozygous for acbp1 and heterozygous for acbp2) and ACBP1+/−acbp2 (i.e., heterozygous for acbp1 and homozygous for acbp2) plants were subsequently generated. The self-fertilized F3 seeds of acbp1ACBP2+/− or ACBP1+/−acbp2 plants were compared to WT by light microscopy, the percentages of aborted ovules in open siliques from WT and acbp1ACBP2+/− or ACBP1+/− acbp2 plants were calculated and their whole-mount embryo development observed. For complementation testing, transgenic line acbp1::35S-ACBP1 (cACBP1-2; Xiao et al., 2008b) was crossed to acbp1ACBP2+/− plants and the F1 progenies were used for further analysis.
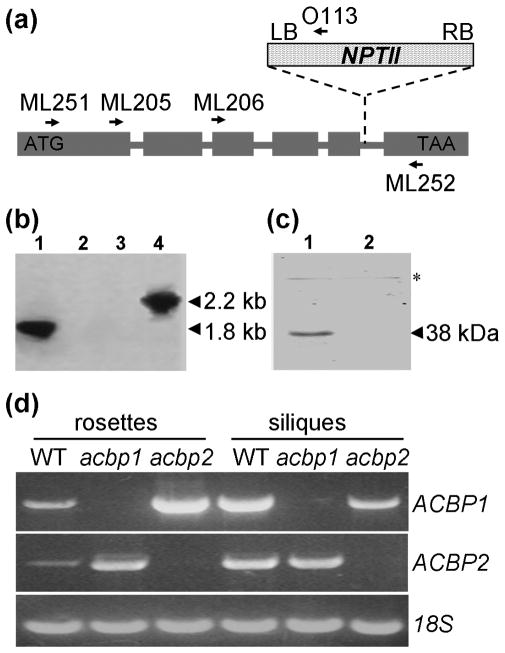
Fig. 3. Characterization of acbp1 and acbp2 knockout mutants.
(a) Diagram of T-DNA insertion into ACBP2, and the primer locations used for PCR genotyping of the acbp2 allele.
(b) Southern blot analysis of PCR products amplified from acbp2 mutant DNA (lanes 1 and 2) or wild-type Arabidopsis DNA (lanes 3 and 4) using primer pair, ML205 and O113 (lanes 1 and 3) or ML251 and ML252 (lanes 2 and 4). The gel was blotted on Hybond-N and hybridized to 32P-labeled ACBP2 cDNA.
(c) Western blot analysis of total protein from the silique-bearing acbp2 mutant (lane 1) or wild-type Arabidopsis (lane 2) using the ACBP2-specific antibodies. Ten μg of total proteins were loaded each lane on a 10% SDS- polyacrylamide gel. Asterisk indicates the position of a nonspecific immunoreacting band.
(d) Semi-quantitative reverse transcription (RT)-PCR analysis on the expression of ACBP1 and ACBP2 in wild type, acbp1 and acbp2 mutants. Total RNA was extracted from rosettes and siliques of 7-week-old wild-type, acbp1 and acbp2 plants. The 18S transcript was used as a control.
Microscopy
Developing seeds or excised embryos were placed in Herr’s solution (Herr, 1971) composed of 85% lactic acid, chloral hydrate, phenol, clove oil and xylene (2:2:2:2:1), for 2 h to overnight. Slides were viewed subsequently using a Leitz photomicroscope using differential interference contrast optics.
Callus induction
Callus induction was carried out according to Liu et al. (2004). Aborted embryos of 4-DAF acbp1ACBP2+/− plants were excised under the microscope, placed onto callus-inducing medium containing 2,4-D (0.5 mg L−1), indoleacetic acid (2 mg L−1), and 2-iP(N6-[2-isopentenyl]adenine; 0.5 mg L−1), and were cultured for 3 weeks at 22°C in the dark. Embryos from wild-type, acbp1 and acbp2 plants at similar developmental stage were excised and grown on callus-inducing medium under the same conditions.
Lipid and acyl-CoA profiling
Total lipid extraction was carried out according to Welti et al. (2002) and lipids were dissolved in chloroform for analysis. The profiles of membrane lipids were measured by an automated electrospray ionization tandem mass spectrometry method (Devaiah et al., 2006).
Acyl-CoA profiling was carried out as described previously (Larson & Graham, 2001; Larson et al., 2002). Briefly, acyl-CoAs from the frozen samples of 7-week-old rosettes and siliques were extracted, dried under vacuum at 40°C and subsequently reacted with 50 μl buffered chloroacetaldehyde reagent to form fluorescent acyl etheno CoA derivatives. The acyl-CoA standards purchased from Avanti (Avanti Polar Lipids, Inc. USA) were similarly treated. The derived standards and acyl-etheno-CoA samples were separated and quantified by a reversed-phased HPLC on a LUNA phenyl-hexyl column (Phenomenex, 150×2.0 mm, 5 μm) together with a 4×2 mm phenyl-propyl guard column. The solvent system was identical with the longer gradient conditions reported by Larson & Graham in 2001..
Lipid binding assay
(His)6-ACBP1 and (His)6-ACBP2 were expressed in the soluble fraction and inclusion bodies, respectively, of E. coli extracts and were each purified through an affinity column of Ni-NTA Agarose (Qiagen, Valencia, CA, USA) as described (Chye, 1998; Chye et al., 2000). Binding of (His)6-ACBP1 and (His)6-ACBP2 to various lipids on filters was carried out according to Chen et al. (2008).
To further confirm the interactions of (His)6-ACBP1 and (His)6-ACBP2 with PC, the Lipidex 1000 competition assay was carried out to determine if PC was capable of competing with [14C]linoleoyl-CoA (American Radiolabelled Chemicals, https://www.arcincusa.com) in binding (His)6-ACBP1 and (His)6-ACBP2 (Rosendal et al., 1993; Gao et al., 2009). In the Lipidex 1000 binding assay, the incubation medium contains both ACBP1/ACBP2 and radiolabelled acyl-CoA. Subsequently, unbound radiolabelled acyl-CoA was removed from the incubation medium by Lipidex 1000. The bound radiolabelled acyl-CoA remain in the supernatant and was detected by measurement of radioactivity counts. In Lipidex 1000 competition assays, PC liposome was further added to the incubation medium containing both ACBP1/ACBP2 and radiolabelled acyl-CoA. If PC liposome successfully competes with acyl-CoA in binding to ACBP1/ACBP2, bound radiolabelled acyl-CoA will decrease and a decline in radioactivity count would result.
PC liposome was prepared according to Sano et al. (1998). Different concentrations of PC liposome (0–5 μM) were mixed with 0.8 μM [14C]linoleoyl-CoA and 0.2 μM (His)6-ACBP1 or (His)6-ACBP2. Each mixture was incubated for 30 min at 37°C, and 400 μl of ice-cold 50% slurry of Lipidex 1000 (PerkinElmer, http://www.perkinelmer.com) and binding buffer were added. Samples were centrifuged at 12,000 × g for 5 min at 4°C and a 200-μl aliquot of the supernatant was taken for analysis of radioactivity counts using a LS 6500 liquid scintillation counter (Beckman). Assays were performed in triplicates, with blanks, at each concentration of PC liposome.
Results
Expression of ACBP1 and ACBP2 mRNAs during Arabidopsis development
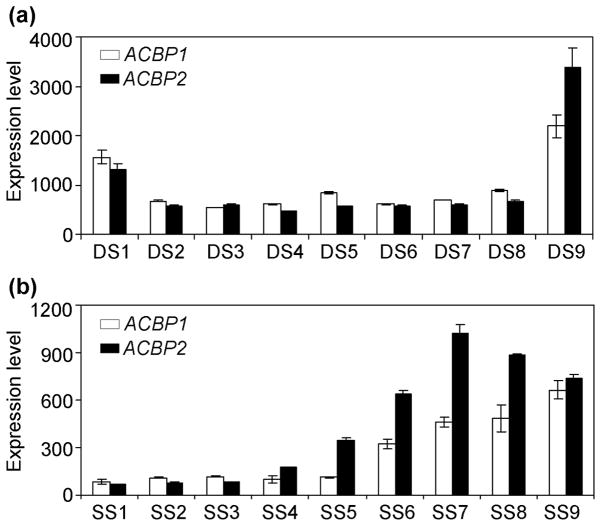
Our previous studies have indicated that ACBP1 (Chye, 1998) and ACBP2 (Chye et al., 2000; Gao et al., 2009) mRNAs are expressed in all plant organs while ACBP1 protein accumulates in developing seeds (Chye, 1998), and ACBP2, in flowers and siliques (Kojima et al., 2007). To further understand the dynamic expression patterns of ACBP1 and ACBP2 during Arabidopsis development, data of their expression at various stages in development were retrieved from the microarray database Gene Chronologer (Zimmermann et al., 2004; https://3.met.genevestigator.com/at/index.php?page=home). Expression of both ACBP1 and ACBP2 was relatively high in early developmental stage 1 (DS1; refers to seedlings aged 1 to 5.9 days post-germination). Expression decreased from stages DS2 to DS8, spanning day 6 to day 44.9 (Fig. 1a). However, at DS9 (representing the seed set developmental stage), both genes again show high expression (Fig. 1a). Further investigation on ACBP1 and ACBP2 expression during seed development was performed by analyses of their expression at different seed stages (SS) using the e-FP Browser (Winter et al., 2007; http://www.bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi) database. Results suggest that the expression of ACBP1 and ACBP2 increased from stages SS5 (walking-stick stage of development) to SS7 (late cotyledonary stage) or SS8 (green cotyledonary stage) and were relatively high in dry seeds (SS9) (Fig. 1b).
Fig. 1. Microarray data showing the expression patterns of ACBP1 and ACBP2 mRNAs.
(a) Expression of ACBP1 and ACBP2 at various developmental stages (DS). DS1, seedlings of 1–5.9 days; DS2, seedlings of 6–13.9 days; DS3, seedlings of 14–17.9 days; DS4, plants of 18–20.9 days; DS5, plants of 21–24.9 days; DS6, plants of 25–28.9 days; DS7, plants of 29–35.9 days; DS8, plants of 36–44.9 days; DS9, 45–50 days (seeds).
(b) Expression of ACBP1 and ACBP2 at various seed stages (SS). SS1, globular; SS2, early heart; SS3, late heart; SS4, torpedo; SS5, walking-stick; SS6, early curled cotyledons; SS7, late cotyledons; SS8, green cotyledons; SS9, dry seeds.
Immunolocalization of ACBP2 in Arabidopsis embryos using light microscopy
Since immunolocalization using anti-ACBP1 polyclonal antibodies has revealed that ACBP1 protein accumulates in the cotyledonary cells of embryos in developing seeds (Chye et al., 1999), we were interested to investigate if ACBP2 accumulates in seeds. Longitudinal sections of siliques at various stages of development were immunolocalized using anti-ACBP2 specific antibodies (Chye et al., 2000; Li & Chye, 2003) followed by light microscopy. Results revealed that embryos at globular (Fig. 2a), heart (Fig. 2b), heart-to-torpedo transition (Fig. 2c) and mature cotyledonary (Fig. 2d) stages were immunostained and express ACBP2. In contrast, similar stage embryos using preimmune serum from the same rabbit as a negative control did not show any staining (Figs. 2e to 2h).
Fig. 2. Immunolocalization of ACBP2 using anti-ACBP2 antibodies in developing seeds of various embryo stages.
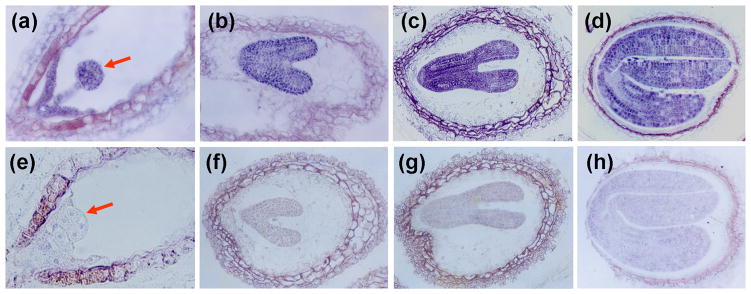
(a) to (d) Longitudinal sections of seed at globular (a, indicated by arrow), heart (b), heart to torpedo transition (c) and cotyledon (d) stages showing the embryos stained with anti-ACBP2 primary antibody followed by biotinylated goat anti-rabbit antibody and reacted with streptavidin-biotinylated alkaline phosphatase.
(e) to (h) Longitudinal section of seed at globular (e, indicated by arrow), heart (f), heart to torpedo transition (g) and cotyledon (h) stages showing embryos stained with preimmune serum followed by biotinylated goat anti-rabbit antibody and reacted with streptavidin-biotinylated alkaline phosphatase.
Characterization of an acbp2 knockout mutant
In this study, an acbp2 T-DNA insertional mutant was isolated from CS19943 (a T-DNA mutagenized Arabidopsis seed pool generated by Thomas Jack; ecotype Col-6) using a combination of gene-specific and T-DNA border-specific primers (Winkler et al., 1998). The location of the T-DNA insertion in ACBP2 was confirmed by PCR using ACBP2 gene-specific primers ML251 and ML252, as well as gene-specific forward primer ML205 and reverse primer Oligo113 (O113) that maps to the T-DNA left border (Figs. 3a and 3b). By further DNA sequencing analysis, the insertion site was mapped to intron 5 on ACBP2 (Fig. 3a). Western blot analysis using ACBP2-specific antibodies (Fig. 3c) and RT-PCR analysis using gene-specific primers ML206/ML192 (Fig. 3d) confirmed the mutant to be a functional knockout line. Since we have previously characterized a T-DNA insertional knockout mutant of ACBP1 (Xiao et al., 2008b), we were able to compare the expression of ACBP1 in the acbp2 mutant and that of ACBP2 in the acbp1 mutant. Semi-quantitative RT-PCR results show that in rosettes, both of the expression of ACBP1 in the acbp2 mutant and the expression of ACBP2 in the acbp1 mutant were up-regulated in comparison to wild type (Fig. 3d). Consistent with microarray data (Fig. 1), the expression levels of ACBP1 and ACBP2 in wild-type siliques were significantly higher than rosettes, while up-regulation of ACBP1 in the acbp2 mutant and vice verse was not evident (Fig. 3d).
Growth and development of the acbp2 and the acbp1 mutants were compared. At early development (2-week-old seedling) and late development (5-week-old plants), both acbp1 and acbp2 mutants were no different from wild type under normal growth conditions (Fig. S1a and b). To gain a better insight into the functions of ACBP1 and ACBP2 in seed development, embryos from the acbp1 and acbp2 mutants were carefully analyzed. Siliques of the acbp1 and acbp2 mutants produced normal seeds, similar to wild type (Fig. S1c). Whole-mount examination of embryo development in wild-type, acbp1 and acbp2 siliques (Fig. S1d) indicated that acbp1 and acbp2 embryos reached heart stage at 4 days after fertilization (DAF) and bent-cotyledonary stage at 6 DAF. These results suggest that the acbp1 and acbp2 single mutants displayed no obvious morphological changes in embryonic and seed development in Arabidopsis and are indistinguishable from wild type.
The acbp2acbp2 double mutant is embryo lethal
Since ACBP1 and ACBP2 are highly conserved (82% identity) and are both expressed in embryos, we next investigated if they function redundantly in lipid metabolism during seed development. To test this possibility, a combination of acbp1 and acbp2 mutations would be necessary. Hence, we crossed the two single acbp1 and acbp2 mutants, generated the F1 progeny, and subsequently screened their F2 populations for acbp1acbp2 double mutants. However, following genotyping of more than 200 F2 plants, acbp1acbp2 double mutants were not encountered. We were only able to identify genotypes of either acbp1ACBP2+/− (i.e., homozygous for acbp1 and heterozygous for acbp2; lane 1 in Fig. 4a) or ACBP1+/−acbp2 (i.e., heterozygous for acbp1 and homozygous for acbp2; lanes 6, 9 and 11 in Fig. 4a).
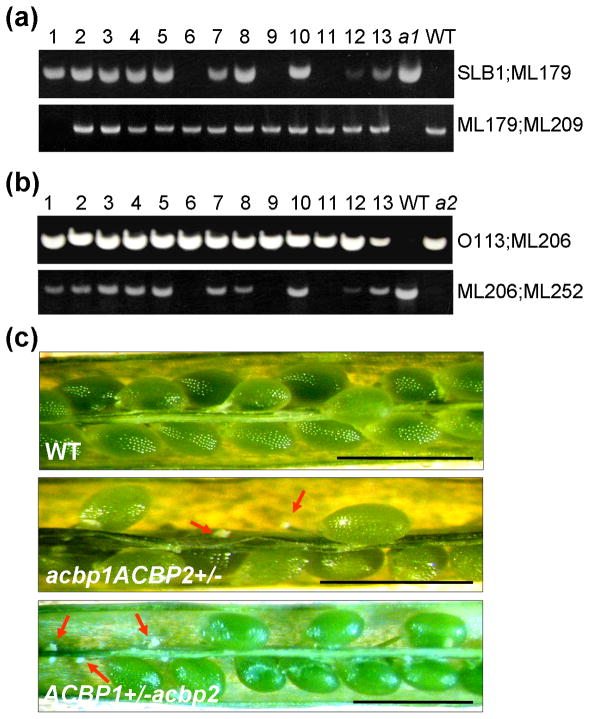
Fig. 4. Genotyping and initial characterization of acbp1acbp2 double mutant.
(a) and (b) Specificity of the primer combinations (at right) used in PCR to screen the acbp1acbp2 double mutant from the kanamycin resistant (for acbp2 allele) F2 population. ML179 and ML209 are ACBP1 gene-specific primers and SLB1 is T-DNA-sepcific primer used for genotyping the acbp1 allele (Xiao et al., 2008b). No acbp1acbp2 double mutants were obtained from over 200 F2 plants screened, so acbp1ACBP2+/− (i.e., homozygous for acbp1 and heterozygous for acbp2; lane 1 in a) and ACBP1+/− acbp2 (i.e., heterozygous for acbp1 and homozygous for acbp2; lanes 6, 9 and 11 in b) were subsequently generated.
(c) The self-fertilized F3 seeds of acbp1ACBP2+/− and ACBP1+/−acbp2 plants were compared with WT by light microscopy (top). Ovules in open siliques from acbp1ACBP2+/− plants showing aborted ovules (indicated by arrows).
WT, wild type. Bars: 1mm.
The morphologies of seeds produced from the self-pollinated acbp1ACBP2+/− and ACBP1+/−acbp2 plants were further examined. As shown in Fig. 4c, in contrast to wild-type siliques which contain almost all normal seeds that were large and green, the siliques of acbp1ACBP2+/− and ACBP1+/−acbp2 plants contain both normal seeds which were large and green, as well as a portion of aborted seeds (Fig. 4c). The percentage of aborted seeds in acbp1ACBP2+/− and ACBP1+/−acbp2 plants of around 35% (Table 1), correlated well with a 3:1 segregation ratio. However, when the ACBP1 full-length cDNA was introduced into an acbp1ACBP2+/− background by crossing acbp1ACBP2+/− with cACBP1-2 (previously verified in Xiao et al., (2008b) to be a complemented line for the acbp1 mutant), only 5.7% of seeds were aborted (Table 1). This indicates that acbp1acbp2 embryos could be rescued by introduction of an ACBP1 cDNA. Also, by taking advantage of the kanamycin resistance phenotype conferred by the T-DNA in the acbp2 mutant, the ratios of resistant: sensitive progenies from both acbp1ACBP2+/− and ACBP1+/−acbp2 were about 2: 1 when grown on MS medium containing kanamycin (Table 2). These results confirm that a combination of acbp1 and acbp2 mutations affect embryo development.
Table 1.
Segregation and complementation analyses of acbp1ACBP2+/− plants.
| Normal seeds | Aborted seeds | x2 (hypothesis) | |
|---|---|---|---|
| ACBP1+/−acbp2 | 291 | 102 | 0.25 (3:1) |
| acbp1ACBP2+/− | 252 | 90 | 0.39 (3:1) |
| acbp1ACBP2+/− x cACBP1 | 368 | 21 | 0.35 (15:1) |
| wild type | 282 | 8 | - |
Seeds were analysed from ten siliques of single plants in each genotype. The genotypes of each plant were confirmed by PCR before analysis. Chi-square values were calculated for the hypothesized segregation ratio. The x2 values showed no significant deviation (P > 0.05) from the hypothesized ratio.
Table 2.
Ratio of Kanr: Kans for the progeny from self-pollinated ACBP1+/−acbp2 and acbp1ACBP2+/− plants.
| Kanr | Kans | x2 (hypothesis) | |
|---|---|---|---|
| ACBP1+/−acbp2 | 316 | 166 | 0.32 (2:1) |
| acbp1ACBP2+/− | 378 | 180 | 0.24 (2:1) |
Chi-square values are shown for the hypothesized segregation ratio and the x2 values show no significant deviation (P > 0.05) from the hypothesized ratio.
To further study the cause for embryo lethality in the acbp1acbp2 double mutant, embryo development at various stages in acbp1ACBP2+/− and wild-type plants were investigated. In the siliques of wild type, most embryos reached globular (Fig. 5g) and heart (Figs. 5h and 5i) stages by 3 DAF (days after fertilization). However, significant variation in embryo development was observed in different seeds derived from a single silique of the acbp1ACBP2+/− line. As shown in Fig. 5, in addition to “normal” seeds in acbp1ACBP2+/− siliques which develop to globular (Fig. 5d) and heart (Figs. 5e and 5f) stages, there were also aborted ovules arrested at either zygote (Fig. 5a), 2-cell (Fig. 5d) or 8-cell (Fig. 5c) stage in the acbp1ACBP2+/− siliques. Further analysis revealed that 161 of 225 (72%) embryos in acbp1ACBP2+/− siliques reached globular and heart stages at 3 DAF, while the remaining 64 embryos (28%) were aborted early at either zygote, 2-cell or 8-cell stage (Fig. 5j). The percentage of aborted embryos in siliques of acbp1ACBP2+/− plants would be in agreement with an expected lethality in the acbp1acbp2 double mutation. In summary, these results demonstrate that ACBP1 and ACBP2 are essential for embryo and seed development and embryos of the acbp1acbp2 double mutant were aborted at early embryo development.
Fig. 5. A combination of acbp1 and acbp2 mutations affects embryo development.
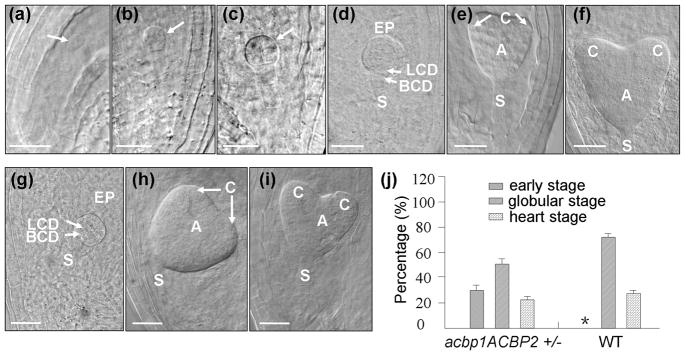
(a) to (i) Embryo development of 3-DAF (days after fertilization) ovules of acbp1ACBP2+/− (a–f) and WT (g–i) plants. At this stage, most of WT embryos have reached globular (g) or heart stages (h and i), while in the acbp1ACBP2+/− siliques, aborted ovules were arrested at zygote (a), 2-cell (b) or 8-cell (c) stage. Normal seeds in the same acbp1ACBP2+/− siliques develop to globular (d) and heart (e and f) stages. Arrows in (a)–(c) indicate arrested embryos.
(j) Percentage of different embryo stages (early stage including zygote, 4-cell or 8-cell stage, globular stage and heart stage) in acbp1ACBP2+/− (28%, 50% and 22%, respectively) and WT (3%, 73% and 24%, respectively) plants.
WT, wild type; A, axis; BCD, basal cell descendant; C, cotyledon; LCD, lens cell descendant; S, suspensor; Bars: 25 μm.
Aborted embryos from acbp1ACBP2+/− are defective in callus induction
To further investigate the nature of embryo aberration in the acbp1ACBP2+/− plants, callus induction was performed. To this end, the aborted embryos from 4-DAF acbp1ACBP2+/− plants were excised and subsequently cultured on a callus induction medium. As controls, embryos from wild type, acbp1 and acbp2 at a similar developmental stage were also excised and cultured. As shown in Fig. S2, wild-type, acbp1, and acbp2 embryos formed callus tissue after a 3-week induction period, while the aborted embryos, which should be acbp1acbp2 double mutants, failed to form calli.
Lipid and acyl-CoA profile changes in the acbp1 and acbp2 mutants
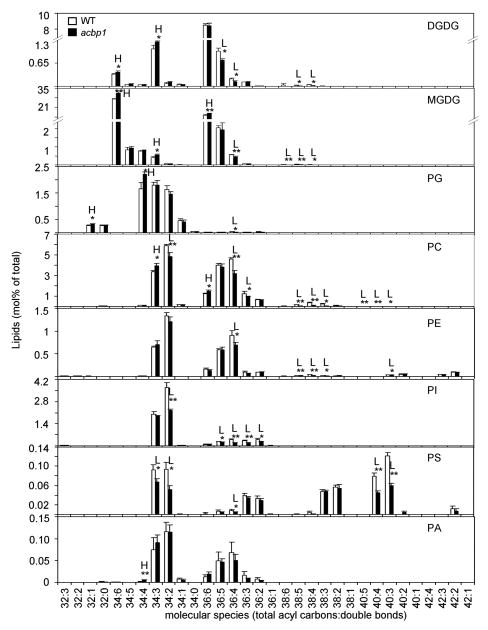
To investigate whether the altered expression of ACBP1 or ACBP2 in the mutants affect plant lipid metabolism, lipid composition in both rosettes and siliques of 7-week-old wild type (Col), acbp1 and acbp2 mutants were analyzed using electrospray ionization tandem mass spectrometry (ESI MS/MS; Welti et al., 2002; Devaiah et al., 2006). From lipid profiling data, we observed that wild-type plants showed different patterns of lipid profiles in rosettes and siliques (Table 3). Specifically, the total contents of galactolipids digalactosyldiacylglycerol (DGDG) and monogalactosyldiacylglycerol (MGDG) were significantly lower in siliques while those of phospholipids such as phosphatidylcholine (PC), phosphatidylethanolamine (PE) and phosphatidylinositol (PI), as well as lysoPC and lysoPE were significantly higher in comparison to rosettes (Table 3). There were little differences between rosettes of wild type and the single acbp1 or acbp2 knockout mutant in the total amounts of all lipid species analyzed including DGDG, MGDG, phosphatidylglycerol (PG), PC, PE, PI, phosphatidylserine (PS), phosphatidic acid (PA), lysoPC, lysoPE and lysoPG (Table 3). However, in comparison to wild type siliques, significantly higher levels of MGDG (P < 0.01) and significantly lower amounts of the phospholipids PC, PI and PS (P < 0.05 or P < 0.01) were observed in the acbp1 mutant, but not the acbp2 mutant (Table 3). Further, analyses of the lipid profiles of wild type and acbp1 mutant siliques revealed that galactolipid species such as 34:6- and 34:3-MGDG, DGDG, and 36:6-MGDG significantly increased in the acbp1 mutant, while those of 36:4-, 38:5- and 38:4- MGDG, DGDG and 38:6-MGDG, 36:5-DGDG significantly decreased in comparison to wild type (Fig. 6). In contrast, most of the polyunsaturated species of phospholipids such as 34:2-, 36:4-, 36:3-, 38:5-, 38:4-, 38:3-, 40:5-, 40:4-, 40:3-PC; 36:4-, 38:5-, 38:4-, 38:3-, 40:3-PE, and 34:2-, 36:5-, 36:4-, 36:3- and 36:2-PI were significantly lower than wild type, except for the species of 34:3- and 36:6-PC which were higher in the acbp1 mutant siliques (Fig. 6).
Table 3.
Lipid profiles in rosettes and siliques of 6-week-old wild type (Col), acbp1 and acbp2 mutants grown at 16 h light (23°C)/8 h dark (21°C). Values are means ± SD (% of total polar glycerolipids analyzed; n = 3). Significant differences in the siliques of wild type from that of rosettes or in the acbp1 mutant from that of wild type are bolded (* P < 0.05; ** P < 0.01).
| Lipid class | Rosettes | Siliques | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| WT | acbp1 | acbp2 | WT | acbp1 | acbp2 | |
| DGDG | 13.52±0.65 | 13.84±0.47 | 14.43±0.53 | 11.46±0.19**b | 11.33±0.38 | 12.01±0.14 |
| MGDG | 66.84±1.40 | 68.29±0.14 | 66.71±0.71 | 47.57±0.49**b | 52.91±1.39**c | 48.46±1.02 |
| PG | 5.55±0.06 | 5.59±0.48 | 5.55±0.66 | 6.23±0.47 | 6.55±0.44 | 5.47±0.25 |
| PC | 7.68±0.48 | 7.73±0.05 | 7.79±0.37 | 22.16±0.71**a | 19.35±1.47*d | 20.83±0.83 |
| PE | 1.50±0.24 | 1.30±0.11 | 1.42±0.11 | 4.18±0.25**a | 3.77±0.35 | 4.39±0.09 |
| PI | 2.78±0.18 | 2.77±0.06 | 3.48±0.28 | 7.25±0.58**a | 5.15±0.19**d | 7.70±0.03 |
| PS | 1.82±2.83 | 0.18±0.01 | 0.18±0.04 | 0.60±0.04 | 0.41±0.04**d | 0.62±0.04 |
| PA | 0.25±0.05 | 0.24±0.04 | 0.36±0.32 | 0.36±0.10 | 0.35±0.06 | 0.28±0.06 |
| LysoPC | 0.014±0.002 | 0.013±0.001 | 0.019±0.004 | 0.060±0.007**a | 0.061±0.013 | 0.078±0.011 |
| LysoPE | 0.046±0.011 | 0.041±0.008 | 0.059±0.019 | 0.124±0.012**a | 0.108±0.017 | 0.142±0.002 |
| LysoPG | 0.005±0.001 | 0.008±0.006 | 0.005±0.005 | 0.005±0.004 | 0.014±0.011 | 0.017±0.026 |
Value increase in siliques of wild type when compared rosettes of wild type in the same experiment.
Value decrease in siliques of wild type when compared rosettes of wild type in the same experiment.
Value higher in siliques of acbp1 mutant when compared to siliques of wild type in the same experiment.
Value lower in siliques of acbp1 mutant when compared to siliques of wild type in the same experiment.
Fig. 6. Membrane lipid content (% of total glycerolipids analysed) in siliques of 7-week-old wild type (WT) and acbp1 mutant.
H indicates value of acbp1 mutant higher than WT; L indicates value of mutant lower than WT (* P < 0.05 or ** P < 0.01). Values are the means ± SD (n = 3).
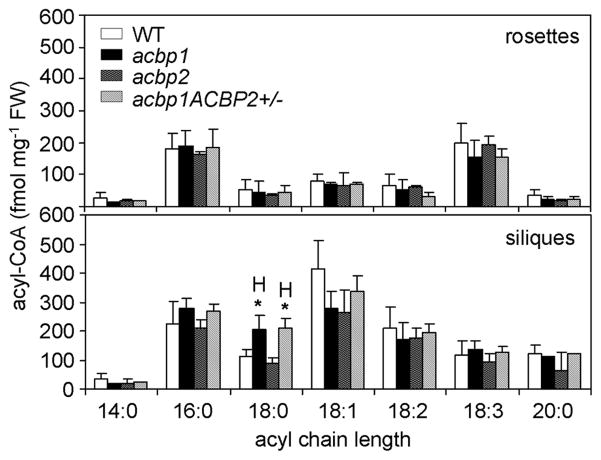
Analyses of acyl-CoA content of rosettes from both acbp1 and acbp2 single knockout mutants did not reveal any differences in acyl-CoA profile in comparison to wild type (Fig. 7, upper column). In siliques, only 18:0-CoA in the acbp1 mutant was higher than wild type (Fig. 7, bottom column). The acyl-CoA profile of rosettes and siliques from the acbp1ACBP2+/− plants resembled the acbp1 mutant (Fig. 7).
Fig. 7. Acyl-CoA content (fmole mg−1 FW) in rosettes and siliques of 7-week-old wild type (WT), acbp1, acbp2 and acbp1ACBP2+/− mutants.
H indicates value of acbp1 mutant and acbp1ACBP2+/− plants higher than WT (* P < 0.05). Values are the means ± SD (n = 3).
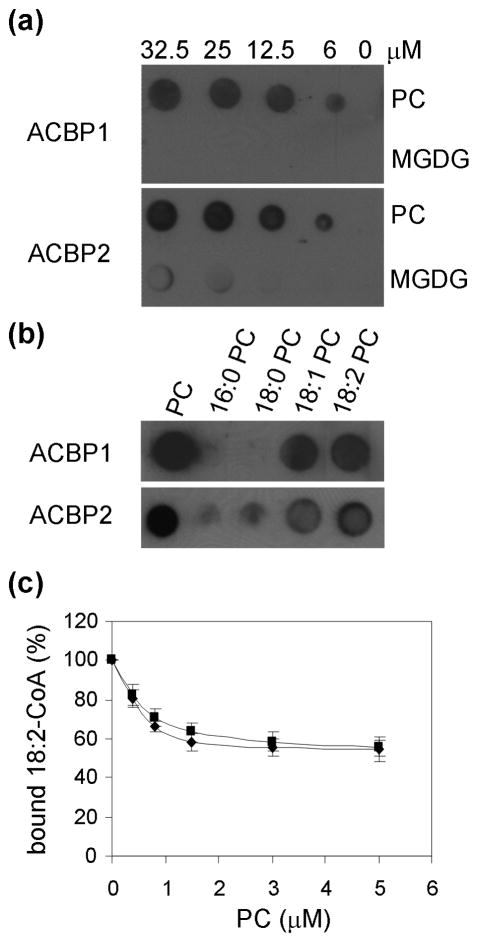
His-tagged ACBP1 and ACBP2 recombinant proteins bind to unsaturated PC and acyl-CoA esters in vitro
We have previously used in vitro filter-binding assays to show that recombinant ACBP4, ACBP5 and ACBP6 binds PC, and not PE, PI, PA and lysoPC (Chen et al., 2008; Xiao et al., 2009). To explore whether recombinant ACBP1 and ACBP2 bind to phospholipids, purified ACBP1 and ACBP2 recombinant proteins (Chye, 1998; Chye et al., 2000; Gao et al., 2009) were similarly tested with various concentrations of PC, PE, PI and PS as well as MGDG. These phospholipids were chosen because their contents had been altered in the siliques of the acbp1 mutant in comparison to wild type (Table 3 and Fig. 6). Results indicated that (His)6-ACBP1 and (His)6-ACBP2 bind PC, but not MGDG in a dose-dependent manner (Fig. 8a). They also do not bind PE, PI and PS (data not shown). As the PC used in Fig. 8a was 1,2-diacyl-sn-glocero-3-phosphatidylcholine, consisting of 33% 16:0, 13% 18:0, 31% 18:1 and 15% 18:2 fatty acids, the binding of several fatty acid species of PC to (His)6-ACBP1 and (His)6-ACBP2 were subsequently tested. Results showed that both (His)6-ACBP1 and (His)6-ACBP2 bind unsaturated species of PC (18:1-PC, 18:2-PC) tested, but unlike (His)6-ACBP4 (Xiao et al., 2009), (His)6-ACBP5 (Xiao et al., 2009), and (His)6-ACBP6 (Chen et al., 2008), they did not bind saturated species of PC (16:0-PC and 18:0-PC) (Fig. 8b).
Fig. 8. Interaction of His-tagged ACBP1 and ACBP2 recombinant proteins to PC.
(a) (His)6-ACBP1 and (His)6-ACBP2 bind to PC on filters. Various concentrations (0, 6, 12.5, 25.0, 32.5 and 50 μM) of PC and MGDG were spotted onto nitrocellulose and incubated with 1 μg ml−1 of purified (His)6-ACBP1 or (His)6-ACBP2 proteins. The (His)6-ACBP/PC binding was detected by immunoblotting with HRP-conjugated anti-penta-His antibodies.
(b) Effect of PC acyl species on (His)6-ACBP/PC binding. Fifty μM lipid (PC, 16:0-PC, 18:0-PC, 18:1-PC or 18:2-PC) spotted onto nitrocellulose was incubated with 1 μg ml−1 of purified (His)6-ACBP1 or (His)6-ACBP2 protein. The (His)6-ACBP/lipid binding was detected by immunoblotting with HRP-conjugated anti-penta-His antibodies. PC, 1, 2-diacyl-sn-glocero-3-phosphocholine; 16:0-PC, 1,2-dipalmitoyl-sn-glycero-3-phosphocholine; 18:0-PC, 1,2-distearoyl-sn-glycero-3-phosphocholine; 18:1-PC, 1,2-dioleoyl-sn-glycero-3-phosphocholine; 18:2-PC, 1,2-dilinoleoyl-sn-glycero-3-phosphocholine.
(c) Displacement of [14C]linoleoyl-CoA by PC liposome in Lipidex competition assays. Different concentrations of PC liposome (0, 0.4, 0.8, 1.5, 3.0, 5.0 μM) were incubated with 0.8 μM [14C]linoleoyl-CoA and 0.2 μM (His)6-ACBP1 or (His)6-ACBP2. The mixtures were mixed with Lipidex-1000. Aliquots (200 μl) of the supernatant were taken for analysis of radioactivity. Assays were performed in triplicates, with blanks, at each concentration of PC liposome. The bound acyl-CoAs in the presence of PC liposome (0–5 μM) were expressed relative to the value obtained from reaction containing 0 μM PC liposome (100%). Bars represent SD (n = 3).
Lipidex assays have previously shown that (His)6-ACBP1 and (His)6-ACBP2 binds [14C]18:2-CoA and [14C]18:3-CoA (Gao et al., 2009). Since they now also bind to 18:2-PC, Lipidex competition assays were used to determine if 18:2-PC liposome can compete with [14C]18:2-CoA in binding (His)6-ACBP1 and (His)6-ACBP2. The binding of [14C]18:2-CoA to both (His)6-ACBP1 and (His)6-ACBP2 (Fig. 8c) were observed to decrease in the presence of 18:2-PC liposome, implying that binding was displaced by the addition of 18:2-PC liposome.
Discussion
Given that both recombinant ACBP1 and ACBP2 proteins have been shown recently to bind linoleoyl-CoA and linolenoyl-CoA esters in vitro, the precursors in phospholipid membrane repair, we have suggested that they may share a role in repair of the phospholipid bilayer membrane following heavy metal stress (Xiao et al., 2008b; Gao et al., 2009). We further demonstrate that Arabidopsis ACBP1 (Chye et al., 1999) and ACBP2 (this study in Figs. 1 and 2) accumulate in the embryos of developing seeds, and they likely play essential roles in embryogenesis, possibly in membrane biogenesis. Northern blot and western blot analyses showed that ACBP1 and ACBP2 mRNAs and proteins are not seed-specific (Chye, 1998; Chye et al., 2000; Gao et al., 2009) and this would support the dual functions of these two ACBPs in both plant development and plant stress response.
Many enzymes/proteins involved in plant lipid metabolism such as acyl-CoA oxidases (ACXs), CoA biosynthetic enzymes (HAL3s), lysophosphatidic acid acyltransferase (LPAAT) and acetyl-CoA carboxylase (ACCase) are essential for either early embryogenesis or seedling development in Arabidopsis (Baud et al., 2003; Rylott et al., 2003; Kim & Huang, 2004; Kim et al., 2005; Rubio et al., 2006). More interestingly, impairment of CoA biosynthesis in the Arabidopsis hal3a-1halb double mutant caused embryo lethality at early/midglobular stage which precedes triacylglycerol deposition, suggesting that CoA is crucial in early embryo development (Rubio et al., 2006). Also, complete breakdown of short-chain acyl-CoA oxidase activity in the Arabidopsis acx3-1acx4-1 double mutant arrested embryo development at a very early stage (Rylott et al., 2003). Possible explanations for the embryo defects observed in the acx3-1acx4-1 double mutant include the accumulation of acyl-CoA or short chain fatty acid to toxic levels, and the failure in CoA transfer to the acyl-CoA pool or to the production of fatty acid- or lipid-based signaling molecules (Rylott et al., 2003). The redundant functions of ACBP1 and ACBP2 in seed development share the characteristics of these previously reported proteins because embryo lethality in the acbp1acbp2 double mutant resembles those of hal3a-1halb and acx3-1acx4-1. Observations of early embryo arrestment and lack of callus induction indicate that the development of the acbp1acbp2 embryo is stalled during embryo morphogenesis which precedes fatty acid accumulation and lipid storage.
During early seed development, one of the major plant membrane lipids, PC serves as a main substrate for desaturation in the ER and plays a central role in assembly of unsaturated TAG (Browse, 1997). An acyl-CoA:lysophosphatidylcholine acyltransferase catalyses the exchange of polyunsaturated fatty acyl chains on sn-2 of PC with linoleoyl-CoA (18:2-CoA) and linolenoyl-CoA (18:3-CoA) in the acyl-CoA pool. Subsequently, glycerol-3-phosphate acyltransferase (GPAT) and lysophophatidic acid acyltransferase (LPAAT) are involved in the formation of polyunsaturated diacylglycerol (DAG) and acyl-CoA:sn-1,2-diacylglycerol acyltransferase in the acylation of the sn-3 position of DAG to produce triacylglycerol (TAG; Browse, 1997). Previous evidence has suggested that acyl-CoA preference of mitochondrial GPAT in young rat liver is regulated by the presence of an ACBP bound to acyl-CoA (Kannan et al., 2003). The 10-kDa Brassica napus ACBP which was the first ACBP identified from plants, is highly expressed in embryos, and has also been demonstrated to activate GPAT activity in vitro (Hills et al., 1994; Brown et al., 1998). A recent study has revealed that the overexpression of BnACBP in Arabidopsis seeds increased polyunsaturated (18:2 and 18:3) and decreased saturated and monounsaturated (16:0, 18:0 and 20:1) fatty acids (Yurchenko et al., 2009). Further, recombinant BnACBP is important for the activity of lysophosphatidylcholine acyltransferase (LPCAT) in the exchange of the acyl group between the acyl-CoA pool and PC (Yurchenko et al., 2009). Given that only one 10-kDa ACBP has been isolated from Brassica so far (Hills et al., 1994; Yurchenko et al., 2009) while a family of six ACBPs exists in Arabidopsis (Xiao & Chye, 2009), it is possible the larger ACBP1 and ACBP2, which are membrane-associated and accumulate in embryos, play a more significant role in this event in Arabidopsis, rather than the 10-kDa ACBP6.
Furthermore, comparison of the lipid profiles in rosettes and siliques of wild-type Arabidopsis has shown that extra-plastidial phospholipids (PC, PE, PI and PS) significantly accumulate in siliques (Table 3), consistent with a previous report (Devaiah et al., 2006), and confirm the crucial role for these phospholipids in embryo development. Our biochemical data have also revealed that siliques (but not rosettes) of the acbp1 mutant show higher levels of galactolipid MGDG and lower levels of polyunsaturated PC, PE, PI and PS species in comparison to wild-type, demonstrating that knockout of one of these two genes affect phospholipid composition in siliques although these changes per se may not effectively stall embryo development. Lack of significant changes in phospholipid in siliques of the acbp2 mutant, indicates that ACBP1 may play a more prominent role in this event than its redundant homologue, ACBP2. Consistently, acyl-CoA profiling revealed that the level of 18:0-CoA was higher in siliques of the acbp1 mutant (but not in the acbp2 mutant), suggesting that similar to BnACBP (Yurchenko et al., 2009), ACBP1 may play a role in the exchange of acyl-CoAs, especially 18:0-CoA, affecting phospholipid content in siliques. This exchange was blocked to a certain extent in the acbp1 mutant and an accumulation of acyl-CoA in acbp1 siliques corresponded to a decline in phospholipids. Also, upregulation of ACBP1 in the acbp2 mutant, and vice verse, occurred only in rosettes (but not siliques), indicating that these two ACBPs may share both redundant and distinct cellular functions during plant development. Moreover, in vitro evidence showed that recombinant ACBP1 and ACBP2 bind unsaturated PC (18:1-PC and 18:2-PC) and acyl-CoAs (18:2-CoA and 18:3-CoA), further supporting their roles in phospholipid metabolism. These results also reinforce ACBP1 and ACBP2 function in phospholipid membrane biogenesis (Chye et al., 1998; Xiao et al., 2008b; Gao et al., 2009) which would be crucial in early embryo development. Taken together, we propose that the combination of acbp1 and acbp2 mutations likely abolish formation of an acyl-CoA pool in the ER or disrupt acyl-CoAs/lipid trafficking between the ER and the plasma membrane during early embryo embryogenesis.
Supplementary Material
Phenotypic analysis of acbp1 and acbp2 single mutants.
Callus induction on the embryos of wild-type, acbp1 and acbp2 plants, as well as aborted embryos from acbp1ACBP2+/− plants.
Acknowledgments
We thank Prof. R. Welti and Ms. M. Roth (Kansas Lipidomics Research Center) for lipid profiling, the Arabidopsis Information Resource (TAIR) for provision of acbp2 mutant seed pools and Prof. S.F. Chen (University of Hong Kong) for provision of HPLC for acyl-CoA analysis. This work was supported by a CRCG grant (10207211), QFC and JYM by university postgraduate studentships and SX by a postdoctoral fellowship from the University of Hong Kong and the University Grants Committee of the Hong Kong Special Administrative Region, China (Project No. AoE/B-07/99). Method development and instrument acquisition at the Kansas Lipidomics Research Center was supported by National Science Foundation (EPS 0236913, MCB 0455318, DBI 0521587), Kansas Technology Enterprise Corporation, Kansas IDeA Network of Biomedical Research Excellence (INBRE) of National Institute of Health (P20RR16475), and Kansas State University.
Footnotes
Additional supporting information may be found in the online version of this article.
References
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C. Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant Journal. 2003;33:75–86. doi: 10.1046/j.1365-313x.2003.016010.x. [DOI] [PubMed] [Google Scholar]
- Baud S, Lepiniec L. Regulation of de novo fatty acid synthesis in maturing oilseeds of Arabidopsis. Plant Physiology and Biochemistry. 2009;47:448–455. doi: 10.1016/j.plaphy.2008.12.006. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brown AP, Johnson P, Rawsthorne S, Hills MJ. Expression and properties of acyl-CoA binding protein from Brassica napus. Plant Physiology and Biochemistry. 1998;36:629–635. [Google Scholar]
- Browse J. Synthesis and storage of fatty acids. In: Vasil I, Larkins B, editors. Advances in Cellular and Molecular Biology of Plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1997. pp. 407–440. [Google Scholar]
- Chen QF, Xiao S, Chye ML. Overexpression of the Arabidopsis 10-kilodalton acyl-Coenzyme A-binding protein ACBP6 enhances freezing tolerance. Plant Physiology. 2008;148:304–315. doi: 10.1104/pp.108.123331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chye ML. Arabidopsis cDNA encoding a membrane-associated protein with an acyl-CoA-binding domain. Plant Molecular Biology. 1998;38:827–838. doi: 10.1023/a:1006052108468. [DOI] [PubMed] [Google Scholar]
- Chye ML, Huang BQ, Zee SY. Isolation of a gene encoding Arabidopsis membrane-associated acyl-CoA-binding protein and immunolocalization of its gene product. Plant Journal. 1999;18:205–214. doi: 10.1046/j.1365-313x.1999.00443.x. [DOI] [PubMed] [Google Scholar]
- Chye ML, Li HY, Yung MH. Single amino acid substitutions at the acyl-CoA-binding domain interrupt 14[C]palmitoyl-CoA binding of ACBP2, an Arabidopsis acyl-CoA-binding protein with ankyrin repeats. Plant Molecular Biology. 2000;44:711–721. doi: 10.1023/a:1026524108095. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE Dα1 knockout mutant. Phytochemistry. 2006;67:1907–1924. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Devic M. The importance of being essential: EMBRYO-DEFECTIVE genes in Arabidopsis. Comptes Rendus Biologies. 2008;331:726–736. doi: 10.1016/j.crvi.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Fan YF, Jiang L, Gong HQ, Liu CM. Sexual Reproduction in Higher Plants I: Fertilization and the Initiation of Zygotic Program. Journal of Integrative Plant Biology. 2008;50:860–867. doi: 10.1111/j.1744-7909.2008.00705.x. [DOI] [PubMed] [Google Scholar]
- Gao W, Xiao S, Li HY, Tsao SW, Chye ML. Arabidopsis thaliana acyl-CoA-binding protein ACBP2 interacts with heavy-metal-binding farnesylated protein AtFP6. New Phytology. 2009;181:89–102. doi: 10.1111/j.1469-8137.2008.02631.x. [DOI] [PubMed] [Google Scholar]
- Harada J. Seed maturation and control of dormancy. In: Larkins BA, Vasil IK, editors. Cellular and Molecular Biology of Plant Seed Development. Dordrecht: Kluwer Academic Publishers; 1997. pp. 545–592. [Google Scholar]
- Herr JM., Jr A new clearing-squash technique for the study of ovule development in angiosperms. American Journal of Botany. 1971;58:785–790. [Google Scholar]
- Hills MJ, Dann R, Lydiate D, Sharpe A. Molecular cloning of a cDNA from Brassica napus L. for a homologue of acyl-CoA-binding protein. Plant Molecular Biology. 1994;25:917–920. doi: 10.1007/BF00028886. [DOI] [PubMed] [Google Scholar]
- Kannan L, Knudsen J, Jolly CA. Aging and acyl-CoA binding protein alter mitochondrial glycerol-3-phosphate acyltransferase activity. Biochimica et Biophysica Acta. 2003;1631:12–16. doi: 10.1016/s1388-1981(02)00367-0. [DOI] [PubMed] [Google Scholar]
- Kim HU, Huang AH. Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiology. 2004;134:1206–1216. doi: 10.1104/pp.103.035832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HU, Li Y, Huang AH. Ubiquitous and endoplasmic reticulum-located lysophosphatidyl acyltransferase, LPAT2, is essential for female but not male gametophyte development in Arabidopsis. Plant Cell. 2005;17:1073–1089. doi: 10.1105/tpc.104.030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Casteel J, Miernyk JA, Thelen JJ. The effects of down-regulating expression of Arabidopsis thaliana membrane-associated acyl-CoA binding protein 2 on acyl-lipid composition. Plant Science. 2007;172:36–44. [Google Scholar]
- Larson TR, Edgell T, Byrne J, Dehesh K, Graham IA. Acyl CoA profiles of transgenic plants that accumulate medium-chain fatty acids indicate inefficient storage lipid synthesis in developing oilseeds. Plant Journal. 2002;32:519–527. doi: 10.1046/j.1365-313x.2002.01440.x. [DOI] [PubMed] [Google Scholar]
- Larson TR, Graham IA. A novel technique for the sensitive quantification of acyl CoA esters from plant tissues. Plant Journal. 2001;25:115–125. doi: 10.1046/j.1365-313x.2001.00929.x. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Mishra G, Chye ML. ACBP4 and ACBP5, novel Arabidopsis acyl-CoA-binding proteins with kelch motifs that bind oleoyl-CoA. Plant Molecular Biology. 2004;55:297–309. doi: 10.1007/s11103-004-0642-z. [DOI] [PubMed] [Google Scholar]
- Leung KC, Li HY, Xiao S, Tse MH, Chye ML. Arabidopsis ACBP3 is an extracellularly targeted acyl-CoA-binding protein. Planta. 2006;223:871–881. doi: 10.1007/s00425-005-0139-2. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Membrane localization of Arabidopsis acyl-CoA-binding protein ACBP2. Plant Molecular Biology. 2003;51:483–492. doi: 10.1023/a:1022330304402. [DOI] [PubMed] [Google Scholar]
- Li HY, Chye ML. Arabidopsis Acyl-CoA-binding protein ACBP2 interacts with an ethylene-responsive element-binding protein, AtEBP, via its ankyrin repeats. Plant Molecular Biology. 2004;54:233–243. doi: 10.1023/B:PLAN.0000028790.75090.ab. [DOI] [PubMed] [Google Scholar]
- Li HY, Xiao S, Chye ML. Ethylene- and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. Journal of Experimental Botany. 2008;59:3997–4006. doi: 10.1093/jxb/ern241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell. 2004;16:5–20. doi: 10.1105/tpc.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends in Plant Science. 2008;13:483–491. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Rosendal J, Ertbjerg P, Knudsen J. Characterization of ligand binding to acyl-CoA-binding protein. Biochemical Journal. 1993;290:321–326. doi: 10.1042/bj2900321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio S, Larson TR, Gonzalez-Guzman M, Alejandro S, Graham IA, Serrano R, Rodriguez PL. An Arabidopsis mutant impaired in coenzyme A biosynthesis is sugar dependent for seedling establishment. Plant Physiology. 2006;140:830–843. doi: 10.1104/pp.105.072066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rylott EL, Rogers CA, Gilday AD, Edgell T, Larson TR, Graham IA. Arabidopsis mutants in short and medium chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-oxidation is essential for embryo development. Journal of Biological Chemistry. 2003;278:21370–21377. doi: 10.1074/jbc.M300826200. [DOI] [PubMed] [Google Scholar]
- Sano H, Kuroki Y, Honma T, Ogasawara Y, Sohma H, Voelker DR, Akino T. Analysis of chimeric proteins identifies the regions in the carbohydrate recognition domains of rat lung collectins that are essential for interactions with phospholipids, glycolipids, and alveolar type II cells. Journal of Biological Chemistry. 1998;273:4783–4789. doi: 10.1074/jbc.273.8.4783. [DOI] [PubMed] [Google Scholar]
- Sellwood C, Slabas AR, Rawsthorne S. Effects of manipulating expression of acetyl-CoA carboxylase I in Brassica napus L. embryos. Biochemical Society Transactions. 2000;28:598–600. [PubMed] [Google Scholar]
- Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, Frye C, Patton D, Meinke D. The Arabidopsis SeedGenes Project. Nucleic Acids Research. 2003;31:90–93. doi: 10.1093/nar/gkg028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, Meinke D. Identification of genes required for embryo development in Arabidopsis. Plant Physiology. 2004;135:1206–1220. doi: 10.1104/pp.104.045179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou HE, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses. Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. Journal of Biological Chemistry. 2002;277:31994–32002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Winkler RG, Frank MR, Galbraith DW, Feyereisen R, Feldmann KA. Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Isolation of mutations in the cytochrome p450 gene superfamily. Plant Physiology. 1998;118:743–750. doi: 10.1104/pp.118.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Chen QF, Chye ML. Light-regulated Arabidopsis ACBP4 and ACBP5 encode cytosolic acyl-CoA-binding proteins that bind phosphatidylcholine and oleoyl-CoA ester. Plant Physiology and Biochemistry. 2009;47:926–933. doi: 10.1016/j.plaphy.2009.06.007. [DOI] [PubMed] [Google Scholar]
- Xiao S, Chye ML. An Arabidopsis family of six acyl-CoA-binding proteins has three cytosolic members. Plant Physiology and Biochemistry. 2009;47:479–484. doi: 10.1016/j.plaphy.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Xiao S, Li HY, Zhang JP, Chan SW, Chye ML. Arabidopsis acyl-CoA-binding proteins ACBP4 and ACBP5 are subcellularly localized to the cytosol and ACBP4 depletion affects membrane lipid composition. Plant Molecular Biology. 2008a;68:571–583. doi: 10.1007/s11103-008-9392-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Gao W, Chen QF, Ramalingam S, Chye ML. Overexpression of membrane-associated acyl-CoA-binding protein ACBP1 enhances lead tolerance in Arabidopsis. Plant Journal. 2008b;54:141–151. doi: 10.1111/j.1365-313X.2008.03402.x. [DOI] [PubMed] [Google Scholar]
- Yanai Y, Kawasaki T, Shimada H, Wurtele ES, Nikolau BJ, Ichikawa N. Genomic organization of 251 kDa acetyl-CoA carboxylase genes in Arabidopsis: tandem gene duplication has made two differentially expressed isozymes. Plant and Cell Physiology. 1995;36:779–787. doi: 10.1093/oxfordjournals.pcp.a078822. [DOI] [PubMed] [Google Scholar]
- Yeung EC, Meinke DW. Embryogenesis in Angiosperms: development of the suspensor. Plant Cell. 1993;5:1371–1381. doi: 10.1105/tpc.5.10.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurchenko OP, Nykiforuk CL, Moloney MM, Ståhl U, Banaś A, Stymne S, Weselake RJ. A 10-kDa acyl-CoA-binding-protein (ACBP) from Brassica napus enhances acyl exchange between acyl-CoA and phosphatidylcholine. Plant Biotechnology Journal. 2009;7:602–610. doi: 10.1111/j.1467-7652.2009.00427.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phenotypic analysis of acbp1 and acbp2 single mutants.
Callus induction on the embryos of wild-type, acbp1 and acbp2 plants, as well as aborted embryos from acbp1ACBP2+/− plants.