Abstract
Existing methodologies for human induced pluripotent stem cell (hiPSC) cardiac differentiation are efficient but require the use of complex, undefined medium constituents that hinder further elucidation of the molecular mechanisms of cardiomyogenesis. Using hiPSCs derived under chemically defined conditions on synthetic matrices, we systematically developed a highly optimized cardiac differentiation strategy, employing a chemically defined medium consisting of just three components: the basal medium RPMI 1640, L-ascorbic acid 2-phosphate, and rice-derived recombinant human albumin. Along with small molecule-based differentiation induction, this protocol produced contractile sheets of up to 95% TNNT2+ cardiomyocytes at a yield of up to 100 cardiomyocytes for every input pluripotent cell, and was effective in 11 hiPSC lines tested. This is the first fully chemically defined platform for cardiac specification of hiPSCs, and allows the elucidation of cardiomyocyte macromolecular and metabolic requirements whilst providing a minimally complex system for the study of maturation and subtype specification.
Keywords: Human induced pluripotent stem cell, differentiation, cardiomyocyte, heart, chemically defined medium, small molecule
Introduction
Human induced pluripotent stem cells (hiPSCs) are increasingly used in various areas of cardiovascular research, including disease modeling, cardiotoxicity screening, drug discovery, and the study of human cardiac development. A major future aim of the cardiac differentiation field is to provide cells suitable for cellular therapy1. Each of these objectives requires large numbers (107–109) of cells to be made in a scalable, cost-effective, and highly reproducible fashion, ideally under chemically defined conditions in which all constituents are chemically known and free of animal-derived products (e.g., bovine serum albumin (BSA), Matrigel, or B27).
Differentiation techniques have progressed from early inefficient and variable fetal bovine serum (FBS)-based embryoid body methods, to multiple methodologies that have been shown to produce highly pure cardiac troponin T (TNNT2)+ cells with relative ease2–6. Improvements in these methods have largely concentrated on mimicking the embryonic developmental signals that control mesoderm induction: activin/NODAL, BMP, Wnt, and FGF2, 3, 5, 7–10, and subsequent cardiac specification using inhibition of Wnt8, BMP3, and TGFβ3, 11 pathways. Despite this progress, little is known of the pathways and macromolecules required for in vitro cardiac differentiation due to the complexity of proprietary media used, the somewhat autonomous nature of in vitro cardiac differentiation, and the complex secretome involved12.
The most efficient protocols to date rely on the basal medium RPMI 1640 (which is chemically defined13) supplemented with ‘B27’, a complex mix of 21 components (many of animal origin), originally designed for the culture of hippocampal neurons14. It is unknown whether B27 components influence differentiation reproducibility, maturation, or subtype specification. Therefore, we sought to develop a novel, optimized, and low-cost cardiac differentiation protocol (without undefined/proprietary medium components) that would provide highly reproducible differentiation and allow further understanding of the macromolecules required for cardiac differentiation. This protocol was demonstrated to reproducibly and efficiently differentiate 11 hiPSC lines that were generated under chemically defined conditions when tested repeatedly from p20 to p83 representing >600 differentiations. Cardiomyocytes could be produced at >85% purity and enriched to >95% using chemically defined metabolic selection. To our knowledge, this methodology is the ‘first of its kind’ fully chemically defined differentiation system for any pluripotent cell derived lineage.
Results
Development of a defined cardiac differentiation platform
We first generated 11 pluripotent hiPSC lines under chemically defined conditions (Supplementary Fig. 1) on a chemically synthesized vitronectin peptide substrate, with non-enzymatic passaging (Supplementary Fig. 2). Early experiments demonstrated that previous monolayer cardiac differentiation protocols6 could be adapted to function with hiPSCs grown under chemically defined conditions (Supplementary Fig. 3). To formulate a chemically defined differentiation protocol, we concentrated on the observation that three unrelated medium formations are capable of supporting growth factor-based monolayer cardiac differentiation: RPMI+B27-ins15, supplemented StemPro-3416, and LI-APEL9 (Supplementary Table 1). Initial experiments demonstrated that of the three medium formulations examined, RPMI+B27-ins resulted in the most efficient small molecule-based cardiac differentiation. Beginning with the 21 components of B27, we subtracted one component at a time and assessed for continued high efficiency differentiation (Supplementary Table 2a–c). We also assessed components from supplemented StemPro-34 and LI-APEL for potential benefits to differentiation (Supplementary Table 2d–f). We concluded that hiPSC cardiac differentiation was successful in a medium consisting of just three components: RPMI 1640 basal medium, L-ascorbic acid 2-phosphate (AA 2-P), and BSA (Supplementary Table 2g).
Optimization of a chemically defined medium
To confirm the full optimization of this formula and protocol, we fine-tuned component concentrations and timing. We showed that total cell death was seen without AA 2-P (Supplementary Fig. 4a). To make the formula chemically defined and xeno-free, we replaced the BSA with recombinant human albumin (rHA) (Supplementary Fig. 4b–c). Although it was possible to differentiate cells without rHA in an entirely protein-free medium and produce ~65% TNNT2+ cells, the cell yield was drastically reduced. Likewise, varying doses of AA 2-P without rHA did not improve yield (Supplementary Fig. 4d). Polyvinyl alcohol (PVA), which prevents shear stress in a similar manner to rHA, combined with varying doses of AA 2-P, did not increase differentiation yield over that of RPMI 1640 with AA 2-P alone. These data suggest that neither the shear-stress prevention nor the antioxidant properties of rHA necessitates its inclusion in this formula. The most effective basal medium was RPMI 1640 (Supplementary Fig. 4e–f). Efficient differentiation was achieved over a broader range (0.8–1.4 × 104 cells/cm2) of initial seeding densities than those for RPMI+B27-ins (1.2–1.4 × 104 cells/cm2, Supplementary Fig. 4g and 3d), suggesting improved differentiation robustness when using this final medium formulation, which we termed CDM3 (chemically defined medium, 3 components).
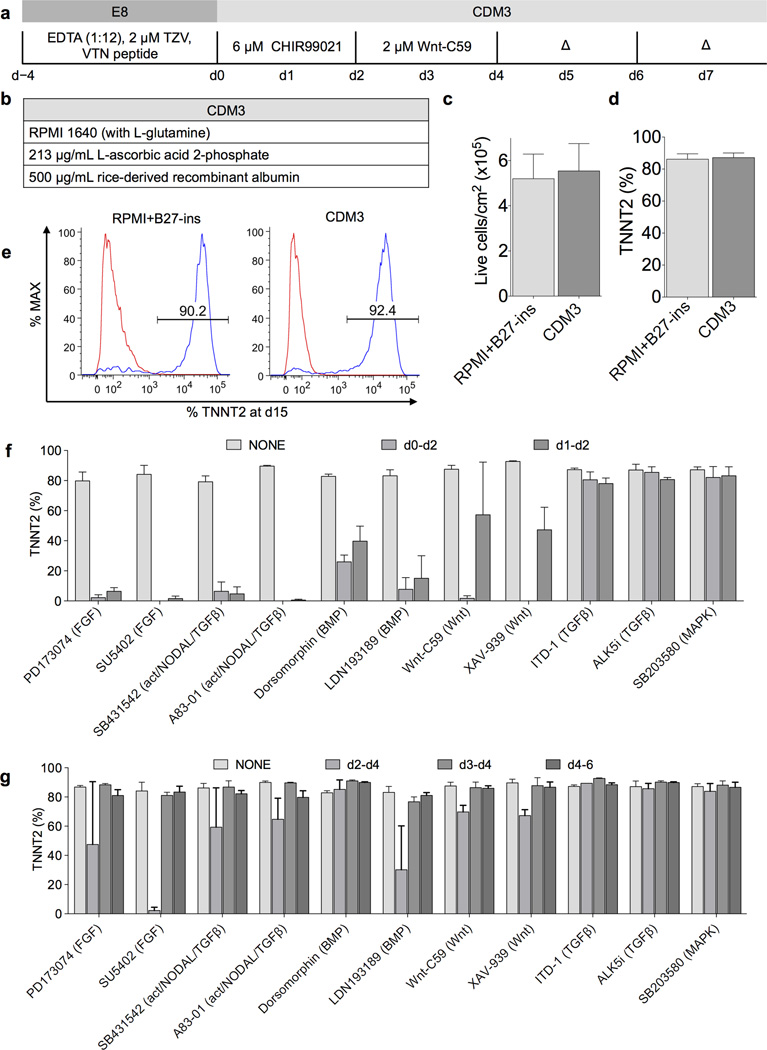
We also assessed a range of small molecules with GSK3B inhibitory activity to identify if any of them had increased mesoderm induction potential in comparison to CHIR99021. Out of six GSK3B inhibitors tested, only BIO and CHIR99021 were successful at inducing any suitable cardiac differentiation, and others were found to be highly toxic (Supplementary Fig. 4h–i). We also assessed alternative small molecule Wnt inhibitors, many of which were found to be similarly effective despite the differences in inhibition mechanism (Supplementary Fig. 4j–k). For example, IWR-1 stabilizes the β-catenin destruction complex, whereas IWP-2 and Wnt-C59 inhibit the palmitoyltransferase PORCN involved in Wnt production. Next we analyzed the importance of the timing of canonical Wnt signaling activation with CHIR99021 and Wnt signaling inhibition with Wnt-C59. We discovered that application of CHIR99021 for 2 days followed by Wnt-C59 for 2 days was optimal (Supplementary Table 3). However, this short biphasic Wnt modulation was not optimal in RPMI+B27-ins (Supplementary Table 3), suggesting that factors modulating the response to CHIR99021 and/or Wnt-C59 may be present in RPMI+B27-ins. The final protocol (Fig. 1a) and formula (Fig. 1b) were demonstrated to produce cardiomyocytes at equivalent yields (Fig. 1c) and efficiencies to those produced using our optimized RPMI+B27-ins protocol (Fig. 1d,e). In CDM3, the yield at day 15 was on average 5.5 × 105 TNNT2+ cells per cm2, but as high as 1.25 × 106, representing a 100-fold increase in cell number over the 1.25 × 104 hiPSCs plated at day −4. Minimal cell death was seen throughout differentiation (Supplementary Fig. 5) and contraction began at days 7–9 (Supplementary Movies 1–3).
Figure 1.
Chemically defined differentiation protocol for efficient cardiac differentiation of hiPSCs. a) Schematic of optimized chemically defined cardiac differentiation protocol. E8, chemically defined pluripotency medium; EDTA, ethylenediaminetetraacetic acid used for clump cell passaging; TZV, thiazovivin, a Rho kinase inhibitor; VTN, vitronectin; CDM3, chemically defined medium 3 components; Δ, medium change. b) Simple three-component formula of CDM3. c) Comparison of total live cell yield from differentiations in RPMI+B27-ins and CDM3, measured by flow cytometry for cardiac troponin T (TNNT2) on day 15 cells (hiPSC line 59FSDNC3 shown), n = 4. d) Comparison of cardiac differentiation efficiency from differentiations in RPMI+B27-ins and CDM3, n = 4. e) Typical TNNT2+ populations in cells produced from differentiations in RPMI+B27-ins medium and CDM3. Left peak represents isotype control. f) Effect of inhibition of signaling pathways during early mesoderm differentiation. Small molecules were added at designated time points (day 0–2 or day 1–2), at 5 µM for all except Wnt-C59, which was used at 2 µM. Normal doses of CHIR99021, Wnt-C59, and medium change timings were maintained, n = 3. g) Effect of inhibition of signaling pathways post mesoderm induction. The same as f) but assessing cardiac induction time points (day 2–4, day 3–4, or day 4–6). Slight reduction of cardiac differentiation efficiency with Wnt-C59 on day 2 to day 4 was likely due to the combined total dose (4 µM) being suboptimal (Supplementary Fig. 4i), n = 3. All error bars represent S.E.M.
Assessment of essential developmental pathways
We previously proposed that Wnt signaling initiates a paracrine feedback loop via FGF, BMP, Wnt, and activin/NODAL signaling, and may be responsible for mesoderm induction in hiPSCs4. To test this hypothesis and to determine if more specific signaling pathway control could improve differentiation robustness, we inhibited the six major pathways associated with in vivo cardiac differentiation: FGF, activin/NODAL, BMP, Wnt, TGFβ, and MAPK17 (Fig. 1f). Inhibitors of each of these pathways (11 total) were added at differentiation day 0–2 or day 1–2. Our results demonstrated that FGF, activin/NODAL, BMP, and Wnt signaling were all essential for mesoderm induction, as inhibition of these pathways hindered efficient cardiac differentiation. By contrast, TGFβ and p38 MAPK were dispensable (Fig. 1f). To determine whether inhibition of these pathways could improve the robustness of subsequent cardiac specification, we added each of the eleven above inhibitors during differentiation days 2–4, 3–4, and 4–6. We found that by days 3–4, none of the six pathways was required, because inhibition did not diminish or enhance differentiation efficiency (Fig. 1g).
Suitability of defined matrices for CDM3 differentiation
We found that when using the synthetic vitronectin peptide18 matrix the highly motile cardiomyocyte monolayers would detach from the surface at ~day 15 (Supplementary Fig. 6), irrespective of vitronectin peptide concentrations, reducing the yield and increasing variability. This lack of long-term adhesion was present could be resolved by passaging and replating onto new matrix, but we sought to find a more suitable solution. We first assessed chemically defined pluripotent culture on other defined matrices: recombinant human (rH) E-cadherin19, rH vitronectin20, rH laminin-52121, truncated rH laminin-51122, 23, human fibronectin, and a fibronectin mimetic (Supplementary Fig. 7). Laminin-based matrices demonstrated higher growth rates than the vitronectin peptide when surface densities were correctly optimized (Supplementary Fig. 8), potentially due to the established role of laminin-511 and −521 interacting with α6β1 integrin and activating the PI3K Akt pathway24. Fibronectin-based matrices did not support pluripotent growth. All five suitable matrices supported efficient differentiation (Supplementary Fig. 6, Supplementary Movies 4–10) and no differences in differentiation efficacy were seen between cells cultured long-term (>6 passages) on defined matrices and cells transferred to the specific matrix just prior to CDM3 differentiation (Supplementary Fig. 9). Only the laminin-based matrices were successful for maintaining long-term adhesion (>15 days) during chemically defined cardiac differentiation (Supplementary Fig. 6d), but these matrices were prohibitively expensive for large-scale application therefore all further characterization was performed on vitronectin peptide.
Efficacy in multiple hiPSC lines and passage numbers
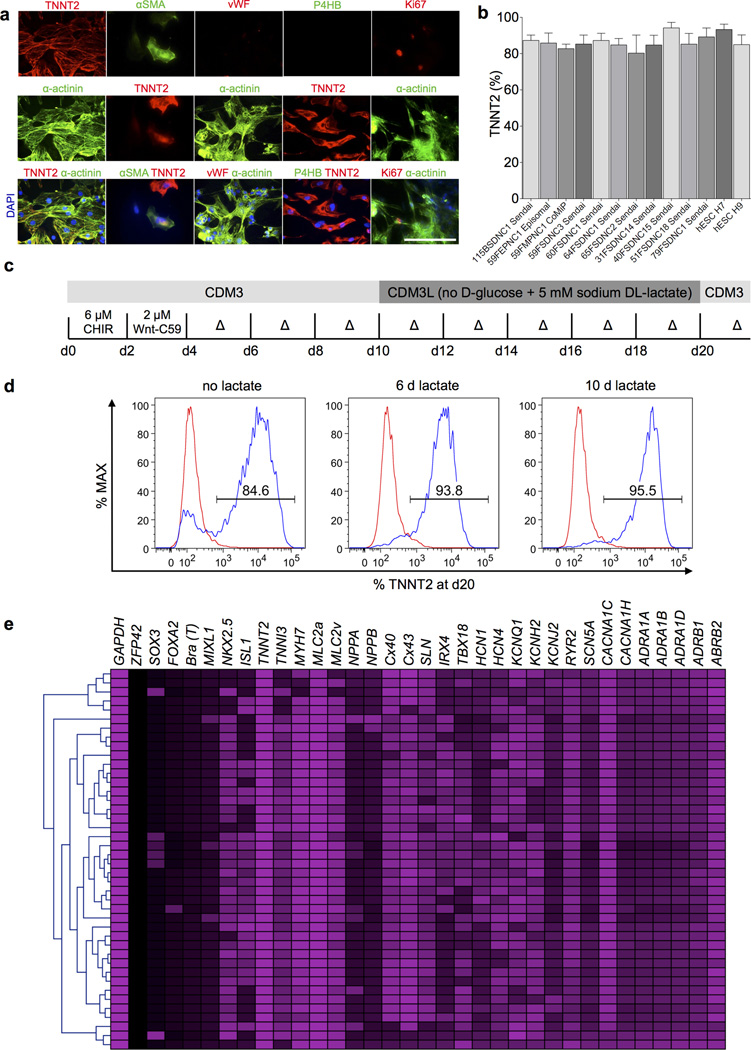
Cardiomyocytes produced by CDM3 differentiation were positive for cardiac markers (TNNT2 and α-actinin) and for the immature cardiomyocyte marker alpha smooth muscle actin (αSMA), and were negative for the endothelial marker Von Willebrand factor (vWF) and fibroblast marker P4HB. A small proportion of cells were positive for the proliferation marker Ki67 (Fig. 2a). One criticism of several established cardiac differentiation protocols is the lack of efficacy across multiple hESC and hiPSC lines. To address this issue, we tested our protocol with a variety of hESC and hiPSC lines (described in Online Methods) repeatedly differentiated from passage <25 to >80; all demonstrated 80–95% differentiation efficiency, as assessed by flow cytometry for TNNT2 (Fig. 2b), and produced yields of greater than 3 × 105 cardiomyocytes per cm2. Cardiomyocytes could be generated and maintained in these chemically defined conditions for >200 days, suggesting minimal macromolecular requirements for long-term cardiomyocyte maintenance.
Figure 2.
Characterization and purification of cardiomyocytes produced by chemically defined differentiation. a) Immunofluorescence staining for TNNT2 and α-actinin (cardiomyocyte structural markers), α-smooth muscle actin (primitive cardiomyocytes), vWF (endothelial cells), P4HB (fibroblasts), and Ki67 (proliferating cells). Scale bar, 12.5 µm. b) Efficiency of chemically defined cardiac differentiation measured by flow cytometry for TNNT2+ on day 15 cells in a range of peripheral blood or fibroblast-derived hiPSC generated with Sendai virus, ‘Yamanaka’ episomal plasmids, or a mini intronic plasmid (CoMiP). All hiPSC and hESC lines were cultured under chemically defined conditions and differentiated in CDM3, n ≥ 3. Error bars represent S.E.M. c) Schematic of chemically defined cardiac differentiation including purification by metabolic selection. CDM3L: CDM3 without D-glucose and supplemented with 5 mM sodium DL-lactate. d) Effect of chemically defined metabolic purification measured by flow cytometry for TNNT2 on day 20 cells. Showing cells with no lactate treatment, 6 days of lactate treatment (day 10–16), and 10 days lactate treatment (day 10–20). e) Assessment of gene expression heterogeneity using single cell real time RT-PCR of day 20 cardiomyocytes differentiated in CDM3 without metabolic purification.
Purification of cardiomyocytes using metabolic selection
Culturing hiPSC-derived cardiomyocytes in glucose-free MEMα supplemented with FBS and lactate has been shown to enrich cells from 4.5% to 98.0% α-actinin positive25. Because the B27 supplement contains a source of glucose (D(+)-galactose), medium formulas with this supplement are not suitable for this methodology. By replacing the RPMI 1640 in our CDM3 formula with RPMI 1640 without glucose and supplementing with sodium DL-lactate (Fig. 2c), we found that 6–10 days of glucose deprivation (by days 10–16 or days 10–20) was able to purify cardiomyocytes from ~85% TNNT2+ to >95% (Fig. 2d). This simple methodology can be used to provide highly pure hiPSC-derived cardiomyocytes with minimal experimental complexity, although the effect of metabolic manipulation of cardiomyocytes has not yet been established.
Characterization of cardiomyocyte subtypes produced
To characterize cardiomyocytes produced under chemically defined conditions, samples were taken every day during differentiation and assessed by real-time RT-PCR (Supplementary Fig. 10). Gene expression patterns were consistent with those from previous differentiation strategies2, 26 and with embryonic development. The pluripotency marker POU5F1 was rapidly downregulated at day 2 with concurrent upregulation of the mesoderm markers T and MIXL1, followed by the cardiac mesoderm marker MESP1. Early cardiomyocyte markers (KDR, ISL1, and GATA4) became highly expressed from days 5–6, with later markers (NKX2-5, TBX5, and MEF2C) peaking at days 8–9. Finally, the expression of the cardiomyocyte myofilament genes TNNT2 and MYH6 peaked at days 8–10, and as previously noted2, 8, the expression subsequently decreased, likely due to the comparatively slow turnover (~3–5 days27) of these proteins once the myofilaments were established. Single cell real-time RT-PCR on day 20 cardiomyocytes (not metabolically selected) showed substantial homogeneity among cells (Fig. 2e), although more variation was noted in the expression of the ion channels HCN1, HCN4, KCNQ1, and KCNH2. Many cells co-expressed markers of human atrial (NPPA [ANP], CX40, and SLN), ventricular (MLC2V and IRX4)28, and nodal cells (TBX18), suggesting that these day 20 cells are comprised of, or still contain, unspecified cardiomyocytes without a fully defined subtype.
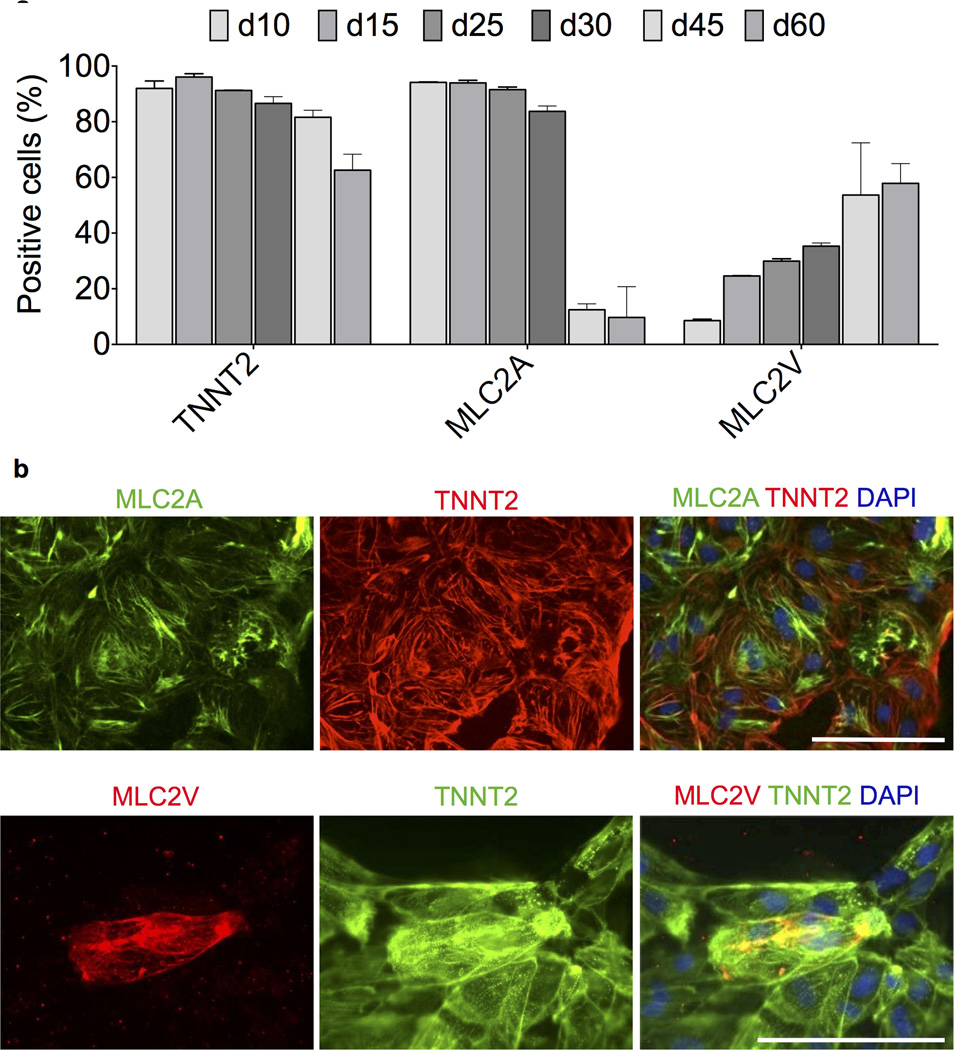
To assess the cardiomyocyte subtypes of cells derived under chemically defined conditions, we next used flow cytometry to examine cells on day 10, 15, 25, 30, 45 and 60 of differentiation (Fig. 3a and Supplementary Figs. 11 and 12). The expression of the two major isoforms of MLC2 (MLC2A and MLC2V) has been used previously to describe the atrial vs. ventricular specification of hPSC-derived cardiomyocytes5, 6, 29. In humans, MLC2A expression is detected in both the atria and ventricles during development30 and in adult cardiomyocytes31. By contrast, MLC2V expression is restricted to the ventricles throughout development and persists into adulthood30. Therefore, for assessment of subtype specification, these markers were be used comparatively. At day 10 of differentiation, cells demonstrated a TNNT2+MLC2A+MLC2V− phenotype, likely representing unspecified cardiomyocyte precursors. The presence of cells that stained positive for MLC2A progressively decreased during the time course, with a dramatic drop to ~10% after day 30, whereas the MLC2V positivity consistently increased during the time course, reaching 60% at day 60 (Fig. 3a). Antibody specificity was confirmed by immunofluorescence staining (Fig. 3b). Similar flow cytometry results were seen in cells differentiated in RPMI+B27-ins (Supplementary Fig. 11b), suggesting a minimal effect of medium on populations of cells that stain positive for MLC2A and MLC2V.
Figure 3.
Characterization of atrial vs. ventricular profile of cardiomyocytes produced under chemically defined conditions. a) Flow cytometry assessment of expression of cardiac troponin T (TNNT2), atrial myosin light chain 2 (MLC2A), and ventricular myosin light chain (MLC2V) in cardiomyocytes derived and maintained in CDM3 at differentiation from day 10 through day 60, n = 3. Error bars represent S.E.M. Additional data is provided in Supplementary Figs. 11 and 12. b) Immunofluorescence staining of day 20 cardiomyocytes with the same antibodies used for flow cytometry to demonstrate specificity. Scale bar, 12.5 µm.
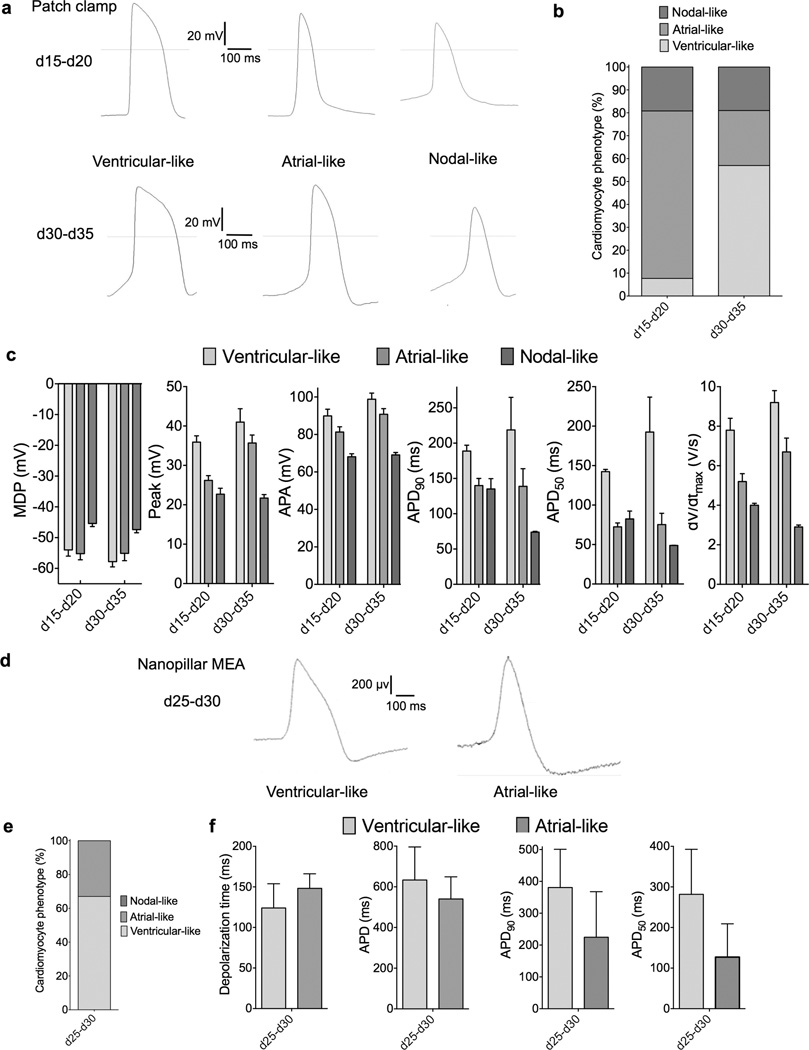
Electrophysiological analysis was performed using two independent techniques. Patch clamp demonstrated that at days 15–20, cells had a somewhat atrial-like action potential with a low average maximum diastolic potential (MDP) of −55.2±2.0 mV (Fig. 4a–b). These cells likely represented unspecified cardiomyocyte precursors in the heart tube during development and not atrial specification. At days 30–35, cardiomyocytes demonstrated a more heterogeneous phenotype, with ventricular-like cells being the predominant phenotype (57%, with an MDP of −57.8 mV) along with atrial-like and nodal-like cells (Fig. 4c–d). A second electrophysiological technique, nanopillar electroporation32 confirmed that day 30 cells possessed a predominantly ventricular-like phenotype (Fig. 4g–i). Taken together, these electrophysiological analysis data were consistent with our flow cytometry (Fig. 3a), immunofluorescence (Fig. 3b), and single cell real-time RT-PCR (Fig. 2e) findings, demonstrating that cells progressed from an unspecified cardiomyocyte precursor phenotype to an immature, predominantly ventricular phenotype.
Figure 4.
Electrophysiological characterization of cardiomyocytes produced under chemically defined conditions. a) Representative action potential (AP) recordings using whole cell patch of three cardiomyocyte subtypes produced from day 15–20 cells and day 30–35 cells. Cells exhibit AP morphologies that can be categorized as atrial-, nodal-, or ventricular-like. b) Proportions of cardiomyocyte subtypes at day 15–20, n = 21 and day 30–35, n = 13. c) Patch clamp recordings of differentiation day 15–20 and day 30–35 cells, demonstrating MDP, maximum diastolic potential; peak voltage; APA, action potential amplitude; AP duration at different levels of repolarization (i.e., 90 or 50%); and dV/dtmax (maximal rate of depolarization). d) Assessment of cells differentiated in CDM3 at day 30 of differentiation using MEA-based nanopillar, representative trace of ventricular-like and atrial-like subtypes. e) Percentages of ventricular-like and atrial-like cells, n = 20. f) Depolarization time; APD, action potential duration; AP duration at different levels of repolarization (i.e., 50 or 90%) and AP morphology used to classify cardiomyocyte subtype. Error bars represent S.E.M.
Discussion
A key surprising finding of our studies is how few medium components are required to complete the progression from pluripotent cell to cardiomyocyte. Of the 21 components in B27, only albumin was required and the simple CDM3 medium consisting of 3 components (recombinant albumin, ascorbic acid, and the basal medium RPMI 1640) supported cardiac differentiation at the same efficiency as RPMI+B27-ins.
Albumin can fulfill multiple roles in medium formulations, including acting as a detoxifier/buffer by binding lipids and excessive proteins, binding hormones and growth peptides to keep them stable, and binding free radicals to reduce oxidative damage to cells33. In this protocol, we used a rice-derived recombinant albumin and can therefore rule out the presence of any contaminating mammalian albumin–associated proteins/lipids/small molecules. Intriguingly, the removal of recombinant albumin from CDM3 did not result in cell death (as with removal of AA 2-P), but merely in a reduction in cardiomyocyte yield. Ascorbic acid is an antioxidant with a well-established role in improving cardiac differentiation2, 34, potentially via increasing collagen synthesis that results in greater proliferation of cardiovascular progenitor cells35. The utility of ascorbic acid in E8 pluripotent medium and during reprogramming raises the question of whether ascorbic acid is merely acting as an antioxidant in the cardiac differentiation system. Recently, ascorbic acid has been shown to produce widespread but targeted Tet-dependent DNA demethylation36, that may have a role during differentiation. RPMI 1640 is one of the simplest classical basal media, related to McCoy’s 5A and consisting of 20 amino acids, inorganic salts, the eight B vitamins, and the antioxidant glutathione. RPMI 1640 does not contain any lipids, iron or zinc, and the exact reason why this basal medium is superior in this protocol awaits further extensive experimentation to elucidate.
In the optimization of this system, we found very tight specification for pluripotent growth was required for successful subsequent differentiation. From repeatedly differentiating 11 hiPSC lines over a range of >60 passages, it became clear that the pluripotent state governs the differentiation efficiency. The matrix to which the pluripotent cells are attached plays an integral role in differentiation. In our study, recombinant laminin-521 and truncated recombinant laminin-511 were found to be most successful for both the support of pluripotent hiPSC growth and long-term adherence of CMs as seen with human adult cardiomyocytes31. It has been demonstrated that hESC-derived cardiomyocytes express integrin α3, α5, α6, α7, αV, β1, and β537, very similar to the predominant expression of α5, α6, αV, β1, and β5 in pluripotent hESCs20. hESCs adhere to laminin-511, −521, and Matrigel using the α6β1 integrin22, likely explaining the similar long-term adherence performance. By contrast, pluripotent cells adhere to vitronectin using αVβ5 integrin18, 20, which has a lower binding affinity for its preferred substrates than does α6β123, and along with potential lower expression of αVβ5 on hiPSC-CM, could explain the differences in long-term cardiomyocyte adherence.
In summary, this chemically defined differentiation methodology provides a much needed reproducibility, scalability, and control of hiPSC-derived cardiomyocyte generation required in disease modeling, drug discovery, and regenerative medicine applications. The question of how to produce populations of mature atrial, ventricular, and nodal cells remains, but the chemically simple nature of CDM3, without factors that could skew subtype specification38–40, provides an excellent inert platform for further study of this subject. Finally, as the CDM3 medium is highly defined, it should also help facilitate the transfer of basic research on human pluripotent stem cells to the clinic by providing significant insights into the macromolecular requirements of cardiovascular progenitor cells and hiPSC-derived cardiomyocytes.
Online Methods
All pluripotent and reprogramming cultures were maintained at 37 °C in a New Brunswick Galaxy 170R humidified incubator (Eppendorf), with 5% CO2 and 5% O2 controlled by the injection of carbon dioxide and nitrogen. Primary cell and differentiation cultures were maintained at 5% CO2 and atmospheric (21%) O2.
Human induced pluripotent cell derivation
Protocols were approved by the Stanford University Human Subjects Research Institutional Review Board. With informed written consent, two 2 mm skin punch biopsies were taken from each volunteer, diced with a scalpel, digested with 1 mg/mL collagenase IV (Life Technologies) for 2 h at 37 °C. Fibroblasts were then grown in DMEM with GlutaMAX (Life Technologies) supplemented with 10% fetal bovine serum (FBS, US origin, Life Technologies) on 6-well plates (Greiner) coated with a 1:200 dilution of growth-factor reduced Matrigel (9 µg/cm2, Corning). Medium was changed every other day. When confluent, fibroblasts were passaged with TrypLE Express (Life Technologies) onto Matrigel-coated T225 flasks (Nunc).
For Sendai reprogramming (Supplemental Fig. 1a), early passage (p2-p3) fibroblasts were seeded at 40,000 cells per well on Synthemax II-SC18 (625 ng/cm2, Corning)-coated 6-well plates. After 24 h, medium was changed to E841. The E8 formula was modified to replace human-derived transferrin with an Oryza sativa-derived recombinant version, to make the formula completely chemically defined. The successful application of Synthemax II-SC at the low concentration of 625 ng/cm2 is in line with reports42 that the minimal surface density of vitronectin protein is very low at 250 ng/cm2. E8 medium consisting of DMEM/F12 (10-092-CM, Corning), 20 µg/mL E. coli-derived recombinant human insulin (Dance Pharmaceuticals/CS Bio), 64 µg/mL L-ascorbic acid 2-phosphate sesquimagnesium salt hydrate (Sigma-Aldrich), 10.7 µg/mL Oryza sativa-derived recombinant human transferrin (Optiferrin, Invitria/Sigma-Aldrich), 14 ng/mL sodium selenite (Sigma-Aldrich), 100 ng/mL recombinant human FGF2 (154 amino acid, E. coli-derived, Peprotech), and 2 ng/mL recombinant human TGFβ1 (112 amino acid, HEK293-derived, Peprotech). To the medium was added four OSKM CytoTune-iPS Sendai Reprogramming Kit viral particle factors (Life Technologies)43 diluted ~1/5 based on manufacturer’s recommendations (3 × 105 cell infectious units (CIU) of each particle per well, multiplicity of infection (MOI) = 7.5). Medium was changed after 24 h and, thereafter once every day. For the first 7 days, cultures were maintained in E8 supplemented with 100 nM hydrocortisone (Sigma-Aldrich) and 200 µM sodium butyrate (Sigma-Aldrich)44. At day 7 medium was swapped to E7N (E8 minus TGFβ1; supplemented with 200 µM sodium butyrate). Medium switched to E8 at day 20.
For plasmid-based reprogramming45, pCXLE-hSK (27078), pCXLE-hUL (27080), and pCXLE-hOCT4-shp53 (27077) plasmids were obtained from Addgene. The OSKM codon optimized mini-intronic plasmid (CoMiP) was generated by S. Diecke. Plasmid-containing E. coli were grown in Miller’s LB (Life Technologies), and purified using Plasmid Maxi Kit (QIAGEN) following manufacturer’s instructions, and quantified using a NanoDrop 2000 (Thermo Scientific). 1 × 106 cells were electroporated with 6 µg total DNA (2 µg of each for 3 plasmid-based systems, 6 µg for CoMiP) using a Neon Transfection System (Life Technologies), with the settings: 1650 V, 3 pulses, 10 ms, and 100 µL tips, and Buffers R and E2. Cells were plated on Synthemax II-SC-coated 6-well plates.
For peripheral blood mononuclear cell (PBMC) reprogramming, 20 mL of blood was collected in EDTA-containing Vacutainer tubes (BD Biosciences). PBMCs were isolated using a Ficoll-Paque PLUS (1.077 g/mL) gradient (GE Healthcare) and plated at 1 million cells per mL in 2 mL of a humanized version of blood medium46 comprised of 50:50 IMDM:F12 (both Life Technologies), 2 mg/mL recombinant human albumin, 1% v/v chemically defined lipid concentrate (Life Technologies), 10 µg/mL recombinant human insulin, 100 µg/mL recombinant human transferrin, 15 ng/mL sodium selenite, 64 µg/mL L-ascorbic acid 2-phosphate, 450 µM 1-thioglycerol (Sigma-Aldrich), 50 ng/mL SCF (Peprotech), 10 ng/mL IL3 (Peprotech), 2 U/mL EPO (EMD Millipore), 40 ng/mL IGF1 (Peprotech), and 1 µM dexamethasone (Sigma-Aldrich). Cells were cultured for 9 days with 50% medium changes every other day. After 9 days, 1 × 106 were plated in blood medium with Sendai virus, as above. Medium was changed every other day and were transferred to E7N in a Synthemax II-SC-coated 6-well plate at d3.
For all reprogramming methods, individual colonies with hESC morphology were picked into 1 well of a 12-well plate (1 colony per well) at d17-d25 in E8 with 2 µM thiazovivin for 24 h after picking. Subsequently, cells were expanded into 6-well plates by passaging 1:1, 1:4, 1:6, 1:8, and finally 1:12, using 0.5 mM EDTA (Life Technologies) in D-PBS without CaCl2 or MgCl2 (Life Technologies) for 7 min at RT.
Human pluripotent stem cell culture
Cells were routinely maintained in E8 (made as above) on either Synthemax II-SC (625 ng/cm2) or 1:200 growth factor-reduced Matrigel (9 µg/cm2) and passaged every four days using 0.5 mM EDTA (as above). 2 µM thiazovivin (Selleck Chemicals) was added for the first 24 h after passage. Control hESC lines H7 (WA07) and H9 (WA09)47 were supplied by WiCell Research Institute. All hESC and hiPSC lines were converted to E8/EDTA-based culture for at least 5 passages before beginning differentiation. For growth comparison experiment cells were grown in mTeSR1 (Stemcell Technologies). Cell lines were used between passages 20 and 83. All cultures (primary, pluripotent and differentiation) were maintained with 2 mL medium per 9.6 cm2 of surface area or equivalent. All pluripotent cultures were routinely tested for mycoplasma using a MycoAlert Kit (Lonza).
Growth assays
To assess cell growth, cells were counted using a Countess automated cell counter (Life Technologies). For growth rate calculation, cells were seeded at 1.25 × 104 cells per cm2 (120,000 cells per well of a 6-well plate) and grown for 96 h. Cumulative population doublings was calculated using the formula n = 3.32 [log10(N/N0)].
Teratoma analysis
Three confluent wells of pluripotent cells were dissociated with EDTA, centrifuged, resuspended in 100 µL of growth factor-reduced Matrigel, and injected into the kidney capsule of female NOD-SCID mice (NOD.CB17-Prkdcscid/NcrCrl Strain code 394, Charles River). After 4–6 weeks, teratomas were removed, fixed in 4% PFA, embedded in paraffin wax, sectioned and hematoxylin and eosin (H&E) stained by the Stanford Tissue Bank. Slides were imaged and analyzed by a qualified clinical pathologist.
Single nucleotide polymorphism karyotyping
A single well of >p20 pluripotent cells was dissociated with 0.5 mM EDTA, pelleted, and snap frozen in liquid nitrogen. Genomic DNA was extracted using a Blood and Tissue DNA extraction kit (QIAGEN), following the manufacturer’s directions. SNP karyotyping was performed using a Genome-Wide CytoScan HD Array (Affymetrix) covering 2.7 million markers and 750,000 SNPs, and was analyzed using Chromosome Analysis Suite (ChAS, Affymetrix).
Cardiac differentiation of hiPSCs
Both hiPSCs and hESCs (>p20) were split at 1:10 or 1:12 ratios, using EDTA as above and grown for 4 days, at which time they reached ~85% confluence. Medium was changed to CDM3, consisting of RPMI 1640 (11875, Life Technologies), 500 µg/mL Oryza sativa-derived recombinant human albumin (A0237, Sigma-Aldrich, 75 mg/mL stock solution in WFI H2O, stored at −20 °C), and 213 µg/mL L-ascorbic acid 2-phosphate (Sigma-Aldrich, 64 mg/mL stock solution in WFI H2O, stored at −20 °C). Medium was changed every other day (48 h). For d0-d2, medium was supplemented with 6 µM CHIR99021 (LC Laboratories). On d2, medium was changed to CDM3 supplemented with 2 µM Wnt-C59 (Selleck Chemicals). Medium was changed on d4 and every other day for CDM3. Contracting cells were noted from d7.
Other additions to cardiac differentiation media tested were 10.7 µg/mL recombinant human transferrin, 14 µg/mL sodium selenite, 1 µg/mL linoleic acid, 1 µg/mL linolenic acid, 2 ng/mL triiodo-l-thyronine, 2 µg/mL L-carnitine, 1 µg/mL D,L-alpha-tocopherol acetate, 100 ng/mL retinol acetate, 1 µg/mL ethanolamine, 20 ng/mL corticosterone, 9 ng/mL progesterone, 47 ng/mL lipoic acid, 100 ng/mL retinol, 1 µg/mL D,L-alpha-tocopherol, 100 ng/mL biotin, 2.5 ug/mL catalase, 2.5 µg/mL glutathione, 2.5 µg/mL superoxide dismutase, 2 µg/mL L-carnitine, 15 µg/mL D(+)-galactose, 16.1 µg/mL putrescine, 450 µM 1-thioglycerol, 55 µM 2-mercaptoethanol, and 64 µg/mL L-ascorbic acid 2-phosphate (all from Sigma-Aldrich).
Basal media assessed were DMEM (catalogue #11965), DMEM/F12 (11330), IMDM (12440), IMDM/F12 (12440/11765), RPMI 1640 (11875), McCoy’s 5A (16600), M199 (with Earle’s Salts, 11150), MEMα (with Earle’s Salts, no nucleosides, 12561), and MEM (with Earle’s Salts, 11095) (all from Life Technologies). RPMI 1640 media assessed were RPMI 1640 with L-glutamine (catalogue number 11875), RPMI 1640 with L-glutamine and HEPES (22400), RPMI 1640 with GlutaMAX (61870), and RPMI with GlutaMAX and HEPES (72400) (all from Life Technologies).
Albumin sources assessed were human serum albumin (A1653, Sigma-Aldrich), Oryza sativa-derived recombinant human albumin (A0237, Sigma-Aldrich), Saccharomyces cerevisiae-derived recombinant Albucult (Novozymes Biopahrma/A6608, Sigma Aldrich), Oryza sativa-derived recombinant Cellastim (Invitria/A9731, Sigma Aldrich), and embryo-grade bovine serum albumin (A3311, Sigma Aldrich).
Wnt inhibitors assessed were IWP-2, IWR-1 (both Sigma-Aldrich), XAV-939, ICG-001 (Selleck Chemicals), IWP-4 (Stemgent), and Wnt-C59 (Selleck Chemicals). GSK3B inhibitors assessed were CHIR99021 (LC Laboratories), BIO, TWS119 (Selleck Chemicals), 1-azenkenpaullone, TDZD-8, ARA014418, and 3F8 (all Sigma-Aldrich). Inhibitors used for pathway analysis were PD173074, SB203580, LDN193189, SB431542 (all Selleck Chemicals), SU5402, Dorsomorphin, A83-01 (all Tocris), ALK5 inhibitor (Stemgent), and ITD-1 (Xcessbio). All small molecules were resuspended to 10 mM in dimethyl sulfoxide (DMSO) and used at 5 µM except for Wnt-C59 which was used at 2 µM.
For control treatments (0 µM) 0.1% DMSO was used. Comparisons of differentiation media were made to RPMI+B27-ins consisting of RPMI 1640 (11875) supplemented with 2% B27 without insulin (0050129SA, Life Technologies), and StemPro-34 (Life Technologies) supplemented as shown in Supplementary Table 1. LI-APEL low insulin medium and Xeno-free Differentiation Medium were made as described2, 9, 48. For optimization of cardiac differentiation conditions, cells were differentiated in 12-well plates and samples were analyzed at day 15 of differentiation after dissociation with TrypLE Express for 10 min at 37 °C.
Matrices
The following matrices were assessed for the ability to support pluripotent growth and subsequent cardiac differentiation: 9 µg/cm2 growth-factor reduced Matrigel (1:200, Corning) in DMEM/F12; 625 ng/cm2 vitronectin peptide (Synthemax II-SC, 1:320, Corning) in ultrapure water (1:50 also tested); 1 µg/cm2 full length recombinant human vitronectin (1:50, Primorigen) in D-PBS with CaCl2 and MgCl2; 2.5 µg/cm2 laminin-521 (1:80, Biolamina) in DPBS; 2 µg/cm2 truncated recombinant human laminin-511 iMatrix-511 (1:50, Iwai North America, Foster City, CA, USA) in DPBS; 1 µg/cm2 rH E-cadherin (1:25, StemAdhere, Primorigen/Stemcell Technologies); and 10 µg/cm2 fibronectin (1:20, EMD Millipore) in D-PBS. All were used at 2 mL per well of a 6-well (9.6 cm2). Matrices were assessed on both 6-well polystyrene tissue culture plates and untreated plates (both from Greiner). Also tested were Synthemax-T 6-well plates, and fibronectin mimetic plates (both from Corning) and 10 µg/cm2 Pronectin (Sigma-Aldrich).
In vitro purification of cardiomyocytes
To purify cardiomyocytes, a variant of RPMI 1640 without D-glucose (11879, Life Technologies) was used in the medium formula. RPMI without D-glucose was supplemented with 213 µg/mL L-ascorbic acid 2-phosphate (Sigma-Aldrich), 500 µg/mL Oryza sativa-derived recombinant human albumin (Sigma-Aldrich), and 5 mM sodium DL-lactate (L4263, Sigma-Aldrich) and used in place of CDM3 on differentiation days 10–20.
Cryopreservation of purified cardiomyocytes
Differentiation day 15 cardiomyocytes were treated with TrypLE for 10 min at 37 °C and triturated with a P1000 tip. Cells were filtered through a 100 µM cell strainer (Corning), counted using a Countess Cell Counter (Life Technologies) and cryopreserved at 2–50 million cells per vial in CDM3 + 10% DMSO using a BioCision CoolCell.
Immunofluorescence staining
hiPSCs or cardiomyocytes were plated onto Synthemax II-SC-coated Lab-Tek II 8-chamber glass slides (154524, Nunc) and were allowed to grow for 3 days. Cells were fixed with 4% PFA (Electron Microscopy Services) for 15 min at RT, permeabilized with 0.1% Triton-X (Sigma-Aldrich) for 10 min at RT, blocked in 10% goat serum (Sigma-Aldrich) and 0.1% Triton-X for 15 min at RT and stained with 1:200 mouse IgM TRA-1–60 (09–0010), 1:200 mouse IgMk TRA-1–80 (09–0012), 1:200 mouse IgG3 SSEA4 (09-0006, all Stemgent), 1:200 rabbit IgG POU5F1 (sc-9081), 1:200 rabbit IgG NANOG (sc-33759, both Santa Cruz Biotechnology), 1:200 rabbit IgG SOX2 (651901, Biolegend), 1:200 mouse monoclonal IgG1 TNNT2 (13-11, Thermo Scientific), 1:200 rabbit monoclonal IgG α-actinin (H-300, Santa Cruz), 1:400 mouse monoclonal IgG2b MLC2A (MYL7, Synaptic Systems), 1:200 rabbit polyclonal IgG MLC2V (MYL2, ProteinTech), 1:50 mouse monoclonal IgG1 P4HB (Abcam), 1:400 mouse monoclonal IgG2a SMA (Sigma-Aldrich), 1:400 rabbit polyclonal IgG vWF (Abcam), or 1:500 rabbit polyclonal IgG Ki67 (Thermo Scientific) overnight at 4 °C in 1% BSA in D-PBS. Cells were washed 4 times, for 10 min, with 1% BSA in D-PBS-T and then incubated for 1 hour at RT in the dark with secondary antibodies 1:1000 Alexa Fluor 488 goat anti-mouse IgM (µ chain), 1:1000 Alexa Fluor 488 goat anti-mouse IgG (H+L), or 1:1000 Alexa Fluor 594 goat anti-rabbit IgG (H+L) (all Life Technologies) in 1% BSA in D-PBS. Cells were washed again as above, nuclei were stained with NucBlue Fixed Cell Stain (Life Technologies), and coverslips were attached to Superfrost Plus (Thermo Scientific) slides with Vectashield (Vectorlabs) and imaged with an LSM510Meta Confocal Microscope (Zeiss).
Flow cytometry
Cells were transferred to flow cytometry tubes (BD Biosciences) and fixed with 1% PFA for 15 min, permeabilized with 90% methanol for 15 min, and stained using 1:200 rabbit polyclonal IgG POU5F1 (H-134, sc-9081, Santa Cruz Biotechnology), 1:20 mouse monoclonal IgG3 SSEA4-APC (clone MC-813-70, FAB1435A, R&D Systems), 1:200 NANOG rabbit polyclonal IgG (H-155, sc-33759, Santa Cruz Biotechnology), 1:20 mouse monoclonal IgM TRA-1–60-FITC (FAB4770P, R&D Systems), 1:200 mouse monoclonal IgG1 TNNT2 (cardiac troponin T, 13-11, MS-295-P, Thermo Scientific), 1:200 mouse monoclonal IgG1 TNNT2 (cardiac troponin T, clone 1C11, ab8295, Abcam), 1:200 mouse monoclonal IgG2b TNNI (cardiac and skeletal troponin I, clone 2Q1100, T8665-16M, US Biological), 1:400 mouse monoclonal IgG2b MLC2A (MYL7, Synaptic Systems), and 1:200 rabbit polyclonal IgG MLC2V (MYL2, ProteinTech) for 45 min at RT. Isotype controls used were mouse IgG1 (557273, BD Biosciences), mouse IgG2b (16–4724-82, eBiosciences), and rabbit IgG (02–6102, Life Technologies). Secondary staining was performed with 1:1000 Alexa Fluor 488 goat anti-mouse IgG1, 1:1000 Alexa Fluor 488 goat anti-rabbit IgG (H+L), 1:1000 Alexa Fluor 647 goat anti-mouse IgG2b, and 1:1000 Alexa Fluor 647 goat anti-rabbit IgG (H+L) secondary antibodies (all Life Technologies) for 20 min. These conditions were shown to stain negative on human skin fibroblasts. Cells were analyzed using a FACSAria II (BD Biosciences) with a 100 µM nozzle and FACSDiva software. Data were analyzed using FlowJo 8.7 (TreeStar).
Quantitative real-time polymerase chain reaction
To analyze gene expression, cells were dissociated with TryPLE Express, and pellets of cells were snap frozen in liquid nitrogen and stored at −80 °C. RNA was isolated using an RNeasy Plus kit (QIAGEN), cDNA was produced using a High Capacity RNA-to-cDNA kit (Life Technologies), and Real-time PCR was performed using TaqMan Gene Expression Assays (Supplementary Table 5), TaqMan Gene Expression Master Mix, and a 7900HT Real-Time PCR System (all Life Technologies). All PCR reactions were performed in triplicate, normalized to the 18S endogenous control gene, and assessed using the comparative ΔCt method.
Single cell quantitative real-time polymerase chain reaction
To analyze single cell gene expression, cells were dissociated as above, and a 300,000 cells/mL suspension was prepared. The cell suspension was loaded onto a 17–25 µm C1 Fluidigm chip for single-cell capture and treated with a LIVE/DEAD Viability/Cytotoxicity Kit (Life Technologies). The C1 chip was then imaged under phase-contrast and fluorescence microscopes to exclude doublets and dead cells from subsequent PCR analysis. Single-cell lysis, reverse transcription, and pre-amplification were all performed on the C1 chip using the C1 Single Cell Auto Prep (Fluidigm) and Single Cell-to-CT kits (Ambion). Reverse transcription was performed at 25 °C for 10 min, 42 °C for 1 hour, and finally 85 °C for 5 min. Pre-amplification was at 95 °C for 10 min, followed by 18 cycles of 95 °C for 15 sec, and 60 °C for 4 min. Amplified cDNA products were harvested from the C1 chip into 96-well 0.2 mL PCR plates and then loaded onto Biomark 48.48 Dynamic Array chips using the Nanoflex IFC controller (Fluidigm). Quantitative single cell PCR was performed using gene amplification with TaqMan Assays (Life Technologies) (Supplementary Table 6), and threshold cycle (Ct) as a measurement of relative fluorescence intensity was extracted by the BioMark Real-Time PCR Analysis software. All PCR reactions were performed in duplicates or triplicates, and CT values were directly used in data analysis after normalization to the 18S endogenous control gene.
Patch clamp
To record cellular action potentials, cardiomyocytes at d15 were dissociated using TrypLE for 10 min, filtered through a 100 µM cell strainer (BD Biosciences), counted with a Countess Cell Counter, plated as single cells (1 × 105 cells per well of a 24-well plate) on 8 mm No. 1 glass cover slips (Warner Instruments) coated with Synthemax II-SC (625 ng/cm2) in CDM3 supplemented with 2 µM thiazovivin and allowed to attach for 72 h, changing the medium every other day. Cells were then subjected to whole-cell patch-clamp at 36–37 °C, using an EPC-10 patch-clamp amplifier (HEKA) attached to a RC-26C recording chamber (Warner Instruments) and mounted onto the stage of an inverted microscope (Nikon). Sharp microelectrodes were fabricated from standard wall borosilicate glass capillary tubes (BF 100-50-10, Sutter Instruments) using a P-97 Sutter micropipette puller to generate electrodes with tip resistances between 50 and 70 MΩ when backfilled with 3 M KCl. Cell cultures were perfused with warm (35–37 °C) Tyrode’s solution consisting of (mM) 135 NaC1, 5.4 KCl, 1.8 CaCl2, 1.0 MgCl2, 0.33 NaH2PO4, 5 HEPES, and 5 glucose; pH was adjusted to 7.4 with NaOH. Membrane potential measurements were made using the current clamp mode of the Multiclamp 700B amplifier after electrode potential offset and capacitance were neutralized. Data were acquired using PatchMaster software (HEKA) and digitized at 1.0 kHz. The following are the criteria used for classifying observed APs into ventricular, atrial and nodal-like hiPSC-CMs. Ventricular-like: a negative maximum diastolic membrane potential (< −50 mV), a rapid AP upstroke, a long plateau phase, APA > 90 mV, and APD90/APD50 ratio < 1.4. Atrial-like: absence of a prominent plateau phase, a negative diastolic membrane potential (< −50 mV), and APD90/APD50 ratio > 1.7. Nodal-like: a more positive MDP, a slower AP upstroke, a prominent phase 4 depolarization, and APD90/APD50 ratio in between 1.4–1.7.
Nanopillar action potential recordings
Before plating, nanopillar electrode devices32 were cleaned with 5 minutes of oxygen plasma treatment. The culture chamber was coated with 9 µg/cm2 Matrigel for 30–60 minutes. hiPSC-CMs were then plated at a density of 1 × 105 per cm in CDM3 with 2 µM thiazovivin for the first 24 h. The cells were maintained in a standard incubator at 37 °C and 5% CO2. Medium was changed every 24 h. 2–3 days after plating, spontaneous and synchronous beating was observed. A 60-channel voltage amplifier system (Multichannel System, MEA1060-Inv-BC) was used for recording hiPSC-derived cardiomyocytes cultured on nanopillar electrode arrays (9 nanopillars per array) after cells started beating. Recording was performed in the same culture medium at room temperature, using an Ag/AgCl electrode in the medium as the reference electrode. The amplification was 53 and the sampling rate was 5 kHz. For electroporation, 20 biphasic square pulses of 3.5 V amplitude and 400 µs period were applied to a nanopillar electrode. The recording system was blanked during the electroporation period. Electrophysiology recordings were resumed 10–20 s after the electroporation to avoid amplifier saturation. Recordings were taken daily. The recordings were analyzed by a custom MATLAB code. Briefly, the first 20–30 intracellular recorded action potentials were overlaid and averaged. The action potential maximum and resting potentials were then identified. APD50 and APD90 were subsequently computed. Similar to multi-electrode array recordings, nanopillar intracellular recordings register a signal amplitude of 1–2 mV. This is because the seal resistance between the cell and the nanoelectrodes does not rely on achieving a giga-ohm seal. However, we have demonstrated that the signal morphologies recorded by the Nanopillar are identical to those recorded by patch clamp electrophysiology49. Nanopillar electrode devices were reused after removal of the cells by TrypLE Express.
Supplementary Material
Acknowledgements
We thank Dr. J. Odegaard for analysis of teratoma slides. We thank Dr. K. R. Boheler and Dr. R. L. Gundry for their insightful comments on this manuscript. This work was supported by the American Heart Association Postdoctoral Fellowship grant 12POST12050254 (PWB), American Heart Association Established Investigator Award 14420025, Foundation Leducq, the National Institutes of Health U01 HL099776, R01 HL113006, R24 HL117756, and the California Institute for Regenerative Medicine TR3-05556 and DR2-05394 (JCW).
Footnotes
Author Contributions
P.W.B. conceived, performed, and interpreted the experiments and wrote the manuscript; E.M. performed cardiomyocyte immunofluorescence, single-cell RT-PCR, and electrophysiology data assessment; P.S., Z.L., and A.J.O. performed electrophysiology experiments and assessed data; S.D. provided CoMiP reprogrammed cells; B.H. performed teratoma assay; J.M.C. A.D.E, F.L., N.M.M., and J.R.P tested differentiation; B.C., J.D.G. provided experimental advice; and J.C.W. provided experimental advice, manuscript writing, and funding support.
Competing Financial Interests
JCW is a co-founder of Stem Cell Theranostics. Other authors declare no competing financial interests.
References
- 1.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burridge PW, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS One. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, et al. Extracellular matrix promotes highly efficient cardiac differentiation of human pluripotent stem cells: the matrix sandwich method. Circ. Res. 2012;111:1125–1136. doi: 10.1161/CIRCRESAHA.112.273144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lian X, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E1848–1857. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burridge PW, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–938. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 9.Elliott DA, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat. Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 10.Laflamme MA, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat. Biotechnol. 2007;25:1015–1024. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 11.Willems E, et al. Small molecule-mediated TGF-beta type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell Stem Cell. 2012;11:242–252. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Titmarsh DM, et al. Microbioreactor arrays for full factorial screening of exogenous and paracrine factors in human embryonic stem cell differentiation. PLoS One. 2012;7:e52405. doi: 10.1371/journal.pone.0052405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore GE, Gerner RE, Franklin HA. Culture of normal human leukocytes. JAMA. 1967;199:519–524. [PubMed] [Google Scholar]
- 14.Brewer GJ, Cotman CW. Survival and growth of hippocampal neurons in defined medium at low density: advantages of a sandwich culture technique or low oxygen. Brain Res. 1989;494:65–74. doi: 10.1016/0006-8993(89)90144-3. [DOI] [PubMed] [Google Scholar]
- 15.Uosaki H, et al. Efficient and Scalable Purification of Cardiomyocytes from Human Embryonic and Induced Pluripotent Stem Cells by VCAM1 Surface Expression. PLoS One. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter L, et al. Efficient differentiation of human induced pluripotent stem cells generates cardiac cells that provide protection following myocardial infarction in the rat. Stem Cells Dev. 2012;21:977–986. doi: 10.1089/scd.2011.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graichen R, et al. Enhanced cardiomyogenesis of human embryonic stem cells by a small molecular inhibitor of p38 MAPK. Differentiation. 2008;76:357–370. doi: 10.1111/j.1432-0436.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 18.Melkoumian Z, et al. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat Biotechno. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- 19.Nagaoka M, Si-Tayeb K, Akaike T, Duncan SA. Culture of human pluripotent stem cells using completely defined conditions on a recombinant E-cadherin substratum. BMC Dev. Biol. 2010;10:60. doi: 10.1186/1471-213X-10-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braam SR, et al. Recombinant vitronectin is a functionally defined substrate that supports human embryonic stem cell self-renewal via alphavbeta5 integrin. Stem Cells. 2008;26:2257–2265. doi: 10.1634/stemcells.2008-0291. [DOI] [PubMed] [Google Scholar]
- 21.Rodin S, et al. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat Commun. 2014;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- 22.Rodin S, et al. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- 23.Miyazaki T, et al. Laminin E8 fragments support efficient adhesion and expansion of dissociated human pluripotent stem cells. Nat Commun. 2012;3:1236. doi: 10.1038/ncomms2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domogatskaya A, Rodin S, Tryggvason K. Functional diversity of laminins. Annu. Rev. Cell Dev. Biol. 2012;28:523–553. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- 25.Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 26.Paige SL, et al. Endogenous Wnt/beta-catenin signaling is required for cardiac differentiation in human embryonic stem cells. PLoS One. 2010;5:e11134. doi: 10.1371/journal.pone.0011134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin AF. Turnover of cardiac troponin subunits. Kinetic evidence for a precursor pool of troponin-I. J. Biol. Chem. 1981;256:964–968. [PubMed] [Google Scholar]
- 28.Ng SY, Wong CK, Tsang SY. Differential gene expressions in atrial and ventricular myocytes: insights into the road of applying embryonic stem cell-derived cardiomyocytes for future therapies. Am. J. Physiol. Cell Physiol. 2010;299:C1234–1249. doi: 10.1152/ajpcell.00402.2009. [DOI] [PubMed] [Google Scholar]
- 29.Minami I, et al. A small molecule that promotes cardiac differentiation of human pluripotent stem cells under defined, cytokine- and xeno-free conditions. Cell Rep. 2012;2:1448–1460. doi: 10.1016/j.celrep.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 30.Chuva de Sousa Lopes SM, et al. Patterning the heart, a template for human cardiomyocyte development. Dev. Dyn. 2006;235:1994–2002. doi: 10.1002/dvdy.20830. [DOI] [PubMed] [Google Scholar]
- 31.Bird SD, et al. The human adult cardiomyocyte phenotype. Cardiovasc. Res. 2003;58:423–434. doi: 10.1016/s0008-6363(03)00253-0. [DOI] [PubMed] [Google Scholar]
- 32.Xie C, Lin Z, Hanson L, Cui Y, Cui B. Intracellular recording of action potentials by nanopillar electroporation. Nat Nanotechnol. 2012;7:185–190. doi: 10.1038/nnano.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Francis GL. Albumin and mammalian cell culture: implications for biotechnology applications. Cytotechnology. 2010;62:1–16. doi: 10.1007/s10616-010-9263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi T, et al. Ascorbic acid enhances differentiation of embryonic stem cells into cardiac myocytes. Circulation. 2003;107:1912–1916. doi: 10.1161/01.CIR.0000064899.53876.A3. [DOI] [PubMed] [Google Scholar]
- 35.Cao N, et al. Ascorbic acid enhances the cardiac differentiation of induced pluripotent stem cells through promoting the proliferation of cardiac progenitor cells. Cell Res. 2012;22:219–236. doi: 10.1038/cr.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moyes KW, et al. Human embryonic stem cell-derived cardiomyocytes migrate in response to gradients of fibronectin and Wnt5a. Stem Cells Dev. 2013;22:2315–2325. doi: 10.1089/scd.2012.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tribulova N, et al. Enhanced connexin-43 and alpha-sarcomeric actin expression in cultured heart myocytes exposed to triiodo-L-thyronine. J Mol Histol. 2004;35:463–470. doi: 10.1023/b:hijo.0000045945.16046.b5. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, Ouyang J, Xia Z. Effects of triiodo-thyronine on angiotensin-induced cardiomyocyte hypertrophy: reversal of increased beta-myosin heavy chain gene expression. Can. J. Physiol. Pharmacol. 2006;84:935–941. doi: 10.1139/y06-043. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Q, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen G, et al. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap LY, et al. Defining a threshold surface density of vitronectin for the stable expansion of human embryonic stem cells. Tissue Eng Part C Methods. 2011;17:193–207. doi: 10.1089/ten.TEC.2010.0328. [DOI] [PubMed] [Google Scholar]
- 43.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mali P, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. Stem Cells. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okita K, et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 46.Chou BK, et al. Efficient human iPS cell derivation by a non-integrating plasmid from blood cells with unique epigenetic and gene expression signatures. Cell Res. 2011;21:518–529. doi: 10.1038/cr.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 48.Ng ES, Davis R, Stanley EG, Elefanty AG. A protocol describing the use of a recombinant protein-based, animal product-free medium (APEL) for human embryonic stem cell differentiation as spin embryoid bodies. Nat. Protoc. 2008;3:768–776. doi: 10.1038/nprot.2008.42. [DOI] [PubMed] [Google Scholar]
- 49.Lin ZC, Xie C, Osakada Y, Cui Y, Cui B. Iridium oxide nanotube electrodes for sensitive and prolonged intracellular measurement of action potentials. Nat Commun. 2014;5:3206. doi: 10.1038/ncomms4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.