Summary
Innate immune responses are critical for mucosal immunity. Here we describe an innate lymphocyte population, iCD8α cells, characterized by expression of CD8α homodimers. iCD8α cells exhibit innate functional characteristics such as the capacity to engulf and kill bacteria. Development of iCD8α cells depends on expression of interleukin-2 receptor γ chain (IL-2Rγc), IL-15, and the major histocompatibility complex (MHC) class Ib protein H2-T3, also known as the thymus leukemia antigen or TL. While lineage tracking experiments indicated that iCD8α cells have a lymphoid origin, their development was independent of the transcriptional suppressor Id2, suggesting these cells do not belong to the family of innate lymphoid cells. Finally, we identified cells with a similar phenotype in humans, which were profoundly depleted in newborns with necrotizing enterocolitis. These findings suggest a critical role of iCD8α cells in immune responses associated with the intestinal epithelium.
Introduction
The intestinal epithelium is comprised of a monolayer of cells that, among other functions, provides a physical barrier between the antigen-laden lumen of the intestine and the sterile environment beyond the basal layer. The epithelium is populated by a large and diverse community of immune cells, which reflects the complexity of interactions present in an environment such as the intestinal mucosa. The most prevalent and most studied of these cells include the intraepithelial lymphocytes (IEL), which are predominantly T cells and express either the T cell receptor (TCR) αβ or γδ (Olivares-Villagomez and Van Kaer, 2010). TCR+ IEL have diverse roles, such as immunity against pathogens (Lepage et al., 1998; Pope et al., 2001), protection against inflammatory bowel disease (Das et al., 2003), and promotion of tissue homeostasis (Chen et al., 2002; Inagaki-Ohara et al., 2004). In the past few years it has become evident that the IEL compartment also comprises TCR− lymphoid cells. For example, some members of the developmentally related family of innate lymphoid cells (ILC) are in direct association with the intestinal epithelium (Bernink et al., 2013; Fuchs et al., 2013).
Despite the diversity of immune cells that are intimately associated with intestinal epithelial cells (IEC), several subsets of these cell populations exhibit common features such as dependence on interleukin-7 (IL-7) or IL-15 for their maintenance and/or development. However, other features such as expression of the CD8α homodimer have been predominantly associated with TCR+ IEL. The role of CD8αα expression by IEL has not been fully elucidated. Instead of serving as a T cell co-receptor similar to CD8αβ heterodimers on conventional CD8 T cells, it has been postulated that CD8αα acts as a T cell differentiation marker as well as a repressor of TCR signaling (Cheroutre and Lambolez, 2008). CD8αα binds with high affinity to H2-T3 (also known as the thymus leukemia antigen or TL), an MHC class I-like molecule that lacks antigen-presenting properties (Liu et al., 2003; Old and Boyse, 1963; Weber et al., 2002). It has been proposed that the interaction of H2-T3 with CD8αα modulates IEL-mediated immune responses (Leishman et al., 2001; Olivares-Villagomez et al., 2011; Olivares-Villagomez et al., 2008). However, very little is known about TCR− lymphoid cells expressing CD8α homodimers within the intestinal epithelium.
In this report we describe an innate lymphoid population closely associated with the intestinal epithelium. Because of its innate features and the prevalent expression of CD8α homodimers, we refer to this population as innate CD8α (iCD8α) cells. We found that iCD8α cells are involved in innate immunity against bacterial infection. Moreover, we identified cells with a similar phenotype in the human intestinal epithelium that were depleted in necrotizing enterocolitis in neonates.
Results
The intestinal epithelium contains innate CD8αα+ lymphocytes
We have previously shown that H2-T3 expression in IEC is dispensable for the development and maintenance of TCR+ IEL expressing CD8α homodimers, the primary ligand for H2-T3 (Olivares-Villagomez et al., 2008). However, it has not been determined whether H2-T3 expression influences the cellularity and/or function of TCR− IEL. Analysis of mice deficient in cells associated with adaptive immune responses frequently facilitates the study of innate immune cells. Thus, we investigated the presence of CD8αα+ innate immune cells in the intestinal epithelium of Rag2−/− and H2-T3−/−Rag2−/− mice. Mechanical disruption of the small intestinal epithelium of Rag2−/− mice yielded at least two well-defined populations of cells: one containing predominantly epithelial cells, and one populated primarily by IEL that lack TCR expression. These cells were predominantly CD45+, indicating a hematopoietic origin (Figure 1A). This compartment of cells could be subdivided into two distinct fractions, CD8α− and CD8α+ cells. Herein, we refer to the latter population as innate CD8α, or iCD8α cells. Staining with H2-T3-tetramers confirmed that iCD8α cells expressed CD8α homodimers (Figure 1A).
Figure 1. The intestinal epithelium contains a lymphocyte population expression CD8αα.
(A) Definition of iCD8α cells obtained from the intestinal epithelium of Rag2−/− mice. Cells were isolated by mechanical disruption and stained with the cell surface markers indicated. (B) Frequency and numbers of iCD8α cells present in the intestinal epithelium of the small intestine (top) and colon (bottom) of Rag2−/− compared with H2-T3−/−Rag2−/− mice. Data are summarized in the graphs. (C) Surface expression profile of iCD8α (solid line) and CD45+CD8α− cells (dotted line) gated as in (A) and stained with the indicated antibodies. Shaded histograms indicate isotype control staining. **P<0.01; ***P<0.005. See also Figure S1.
iCD8α cells were found in the epithelium of the small intestine and colon, and constituted nearly 50% and 15%, respectively, of all CD45+ cells present within the epithelium of these organs in Rag2−/− mice (Figure 1B). We found that the frequencies of iCD8α cells in the small intestine and colon of H2-T3−/−Rag2−/− mice were significantly reduced when compared with RAG-2−/− mice (Figure 1B). A more dramatic reduction was observed in the total numbers of iCD8α cells in the small intestine (~6 fold) and colon (~4 fold) of H2-T3−/−Rag2−/− mice (Figure 1B).
We next performed a detailed phenotypic analysis of iCD8α cells compared with the CD8α-negative cell fraction within the IEL gate. The latter fraction represented a heterogeneous population of cells (Figure 1C), whereas iCD8α cells were a homogeneous population characterized as CD11bintCD11cint (suggesting a possible relationship with myeloid cells), were positive for an antibody recognizing a common epitope between CD16 and CD32 (suggesting innate effector functions such as ADCC and/or phagocytosis), CD69+Lag-3+ (consistent with an immediate effector function), CD103+g8.8+ (consistent with intraepithelial residency), c-kit− (arguing against a stem cell function and distinguishing them from group 2 ILC, some group 3 ILC, and putative precursors of TCR+ IEL), Sca-1+ (distinguishing them from lymphoid tissue inducer cells) and NK1.1−NKp46− (distinguishing them from NK cells) (Figure 1C). The surface marker profile of iCD8α cells derived from the epithelium of the colon was similar to that observed for iCD8α cells of the small intestine (Supp. Figure 1A and data not shown).
Microscopic analysis of purified iCD8α cells revealed small cells with an abundant nucleus and small cytoplasm that resemble TCR+ IEL, but distinct from most CD8α− cells of the intestinal mucosa, which showed a more prominent cytoplasm and a lobular nucleus typical of myeloid lineage cells (Supp. Figure 1B). Immunohistochemical analysis of the intestines of Rag2−/− mice identified iCD8α cells within the intestinal epithelium, and these cells were primarily associated with the villi and in intimate contact with IEC (Supp. Figure 1C).
We were unable to detect iCD8α-like cells in the lamina propria (LP), mesenteric lymph nodes (MLN) or spleen (Supp. Figure 1D), suggesting that this population, as defined herein, is only prevalent in the intestinal epithelium. Analysis of iCD8α cells during murine ontogeny revealed that this population appeared in the intestinal epithelium around 14 days after birth (Supp. Figure 1E).
A similar population to iCD8α cells was observed in the TCR− fraction of the IEL gate of WT mice (Supp. Figure 1F). This population had a comparable surface marker profile to the one observed for iCD8α cells in Rag2−/− mice. However, for some surface markers (e.g., CD103 and Sca-1) the TCR−CD8α+ population presented with bimodal surface marker staining (Supp. Figure 1F). When Rag2−/− mice were reconstituted with total splenocytes from WT animals, the endogenous iCD8α cell population adopted a surface marker profile resembling that of TCR−CD8α+ cells from WT mice (Supp. Figure 1G). These findings suggested that the iCD8α cells of Rag2−/− mice and the TCR−CD8α+ cells of WT mice are the same cell population but that they may exhibit distinct developmental or activation states due to the presence of T cells in the intestinal mucosa.
iCD8α cell development depends on IL-15 but not IL-7
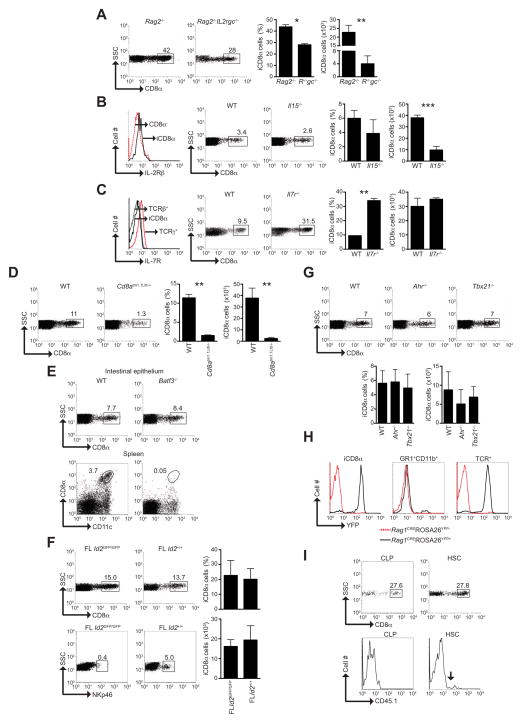
Because all lymphoid cells require the common IL-2 receptor γ chain (IL-2Rγc) for their development (Rochman et al., 2009), we investigated Rag2−/−Il2rg−/− mice for the development of iCD8α cells. We found that the prevalence and numbers of iCD8α cells were significantly reduced in Rag2−/−Il2rg−/− compared with Rag2−/− mice (Figure 2A), indicating that the development of the majority of iCD8α cells is γc-dependent. IL-15 is important for the development of innate TCRαβ+CD8αα+ and TCRγδ+ IEL (Suzuki et al., 1997), as well as other lymphoid cells such as natural killer (NK) and NKT cells (Kennedy et al., 2000; Ohteki et al., 1997). We observed that iCD8α cells expressed the IL-2Rβ chain (CD122), which is required for IL-15 signaling (Figure 2B, histogram). Although the proportions of iCD8α cells remained similar between WT and Il15−/− mice, the total numbers of iCD8α cells in IL-15-deficient mice were dramatically reduced (Figure 2B), indicating a requirement for IL-15 in the development of most iCD8α cells. IL-7 is an important cytokine involved in the maintenance and proliferation of immune cells associated with the intestinal mucosa (Maki et al., 1996; Watanabe et al., 1995). Unlike TCRγδ IEL, iCD8α cells lacked expression of the IL-7 receptor (IL-7R or CD127) (Figure 2C, histogram), and although the frequencies of iCD8α cells were higher in Il17r−/− mice, their numbers were comparable between Il17r−/− and WT mice (Figure 2C). Thus, our results showed that iCD8α cells depend on IL-15 but not IL-7 for their development.
Figure 2. iCD8α cells represent a lymphocyte population dependent on IL-2Rγc, IL-15 and E8I.
(A) Analysis of the frequency and numbers of iCD8α cells in Rag2−/− and Rag2−/−Il2rg−/− mice, as defined in Figure 1A. Summary is represented by bar graphs. (B) Left, IL-2Rβ (CD122) expression by iCD8α cells and TCR−CD8α− cells from WT mice. Right, iCD8α cell frequency and numbers in WT and Il15−/− mice. Summary is represented by bar graphs. (C) Left, IL-7R (CD127) expression by iCD8α cells, TCRαβ and TCRγδ IEL. Right, iCD8α cell frequency and numbers in WT and Il7r−/− mice. Summary is represented by bar graphs. (D) Analysis of the frequency and numbers of iCD8α cells in WT and Cd8atm1.1Litt−/− mice. Summary is represented by bar graphs. (E) Analysis of iCD8α+ cell frequency in WT and Batf3−/− mice. As controls, bottom panels represent the presence of splenic CD11c+CD8α+ DC in the indicated mice. (F) Analysis of the iCD8α cell frequency and numbers in Id2-deficient and -competent fetal liver (FL Id2GFP/GFP and FL Id2+/+, respectively) recipient mice four weeks after transplant. Summary is represented by bar graphs. As control for Id2-deficiency, bottom dot plots indicate ILC reconstitution in the indicated recipients. (G) Analysis of the iCD8α cell frequency and numbers in WT, Ahr−/− and Tbx21−/− mice. Summary is represented by bar graphs. (H) Representative analysis of YFP expression driven by RAG-1 in total TCR+ IEL, iCD8α cells, and Gr1+CD11b+ cells present in the intestinal epithelium. (I) Analysis of iCD8α cell reconstitution by CLP and HSC at four weeks after transplantation into sublethally irradiated Rag2−/−Il2rg−/− mice. As control for CLP reconstitution, blood from recipient mice was stained for myeloid cells (Gr-1) and gated for donor-derived cells (right dot plots, arrow indicates donor-derived cells). At least three mice were analyzed per group and the experiment was repeated two or three times. *P<0.05; **P<0.01; ***P<0.005. SD is indicated in bar graphs. See also Figure S2.
iCD8α cells are distinct from CD8α+ DC
Because CD8α homodimers are expressed by a subset of DC with a lymphoid origin (Seillet and Belz, 2013), we investigated whether iCD8α cells represent a subpopulation of DC. For this purpose, we analyzed Cd8atm1.1Lit−/− mice, which are selectively deficient in the cluster I enhancer (E8I) of the Cd8α gene locus (Ellmeier et al., 2002). E8I is critical for driving expression of CD8α in CD8αα+TCR+ IELs, but not peripheral CD8αβTCR+ cells and CD8α+ DC (Ellmeier et al., 1999). We found a substantial decrease in the percentage as well as the total numbers of iCD8α cells in the intestines of Cd8atm1.1Litt−/− mice when compared with WT animals (Figure 2D). These results indicated that, similar to innate CD8αα+TCR+ IEL in the intestinal epithelium, the E8I enhancer is required for expression of CD8αα homodimers by iCD8α+ cells. In a complementary approach, we analyzed mice deficient in the transcription factor basic leucine zipper transcription factor ATF-like 3 (Batf-3), which is essential for the development of CD8α+ DC (Hildner et al., 2008). Batf3−/− mice had normal proportions and numbers of iCD8α cells but lacked CD8α+ DC (Figure 2E and data not shown).
iCD8α cells represent an Id2-independent lymphoid population
To determine whether iCD8α cells have developmental requirements similar to ILC, we analyzed the role of the transcriptional repressor Id2, which is required for the development of all ILC subsets (Spits and Di Santo, 2011). Using a mouse line in which GFP is knocked-in to the Id2 gene locus, we found that iCD8α cells express Id2 (Supp. Figure 2, histogram). Id2-deficient (Id2GFP/GFP) mice harbored iCD8α cells at higher proportions than WT mice, but had fewer total cell numbers (Supp. Figure 2). However, because Id2GFP/GFP mice suffer from profound growth retardation (Yokota et al., 1999), these findings may be misleading. Therefore, we transferred fetal liver cells from Id2GFP/GFP or Id2+/GFP donors into irradiated WT mice. As shown in Figure 2F, fetal liver cells derived from Id2GFP/GFP and Id2+/GFP mice were able to reconstitute iCD8α cells in recipient animals to similar frequencies and numbers (top dot plots and graphs), whereas reconstitution of NKp46+ ILC was only observed in mice that received fetal liver cells from Id2+/GFP mice (bottom dot plots). Thus, in contrast to conventional ILC, our results indicated that iCD8α cells do not require Id2 for their development.
ILC such as ILC1 and ILC3 that are prevalent in the intestinal mucosa depend on the transcription factors arylhydrocarbon receptor (Ahr) and T-bet for their development and/or maintenance (Klose et al., 2013; Spits et al., 2013). Analysis of the intestinal mucosa of Ahr−/− and Tbx21−/− mice revealed the presence of iCD8α cells in proportions and numbers similar as WT mice (Figure 2G).
Bone marrow-derived lymphoid progenitors, which include common lymphoid progenitors (CLP) and lymphoid-primed multipotent progenitors, transiently express Rag1 and Rag2 (Yang et al., 2011). Thus, fate-based lineage analysis revealed by expression of Rag1 and Rag2 genes in progenitor cells should permit us to discern the immune lineage of iCD8α cells. For this purpose, we employed Rag1CREROSAYFP mice in which cells with a history of RAG-1 protein expression are positive for YFP (Yang et al., 2011). As shown in Figure 2H, in contrast with myeloid Gr1+CD11b+ cells that lacked YFP expression, iCD8α cells expressed YFP similar to TCR+ IEL. These results provide strong evidence that iCD8α cells are lymphoid-derived. To further corroborate the lymphoid lineage development of iCD8α cells, we transplanted CLP or hematopoietic stem cells (HSC) from Rag2−/− CD45.1 donors into sublethally irradiated Rag2−/−Ilr2g−/− CD45.2 recipient mice. CLP and HSC were capable of reconstituting the intestinal epithelium with iCD8α cells at similar proportions (Figure 2I, left plots). Blood reconstitution of myeloid cells (Gr-1+) by HSC but not CLP confirmed the lymphoid lineage of iCD8α cells (Figure 2I, right plots. Arrow indicates donor-derived cells).
In summary, our findings provide evidence that iCD8α cells constitute an Id2-independent lymphoid cell population.
iCD8α cells present a unique global transcriptome
To obtain a better understanding of the lineage relationship and potential effector functions of iCD8α cells, we performed a global transcriptional analysis of FACS-enriched iCD8α cells (for purity refer to Supp. Figure 3), and compared the transcriptome of these cells with two innate immune cell types, CD45+CD8α−NKp46−CD11bhi myeloid cells and CD45+CD8α−NKp46+NK1.1+ group 1 ILC (g1-ILC) associated with the intestinal epithelium. While iCD8α cells uniquely expressed 465 genes, they shared 573 transcripts with g1-ILC and 322 transcripts with CD11b+ myeloid cells (Figure 3A). These results suggested a closer relationship between iCD8α cells and g1-ILC than between iCD8α cells and CD11b+ myeloid cells.
Figure 3. iCD8α cells present a unique gene transcription profile.
(A) Comparison of expressed transcripts between unstimulated iCD8α cells, CD45+CD8α−CD11b+ myeloid cells, and CD45+CD8α−NK1.1+NKp46+ cells (group 1-ILC) isolated from the intestinal epithelium of Rag2−/− mice. (B-K) Comparative analysis of individual gene transcript expression between iCD8α cells, CD11b+ myeloid cells and group 1 ILC (g1-ILC), for the indicated gene groups. Data represent a pool of at least 10 mice. ND = not detected. See also Figure S3.
More detailed analysis revealed that iCD8α cells had lower expression of the lymphoid cell-specific transcription factors Eomes and Tcf7 than g1-ILC (Figure 3B). iCD8α cells and g1-ILC exhibited a comparable chemokine and interleukin expression profile, whereas myeloid cells greatly expressed Cxcl16, Cxcl9, Ccl22, Ccl17, IL1, IL-1m, Il23 and Il1a (Figures 3C and D). Among the three cell populations, iCD8α cells expressed the highest amounts of CD8α, indicating that its product is an appropriate surface marker for identifying these cells (Figure 3E). iCD8α cells also exhibited increased expression of granzymes A and B (Gzma and Gzmb), suggesting a role for these cells in cytotoxicity, and increased expression of the Fc-receptor-like protein (Fcrla), which is suggestive of phagocytic properties (Figure 3E). iCD8α cells showed heterogeneous expression of TNF family members, cytokine receptors and pattern recognition receptors (Figure 3F–H). Analysis of adhesion molecules revealed strong expression of CD146 (Figure 3I), the melanoma cell adhesion molecule (Mcam), which is expressed in a fraction of pathogenic activated T cells (Dagur et al., 2011). As expected, CD11b+ myeloid cells showed strong expression of genes related to MHC class II antigen processing and presentation. Surprisingly, these genes were also expressed, albeit in lower amounts, by iCD8α cells but not by g1-ILC (Figure 3J), suggesting a possible role in antigen presentation. Finally, iCD8α cells showed expression of the cytokine osteopontin (OPN, Spp1) (Figure 3K), which has been implicated in Th1-mediated immunity, chemotaxis and wound healing (Hildner et al., 2008).
In summary, the transcriptome profile of iCD8α cells is clearly distinct from that of CD11b+ myeloid cells and has limited overlap with the transcriptome of g1-ILC.
iCD8α cells present functional properties related to innate immune cells
To determine the effector functions of iCD8α cells, we stimulated these cells in vitro with PMA plus ionomycin and determined their cytokine and chemokine production profile by Luminex technology. iCD8α cells secreted monocyte chemotactic protein-1 (MCP-1 or CCL2), macrophage inflammatory protein-1β (MIP-1β or CCL4), MIP-2 (CXCL-2), interferon-γ (IFN-γ) and regulated on activation normal T cell expressed and secreted (RANTES or CCL5) (Figure 4A), suggesting that these cells are involved in innate immune responses.
Figure 4. iCD8α cells posses innate-like properties.
(A) Cytokine and chemokine expression by iCD8α cells. Supernatants of PMA/ionomycin-stimulated iCD8α cells were analyzed by Luminex technology. Results represent data of two combined experiments, in which cells were pooled from at least 10 mice. (B) Real-time PCR analysis of the indicated cytokine receptors. Cells were FACS-enriched. CD8α− cells represent CD45+CD8α− cells from the intestinal epithelium; IEL represent total TCR+ cells; NK and CD4 T cells represent splenic cells. mRNA expression was compared to the expression observed in IEC. Data represents 3 mice from at least 2 individual experiments. (C) Total cells associated with the intestinal epithelium were cultured in the presence or absence of 10 ng/ml IL-12 for 9 hours followed by surface marker and intracellular staining. Summary is represented by bar graphs. Data represent 3 mice from at least 2 individual experiments. (D) Left, OPN mRNA expression of the indicated populations as in (B); right, intracellular OPN staining of iCD8α cells. Shaded histogram represents secondary antibody staining only. Data represent 3 mice from at least 4 individual experiments. (E) Real-time PCR analysis of PGPR-2 in the indicated populations. CD8α− cells represent CD45+CD8α− cells from the intestinal epithelium. Expression levels are compared to the expression observed in IEC. Data represent 3 mice from at least 2 individual experiments. (F) Phagocytosis and killing assay. To determine phagocytosis, FACS-enriched iCD8α and CD45+CD8α− cells were incubated for 2 hours with C. rodentium-FITC or H. pylori-FITC followed by FACS analysis. For killing assays (bar graphs) cells were incubated with C. rodentium for the indicated times and analyzed as described in the Experimental Procedures section. Data represent the pool of 10 mice and at least two replicas. (G) OPN downregulation assays. Total immune cells associated with the epithelium were cultured in the presence or absence of graded doses of peptidoglycan suspension or heat-killed Listeria monocytogenes bacteria. Four hours after incubation cells were washed and analyzed for extra- and intracellular staining. (H) Summary of (G) including surface staining of LAMP-1 under the conditions specified. OPN was detected in the supernatant after 24 hr incubation. Data represent 3 mice from at least 2 individual experiments. *P<0.05; **P<0.01; ****P<0.001. SD is indicated in bar graphs. See also Figure S4.
iCD8α cells showed substantial expression of IL-12Rβ1 and IL-12Rβ2, as determined by real-time PCR, but expressed low amounts of IL-18R and IL-23R (Figure 4B). To determine the functionality of the IL-12 receptors, we stimulated iCD8α cells with rIL-12 and found that this cytokine induces IFN-γ production by iCD8α cells (Figure 4C), confirming the results observed using Luminex technology.
Our transcriptome analysis indicated that iCD8α cells express OPN transcripts under steady-state conditions (Figure 3K). We confirmed OPN mRNA expression by real-time PCR and compared its expression in iCD8α cells with that in IEC, CD45+CD8α− cells, TCR+ IEL, NK and CD4+ T cells. We found that iCD8α cells expressed more OPN transcripts than any of the other cell populations analyzed, and OPN expression could be detected in iCD8α cells by intracellular staining (Figure 4D).
Because iCD8α cells are located within the epithelium we considered the possibility that these cells express anti-microbial molecules. However, we were unable to detect the anti-bacterial peptides RegIIIβ, RegIIIγ and cathelicidin (Cramp), and the antimicrobial proteins S1006A8 and S100A9 (data not shown). To determine whether iCD8α cells express receptors specific for peptidoglycan, we examined mRNA expression of the peptidoglycan recognition protein (PGRP). As shown in Figure 4E, iCD8α cells, as well as CD45+CD8α− cells, expressed PGRP-2, which, besides recognizing bacterial peptidoglycan, also possesses amidase activity that disrupts the sugar-peptide backbone of this molecule (Gelius et al., 2003).
We considered that iCD8α cells could potentially interact with bacteria that attach to the gastrointestinal epithelium. To test this possibility, we co-cultured FACS-enriched iCD8α cells with FITC-labeled Citrobacter rodentium or Helicobacter pylori bacteria and determined whether these cells were able to phagocytose the microorganisms. As shown in Figure 4F (histograms), iCD8α cells were capable of taking up the microorganisms, an effect abrogated by cytochalasin D, a reversible inhibitor of phagocytosis. The latter finding rules out non-specific binding of the bacteria to the cell membrane of iCD8α cells. The amount of phagocytosis mediated by iCD8α cells was similar to that observed for cells present in the CD8α− fraction (Figure 4F), which largely contains myeloid-derived innate immune cells. Moreover, iCD8α cells were capable of killing phagocytosed C. rodentium bacteria (Figure 4F, bar graph). Thus, iCD8α cells can engulf and kill bacteria.
Because iCD8α cells can interact with bacteria or their products, and express pattern recognition molecules such as PGRP-2, we reasoned that iCD8α cells might respond to peptidoglycan or Gram-positive bacteria by secreting OPN. OPN intracellular staining showed that more than 60% of unstimulated iCD8α cells contained OPN (Figure 4G, top dot plots). After a brief stimulation with graded doses of peptidoglycan, OPN+ iCD8α cell numbers decreased, and similar results were obtained with the Gram-positive bacterium Listeria monocytogenes (Figure 4G). These alterations in intracellular OPN expression correlated with surface expression of LAMP-1, a marker for degranulation (Figure 4H). Although this short incubation period was not sufficient for detection of secreted OPN by iCD8α cells (data not shown), it was readily detected after 24 hrs of incubation (Figure 4H). At the latter time point, OPN was detected in the culture supernatant even in non-stimulated cells, indicating that iCD8α cells secrete OPN in the absence of external stimuli, at least in vitro. Similar results were obtained for iCD8α cells isolated from the colon (Supp. Figure 4A).
In summary, iCD8α cells exhibit features of innate-type immunity, including recognition of pathogens, phagocytosis, bacterial killing, and release of pro-inflammatory cytokines such as IFN-γ and OPN.
iCD8α cells have the capacity to process and present antigen
Because iCD8α cells can take up bacteria, we considered the possibility that these cells can function as antigen-presenting cells. Although iCD8α cells from Rag2−/− mice expressed low amounts of MHC class II transcripts (Figure 3J), we were unable to detect surface expression of these molecules by FACS analysis (Figure 5A). Co-stimulatory molecule expression was also not detected (Figure 5A). However, a fraction of iCD8α cells from WT mice expressed surface MHC class II molecules, albeit in reduced amounts as compared with professional DCs from the Peyer’s patches (Figure 5A). These results suggested the possibility that the presence of TCR+ IEL induces surface expression of MHC class II molecules on iCD8α cells. To test this scenario, we adoptively transferred total IEL from WT mice into Rag2−/− recipient mice. We found that four weeks later recipient-derived iCD8α cells expressed more MHC class II compared with iCD8α cells from untreated Rag2−/− mice (Figure 5B). We also observed that in the presence of bacteria, iCD8α cells had increased MHC class II surface expression (Figure 5C). These results indicated that MHC class II expression by iCD8α cells is an inducible process. However, we cannot rule out the possibility that iCD8α cells snatch MHC class II molecules from APC, as has been previously observed for NK cells (Carlin et al., 2001).
Figure 5. iCD8α cells have the capacity to process and present antigen.
(A) Expression of MHC class II molecules, B7-1 and B7-2 in iCD8α cells and DCs isolated from the indicated mice. Shaded histograms indicate isotype control staining. Data represent 3 mice from 2 individual experiments. (B) MHC class II expression in recipient-derived iCD8α cells from untreated or TCR+ IEL-reconstituted Rag2−/− mice. Data represent 3–4 mice from two individual experiments. (C) MHC class II expression in iCD8α cells derived from Rag2−/− mice incubated for 24 hours with heat-killed Listeria. Data represent 3 mice for two individual experiments. (D) Processing and presentation of antigen by iCD8α cells to CD4+ T cells. Total cells associated with the epithelium were incubated with 50 μg/ml of GFP-Eα chimera protein for three hours. Cells were then stained for surface markers and with Y-Ae antibodies specific for Eα-derived Eα52–68 peptide bound with I-Ab molecules. Left panels indicate GFP staining (uptake) in gated cells. Solid histograms indicate cells incubated in the absence of GFP-Eα. Right panels indicate cell staining for Y-Ae antibody (processed antigen) among GFP+ cells. Solid histograms indicate cells stained only with secondary antibody. Data represent 4 mice in two independent experiments. Mucosal DC were used as controls (lower panels). **P<0.01
To determine whether iCD8α cells can take up, process and present antigen at their cell surface in the context of MHC class II, we cultured iCD8α cells with GFP-labeled class II I-Eα protein (Pape et al., 2007), and stained the cells with an antibody (Y-Ae) specific for Eα-derived Eα52–68 peptide bound with I-Ab class II molecules. We found that iCD8α cells were capable of taking up GFP-Eα, process the antigen, load the Eα52–68 peptide on I-Ab and display this complex at the cell surface, although these cells were less effective than mucosa-derived DC (Figure 5D).
Taken together, our findings indicate that iCD8α cells exhibit functional properties as antigen-presenting cells, in accordance with cells of the innate immune system.
Expression of H2-T3 by IEC does not affect iCD8α cell function
To evaluate the possible influence of H2-T3 in the functionality of iCD8α cells, we cultured iCD8α cells from WT and H2-T3−/− mice in the presence of peptidoglycan (as in Figure 4G–H). We observed that iCD8α cells from either H2-T3 background had similar rates of OPN degranulation (measured as reduction of OPN intracellular staining, see Figure 4H) (Supp. Figure 4B, left). Moreover, culture of iCD8α cells from WT mice in the presence of WT IEC or H2-T3− IEC showed similar intracellular OPN levels (Supp. Figure 4B, right). These results suggest that the residual iCD8α cells in H2-T3-deficient mice have similar functional properties as iCD8α cells from H2-T3-competent mice. In agreement with these results, the surface marker profiles of iCD8α cells isolated from Rag2−/− and H2-T3−/−Rag2−/− mice were indistinguishable (data not shown).
iCD8α cells are involved in immunity against C. rodentium
We have previously shown that H2-T3-deficient mice, compared with WT mice, exhibit increased susceptibility to C. rodentium infection during the first four days after challenge (Olivares-Villagomez et al., 2011). Because innate cells are important players in C. rodentium immunity, we analyzed whether H2-T3−/−Rag2−/− mice, which have reduced numbers of iCD8α cells (Figure 1B), were more susceptible to this infection than H2-T3-competent Rag2−/− mice. Although both groups of mice survived the infection (data not shown), we found that the compound knockout mice lost significantly more weight (Figure 6A). At six days post infection, H2-T3−/−Rag2−/− mice presented with almost a 10-fold higher increase in bacterial colonization and with a reduction in IL-22 production, a cytokine that protects against C. rodentium infection (Figure 6B). However, the injury to the mucosa was similar between the two groups. These differential effects were no longer observed at 14 days after infection (Figure 6C). We were unable to detect a change in the expression of RegIIIβ and RegIIIγ mRNA expression or in intracellular IL-22 production by ILC between Rag2−/− and H2-T3−/−Rag2−/− mice (Supp. Figure 5). However, in vitro culture of iCD8α cells derived from Rag2−/− mice enhanced IL-22 production by NKp46+ ILC (Figure 6D). Interestingly, the residual iCD8α cells isolated from H2-T3−/−Rag2−/− mice also had an enhancing effect over IL-22 production by NKp46+ ILC (Figure 6D). These results suggest that the failure to control C. rodentium colonization observed in H2-T3−/−Rag2−/− mice is due to a lack of adequate iCD8α cell numbers in the intestinal epithelium of these mice. Thus, adoptive transfer of iCD8α cells from Rag2−/− mice into H2-T3−/−Rag2−/− mice reduced bacterial colonization and increased total colonic IL-22 concentration in the recipient animals (Figure 6E).
Figure 6. iCD8α cells contribute to the immune responses against C. rodentium infection.
Rag2−/− and H2-T3−/−Rag2−/− mice were infected orally with C. rodentium and followed for 14 days. (A) Percentage of original weight change during the course of the experiment. (B, C) Groups of mice were analyzed at 6 (B) and 14 days (C) post infection for colonic bacterial colonization, injury score and IL-22 mRNA expression. Results are representative of at least two independent experiments with 6–10 mice per group. SD is indicated in bar graphs. (D) IL-22 production by FACS-enriched NKp46+ ILC derived from the lamina propria of Rag2−/− mice. Cells were incubated for 24 hrs in the presence or absence of enriched iCD8α cells derived from Rag2−/− or H2-T3−/−Rag2−/− mice. IL-23 (10 ng/ml) was added to the cultures to stimulate IL-22 production. Golgi Stop was added 2 hours before harvesting. Summary is represented by bar graphs. Results are representative of at least two independent experiments. Four to five mice were pooled. SD is indicated in bar graphs. (E) A group of H2-T3−/−Rag2−/− mice was reconstituted with 4–9 × 105 FACS-enriched iCD8α cells, and after reconstitution mice were infected with C. rodentium. Six days later colonization and IL-22 expression were determined. Results are representative of at least two independent experiments with 6–10 mice per group. SD is indicated in bar graphs. Results are representative of at least two independent experiments of 3 mice per group. *P<0.05; **P<0.01; ***P<0.005; ****P<0.001. See also Figure S5.
iCD8α cells are present in human intestinal epithelium
To find a potential human correlate for the iCD8α cells we have identified in mice, we analyzed human subjects. We followed a different gating approach to ensure exclusion of T cells. As shown in Figure 7A, we observed that iCD8α cells could be identified in humans as CD3−CD5−TCRαβ−TCRγδ−CD103+CD8α+ cells. In vitro stimulation of iCD8α cells with PMA plus ionomycin induced expression of IFN-γ and OPN, similar to iCD8α cells in mice (Figure 4A and 4G). A greater proportion of iCD8α cells produced more OPN than IFN-γ (Figure 7B). We next determined the status of iCD8α cells in a severe intestinal disorder, necrotizing enterocolitis (NEC). NEC is a common and frequently lethal intestinal disease predominantly affecting preterm infants (Lin and Stoll, 2006). The etiology of NEC is unknown but a large body of evidence suggests that the pathological process is driven by an excessive immune response following invasion of pathogenic bacteria through the immature intestinal barrier (Weitkamp et al., 2013). Interestingly, we found a significant decrease in the proportion of iCD8α cells in the intestinal epithelium of infants with NEC compared with age-matched controls (Figure 7C). These results suggest that the pathology associated with NEC may be contributed in part by a decrease of iCD8α cells in the intestinal mucosa.
Figure 7. iCD8α cells are present in human intestinal epithelium and reduced in NEC patients.

(A, B) Characterization of iCD8α-like cells derived from normal human intestinal epithelium (A). In vitro stimulation with PMA/ionomycin of total cells associated with the epithelium results in IFN-γ and OPN production by iCD8α cells (bottom panel in (A), and summarized in (B)). (C) Percentage of iCD8α cells in control and NEC patients. Each symbol represents a single patient. ****P<0.001.
Discussion
Studies of immune cells that are intimately associated with the intestinal epithelium have placed a significant emphasis on TCR-expressing cells, including both αβ and γδ IEL. In contrast, little is known about innate, TCR− cells that reside in close contact with the intestinal epithelium. In this report we have characterized a population of immune cells, which we have designated iCD8α cells, involved in early innate immune responses in the intestinal mucosa.
In Rag1CREROSAYFP mice, cells that most likely belong to the lymphoid lineage express Rag1 early during development (Igarashi et al., 2002). Some innate lymphoid cells, such as natural helper cells do not express rearranged T or B cell receptors but are positive for YFP driven by the Rag1 gene, suggesting a lymphoid lineage origin (Yang et al., 2011). Similarly, we found that iCD8α cells of Rag1CREROSAYFP mice express YFP. Another indicator that iCD8α cells belong to the lymphoid lineage is that the numbers of these cells are severely reduced in Rag2−/−Il2rg−/− and Il15−/− mice. While the effector function of myeloid-derived cells such as monocytes and DC is affected by IL-15-deficiency, their development is not (Ohteki et al., 2001). Moreover, the prevalence of iCD8α cells was unaffected in Baft-3-deficient mice, which lack CD8α+ DC with a lymphoid origin (Hildner et al., 2008). Finally, we showed that CLP were capable of reconstituting iCD8α cells. Each of these findings indicates that iCD8α cells likely belong to the lymphoid lineage. Nevertheless, we cannot formally exclude the possibility that iCD8α have a mixed lymphoid and myeloid origin.
It has been proposed that the TCR−CD8α+ cell population in the intestinal epithelium of mice, especially RAG-deficient mice, corresponds to precursors of conventional IEL (Page et al., 1998; Rocha et al., 1995). We do not believe this is the case because the putative IEL precursors suggested in these earlier studies exhibited a surface expression phenotype distinct from iCD8α cells: CD45loc-kithiCD16hiCD127hi (Woodward and Jenkinson, 2001). Furthermore, recent studies have provided substantial evidence for thymic development of TCR+ IEL, making it highly unlikely that iCD8α cells are precursors to TCR+ IEL (Lambolez et al., 2007).
Because of the innate qualities of iCD8α cells it may be appropriate to include them as part of the ILC family. However, all subsets of ILC characterized to date are defined by their developmental dependency on the transcriptional regulator Id2 (Spits et al., 2013). Some ILC subsets also depend for their development on transcription factors such as Ahr, T-bet and Rorγt, and we showed that iCD8α cells could develop in mice deficient in Ahr or T-bet, whereas the expression of Rorc in iCD8α cells is very low (data not shown). We further showed that expression of the IL-7R, which is important for the development of group 2 and 3 ILCs (Moro et al., 2010; Spits et al., 2013), is not required for the development of iCD8α cells. Moreover, iCD8α cells lack expression of NK1.1, NKp46, and members of the Ly49 receptor family (data not shown), which are expressed by some subsets of group 1 ILCs. These findings, together with the results of our global transcriptome analyses, indicate that iCD8α cells exhibit a distinctive gene expression profile. We therefore favor the notion that iCD8α cells represent either a new branch of Id2-independent ILC or a new subset of lymphoid cells.
Our RNA-seq analyses showed that CD8α is expressed at ~100- to 600-fold higher levels by iCD8α cells than by group 1-ILC and CD11b+ myeloid cells (Figure 3E and data not shown). Thus, surface CD8αα is currently the most specific marker for these cells. OPN expression was also more prevalent in iCD8α cells over other immune cells in the intestine or elsewhere. OPN is thought to be a key cytokine that sets an environment conducive to the development of Th1 immune responses (Ashkar et al., 2000). In addition, OPN has been implicated in promoting Th17 responses, via CD103− DC, during intestinal inflammation (Kourepini et al., 2014). If iCD8α cells are one of the main producers of OPN in the intestinal epithelium, it is possible that these cells can modulate the immune response of TCR+ IEL and ILC.
We found that the prevalence and numbers of iCD8α cells in H2-T3-deficient mice were reduced. This may be due to a requirement of H2-T3 expression for the development of iCD8α cells, or for maintaining these cells within the epithelium. While this issue remains to be resolved, our results show that iCD8α cells isolated from WT and H2-T3− environments have similar functional responses, at least in the experimental models tested.
We were intrigued by the capacity of iCD8α cells to phagocytose and kill bacteria, as these are features predominantly associated with cells of the myeloid lineage. However, it has been reported that human TCRγδ cells (Wu et al., 2009) and murine RORγt+ ILCs (Hepworth et al., 2013), which play a sentinel role in the immune system, can serve as professional phagocytes and antigen presenting cells. In this regard, we propose that iCD8α cells represent a first line of defense against bacterial pathogens that attach to the intestinal epithelium by rapidly responding during the early stages of an infection. This may explain our observation that deficiency of iCD8α cells results in increased susceptibility to C. rodentium colonization during the first 6 days after infection. Moreover, the capacity of iCD8α cells to process and present antigen suggests that these cells can activate CD4+ T cells in vivo. Because iCD8α cells lack expression of co-stimulatory molecules such as B7-1 and B7-2 (Figure 5A), it is tempting to speculate that iCD8α cells present antigen to T cells such as CD4+ IEL, which are known to be in a “partially” activated state, requiring less co-stimulation that conventional CD4+ T cells (Montufar-Solis et al., 2007).
The presence of iCD8α cells in the intestinal epithelium of humans and their decreased prevalence in infants with NEC suggest that these cells have an important role in human intestinal immunity. Because the etiology of NEC is not well understood, the discovery of iCD8α cells may provide an important clue to a better understanding of this devastating disease.
In summary, we have identified an innate lymphoid population characterized by expression of CD8αα homodimers and primarily residing within the intestinal epithelium. iCD8α cells may represent an important line of defense in the intestinal mucosa in both mice and humans, promoting bacterial clearance, presenting antigens to T cells, and thus regulating innate and adaptive immune responses. In sum, iCD8α cells constitute another exciting cell type in the expanding family of innate immune cells that reside in the intestinal mucosa.
Experimental Procedures
Mice
As wild-type (WT) mice, we used C57BL/6 mice derived from our own colony. B6.129S(Cg)-Id2tm2.1Blh/ZhuJ, B6.129S7-Il7rtm1Imx/J, B6(Cg)-Rag2tm1.1Cgn/J, 129S4-Rag2tm1.1FlvIl2rgtm1.1Flv, and B6.129S6-Tbx21tm1Glm/J mice were purchased from The Jackson Laboratories. C57BL/6-Ahrtm1.2Arte mice were purchased from Taconic. C57BL/6-H2-T3tm1Luc/J-deficient mice and H2-T3−/−Rag2−/− mice on a C57BL/6 background have been previously described (Olivares-Villagomez et al., 2011; Olivares-Villagomez et al., 2008). Il15−/− mice were kindly provided by Dr. Sebastian Joyce, Batf3−/− mice by Dr. Kenneth Murphy, and Cd8atm1.1Litt−/− mice by Dr. Hilde Cheroutre. Drs. Paul Love and LiQi Li kindly donated intestines from Rag1CREROSA26YFP mice. All mice were bred and maintained under similar conditions, and housed in accordance with the Institutional Animal Care and Use Committee at Vanderbilt University.
Isolation of cells associated with the intestinal epithelium, lamina propria, MLN, and spleen
Cells associated with the intestinal epithelium were obtained by mechanical disruption of the epithelium following established protocols for IEL isolation (Olivares-Villagomez et al., 2011). Briefly, after flushing the intestinal contents, the intestines were cut longitudinally and in small pieces. Tissue was shaken for 45 min at 37°C in Hank’s balanced salt solution (HBSS) supplemented with 5% FBS. Supernatant was recovered and passed through a glass wool column. Cells were purified using a discontinuous 40/70 Percoll gradient. LP cells were obtained by collagenase digestion of mechanically disrupted intestinal tissue as described above, followed by a discontinuous 40/70 Percoll gradient. MLN and spleen cells were isolated using conventional methods.
CLP transplants
Common lymphoid precursors (Lin−IL-7R+Thy1−Sca-1loc-kitlo) isolated from the bone morrow of Rag2−/− mice were transplanted into sublethally irradiated Rag2−/−Il2rg−/−mice (1000 cells/mouse) (Kondo et al., 1997). As a control for reconstitution, hematopoietic stem cells (Lin−IL-7R−Thy1−Sca-1hic-kithi) were transplanted into similar recipients. Four weeks later, reconstitution of iCD8α cells was determined in the intestinal epithelium.
Luminex assay
Enriched iCD8α cells were cultured for 4 hours in the presence or absence of PMA (1 ng/ml) and ionomycin (200 ng/ml). The supernatant was recovered and assayed according to the manufacturer’s instructions.
Phagocytosis and bacterial killing assay
Purified iCD8α cells and CD45+CD8α− cells were incubated with FITC-labeled C. rodentium or H. pylori. Briefly, bacteria were cultured in standing culture overnight. Two × 108 bacteria were resuspended in 1 ml of FITC (0.5 mg/ml) in 0.1 M carbonate buffer pH 9, and incubated for 20 min at room temperature in the dark. Bacteria were washed with 1 ml of PBS supplemented with 0.25% bovine serum albumin and 2 mM HEPES and then suspended in 100 μl of PBS. Labeled bacteria were opsonized with FBS at 37°C for 15 min. Bacteria were added to cells at a multiplicity of infection (MOI) of 1:10. Cells were incubated with bacteria for 4 hours. Some wells were also pretreated with cytochalasin D (10 μM, Sigma) before adding bacteria. After 4 hours of incubation, cells were fixed in 0.1% paraformaldehyde and then analyzed by FACS. For bacterial killing assays, C. rodentium bacteria were added to cells at an MOI of 1:10 and incubated for 4 hours. After incubation, 20 μg/ml gentamycin was added and cells were further incubated for 1 hour at 37°C. Cells from some wells were incubated for an additional 24 hours. After incubation, cells were lysed with 1 ml of 0.1% saponin for 30 min. Serial dilutions of lysates were prepared and 10 μl of each dilution was plated on agar plates. Plates were incubated for 48 hours and colonies were counted.
RNA-seq analysis
High quality RNA from FACS-enriched CD45+CD8α+ (iCD8α cells), CD45+CD8α−NKp46+NK1.1+ (group 1-ILC), and CD45+CD8α−CD11b+ cells (myeloid cells) from the intestinal epithelium were sequenced at the Vanderbilt Technologies for Advanced Genomics (VANTAGE) core using an Illumina HiSeq 2500. RAN data alignment was performed by top hat, followed by gene quantification (FPKM) using Cufflinks. Additional read count was generated using HTSeq. Differential expression analysis was carried out using both FPKM and read count-based methods. Pathway and network analyses were performed using Ingenuity.
Statistical analysis
Statistical significance between the groups was determined by application of an unpaired two-tailed Student t test or ANOVA. A p value <0.05 was considered significant. In the experiments involving C. rodentium infection, statistical analyses were performed with Prism version 5.0c, using a Mann-Whitney test. A p value <0.05 was considered significant.
Supplementary Material
Supplemental Figure 1 related to Figure 1. Characterization of iCD8α cells present in the intestinal epithelium. (A) Analysis of CD16 and CD32, CD69 and CD103 surface markers of iCD8α cells present in the epithelium of the colon. Solid line represents iCD8α cells from the small intestine, dotted line iCD8α cells from the colon, and solid histogram represents isotype control staining. Data are from 3 mice. (B) Light microscopy analysis of FACS-enriched iCD8α cells, TCR−CD8α− cells, and conventional TCR+CD8α+ IEL stained with H&E (400X magnification). Two examples are shown for each cell type. (C) Immunohistochemical analysis of iCD8α cells present in the intestine of Rag2−/− mice (left, 200X and right, 400X magnification). Arrows indicate representative iCD8α cells and their intimate relationship with intestinal epithelial cells. Representative analysis of more than 3 mice analyzed independently. (D) Analysis of the presence of iCD8α cells in the indicated organs. (E) Analysis of the ontogeny of iCD8α cells in the intestinal epithelium of 14- and 21-day-old mice. Cells were gated as in Figure 1A. Data represent 3–4 mice from two independent experiments. (F) Immune cells associated with the epithelium of WT mice were isolated by mechanical disruption and gated for iCD8α cells as indicated in the top panels. Bottom panels indicate surface marker staining of iCD8α cells. Solid histograms represent isotype control staining. Data represent more than 5 mice. (G) Rag2−/− mice were reconstituted with total splenocytes from WT mice. After 4 weeks, recipient-derived iCD8α cells were analyzed for expression of the indicated surface markers and compared to cells from untreated Rag2−/− mice (right panels). Left panels indicate the expression level for the surface markers of iCD8α cells from WT mice. Solid histograms represent isotype control staining. Data represent 3 mice per group from two independent experiments.
Supplemental Figure 2 related to Figure 2. Role of Id2 in iCD8α cell development. Left, Id2 expression by iCD8α cells in Id2+/GFP mice, in comparison to TCR+ IEL. Center, iCD8α cell proportions and numbers in Id2GFP/GFP mice. Summary is represented by bar graphs. **P<0.01.
Supplemental Figure 3 related to Figure 3. Purity of iCD8α cells after FACS enrichment. Representative dot plot showing the purity of iCD8α cells post FACS-enrichment isolated from intestinal epithelium of Rag2−/− mice.
Supplemental Figure 4 related to Figure 4. iCD8α cells isolated from the colon behave similarly than cells from the small intestine, and their function is H2-T3-independent. (A) iCD8α cells from colon and small intestine were incubated in the presence of peptidoglycan for four hours. Decrease of intracellular OPN was determined by staining. Results represent 3–4 mice from two independent experiments. (B) Left, enriched iCD8α cells derived from Rag2−/− (H2-T3+) or H2-T3−/−Rag2−/− (TL−) mice were incubated in the presence of peptidoglycan and OPN reduction was measured as in Figure 4G. Right, enriched iCD8α cells derived from Rag2−/− mice were incubated in the presence of TL+ or TL− IEC (1:3 ratio) and treated as indicated above.
Supplemental Figure 5 related to Figure 6. Effects of iCD8α cells on innate immune responses to C. rodentium infection. C. rodentium-infected mice were sacrificed 6 days post treatment. (A) A portion of the distal colon was used to determine RegIIIβ and RegIIIγ expression by real-time PCR. (B) Lamina propria lymphoid cells from 4-days infected mice were isolated and cultured for 4 hrs in the presence of Golgi Stop and stained for the presence of IL-22+ NKp46+ ILC. Summary of the data is represented in the bar graph. Data are from 5–6 mice per group from two independent experiments.
Highlights.
iCD8α cells are a novel innate lymphocyte population in the intestinal epithelium
iCD8α cells depend on IL-2Rγc, IL-15 and H2-T3 for their development and maintenance
iCD8α cells are involved in early innate immune responses
Acknowledgments
We thank Dr. Sebastian Joyce for providing IL-15-deficient mice, Dr. Hilde Cheroutre for Cd8atm1.1Litt-deficient mice, Dr. Kenneth Murphy for Batf3-deficient mice, the NIH Tetramer Core Facility (Emory University) for H2-T3-tetramers, and Dr. Marc Jenkins for the GFP-Eα construct. We also thank Drs. Paul Love and LiQi Li for providing intestines from Rag1CreROSA26YFP mice. This work was supported by a pilot grant from the Digestive Disease Research Center (DDRC) at Vanderbilt University School of Medicine, funded by NIH grant P30DK058404 (D.O-V.); by NIH grants R01AI072471, R01AI070305, R01HL089667, and R01DK081536 (L.V.K.); HD061607 (J.H.W.), and R01AT004821 (K.T.W.), and Merit Review Grants from the Office of Medical Research, Department of Veterans Affairs (H.M.S.A and K.T.W.). V.V.P was supported by a postdoctoral fellowship from the National Multiple Sclerosis Society of America. R.C. was supported by 1K01AT007324. Core Services performed through Vanderbilt University Medical Center’s Digestive Disease Research Center are supported by NIH grant P30DK058404.
Footnotes
Supplemental information includes five figures and Supplemental Experimental Procedures.
Author Contributions
L.V.K. and D.O.-V. designed research; H.M.S.A., K.S., V.V.P., M.J.G., M.B.P., P.M., and R.C. performed research; J.-H.W. and K.T.W. contributed with reagents/samples/analytical tools; L.V.K. and D.O.-V. analyzed the data; L.V.K. and D.O.-V. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, Zawaideh S, Rittling SR, Denhardt DT, Glimcher MJ, Cantor H. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- Bernink JH, Peters CP, Munneke M, te Velde AA, Meijer SL, Weijer K, Hreggvidsdottir HS, Heinsbroek SE, Legrand N, Buskens CJ, et al. Human type 1 innate lymphoid cells accumulate in inflamed mucosal tissues. Nat Immunol. 2013;14:221–229. doi: 10.1038/ni.2534. [DOI] [PubMed] [Google Scholar]
- Carlin LM, Eleme K, McCann FE, Davis DM. Intercellular transfer and supramolecular organization of human leukocyte antigen C at inhibitory natural killer cell immune synapses. J Exp Med. 2001;194:1507–1517. doi: 10.1084/jem.194.10.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A. 2002;99:14338–14343. doi: 10.1073/pnas.212290499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheroutre H, Lambolez F. Doubting the TCR Coreceptor Function of CD8alphaalpha. Immunity. 2008;28:149–159. doi: 10.1016/j.immuni.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Dagur PK, Biancotto A, Wei L, Sen HN, Yao M, Strober W, Nussenblatt RB, McCoy JP., Jr MCAM-expressing CD4(+) T cells in peripheral blood secrete IL-17A and are significantly elevated in inflammatory autoimmune diseases. J Autoimmun. 2011;37:319–327. doi: 10.1016/j.jaut.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Augustine MM, Das J, Bottomly K, Ray P, Ray A. An important regulatory role for CD4+CD8 alpha alpha T cells in the intestinal epithelial layer in the prevention of inflammatory bowel disease. Proc Natl Acad Sci U S A. 2003;100:5324–5329. doi: 10.1073/pnas.0831037100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellmeier W, Sawada S, Littman DR. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu Rev Immunol. 1999;17:523–554. doi: 10.1146/annurev.immunol.17.1.523. [DOI] [PubMed] [Google Scholar]
- Ellmeier W, Sunshine MJ, Maschek R, Littman DR. Combined deletion of CD8 locus cis-regulatory elements affects initiation but not maintenance of CD8 expression. Immunity. 2002;16:623–634. doi: 10.1016/s1074-7613(02)00309-6. [DOI] [PubMed] [Google Scholar]
- Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelius E, Persson C, Karlsson J, Steiner H. A mammalian peptidoglycan recognition protein with N-acetylmuramoyl-L-alanine amidase activity. Biochemical and biophysical research communications. 2003;306:988–994. doi: 10.1016/s0006-291x(03)01096-9. [DOI] [PubMed] [Google Scholar]
- Hepworth MR, Monticelli LA, Fung TC, Ziegler CG, Grunberg S, Sinha R, Mantegazza AR, Ma HL, Crawford A, Angelosanto JM, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- Inagaki-Ohara K, Chinen T, Matsuzaki G, Sasaki A, Sakamoto Y, Hiromatsu K, Nakamura-Uchiyama F, Nawa Y, Yoshimura A. Mucosal T cells bearing TCRgammadelta play a protective role in intestinal inflammation. J Immunol. 2004;173:1390–1398. doi: 10.4049/jimmunol.173.2.1390. [DOI] [PubMed] [Google Scholar]
- Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, Diefenbach A. A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature. 2013;494:261–265. doi: 10.1038/nature11813. [DOI] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kourepini E, Aggelakopoulou M, Alissafi T, Paschalidis N, Simoes DC, Panoutsakopoulou V. Osteopontin expression by CD103- dendritic cells drives intestinal inflammation. Proc Natl Acad Sci U S A. 2014;111:E856–865. doi: 10.1073/pnas.1316447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambolez F, Kronenberg M, Cheroutre H. Thymic differentiation of TCR alpha beta(+) CD8 alpha alpha(+) IELs. Immunol Rev. 2007;215:178–188. doi: 10.1111/j.1600-065X.2006.00488.x. [DOI] [PubMed] [Google Scholar]
- Leishman AJ, Naidenko OV, Attinger A, Koning F, Lena CJ, Xiong Y, Chang HC, Reinherz E, Kronenberg M, Cheroutre H. T cell responses modulated through interaction between CD8alphaalpha and the nonclassical MHC class I molecule, TL. Science. 2001;294:1936–1939. doi: 10.1126/science.1063564. [DOI] [PubMed] [Google Scholar]
- Lepage AC, Buzoni-Gatel D, Bout DT, Kasper LH. Gut-derived intraepithelial lymphocytes induce long term immunity against Toxoplasma gondii. J Immunol. 1998;161:4902–4908. [PubMed] [Google Scholar]
- Lin PW, Stoll BJ. Necrotising enterocolitis. Lancet. 2006;368:1271–1283. doi: 10.1016/S0140-6736(06)69525-1. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Naidenko OV, Liu JH, Zhang R, Joachimiak A, Kronenberg M, Cheroutre H, Reinherz EL, Wang JH. The crystal structure of a TL/CD8alphaalpha complex at 2.1 A resolution: implications for modulation of T cell activation and memory. Immunity. 2003;18:205–215. doi: 10.1016/s1074-7613(03)00027-x. [DOI] [PubMed] [Google Scholar]
- Maki K, Sunaga S, Komagata Y, Kodaira Y, Mabuchi A, Karasuyama H, Yokomuro K, Miyazaki JI, Ikuta K. Interleukin 7 receptor-deficient mice lack gammadelta T cells. Proc Natl Acad Sci U S A. 1996;93:7172–7177. doi: 10.1073/pnas.93.14.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montufar-Solis D, Garza T, Klein JR. T-cell activation in the intestinal mucosa. Immunol Rev. 2007;215:189–201. doi: 10.1111/j.1600-065X.2006.00471.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro K, Yamada T, Tanabe M, Takeuchi T, Ikawa T, Kawamoto H, Furusawa J, Ohtani M, Fujii H, Koyasu S. Innate production of T(H)2 cytokines by adipose tissue-associated c-Kit(+)Sca-1(+) lymphoid cells. Nature. 2010;463:540–544. doi: 10.1038/nature08636. [DOI] [PubMed] [Google Scholar]
- Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1+ T cell receptor-alpha beta+ cell development. J Immunol. 1997;159:5931–5935. [PubMed] [Google Scholar]
- Ohteki T, Suzue K, Maki C, Ota T, Koyasu S. Critical role of IL-15-IL-15R for antigen-presenting cell functions in the innate immune response. Nat Immunol. 2001;2:1138–1143. doi: 10.1038/ni729. [DOI] [PubMed] [Google Scholar]
- Old LJ, Boyse EA. Antigenic Properties of Experimental Leukemias. I. Serological Studies in Vitro with Spontaneous and Radiation-Induced Leukemias. Journal of the National Cancer Institute. 1963;31:977–995. [PubMed] [Google Scholar]
- Olivares-Villagomez D, Algood HM, Singh K, Parekh VV, Ryan KE, Piazuelo MB, Wilson KT, Van Kaer L. Intestinal epithelial cells modulate CD4 T cell responses via the thymus leukemia antigen. J Immunol. 2011;187:4051–4060. doi: 10.4049/jimmunol.1101052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagomez D, Mendez-Fernandez YV, Parekh VV, Lalani S, Vincent TL, Cheroutre H, Van Kaer L. Thymus leukemia antigen controls intraepithelial lymphocyte function and inflammatory bowel disease. Proc Natl Acad Sci U S A. 2008;105:17931–17936. doi: 10.1073/pnas.0808242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares-Villagomez D, Van Kaer L. TL and CD8alphaalpha: Enigmatic partners in mucosal immunity. Immunol Lett. 2010;134:1–6. doi: 10.1016/j.imlet.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page ST, Bogatzki LY, Hamerman JA, Sweenie CH, Hogarth PJ, Malissen M, Perlmutter RM, Pullen AM. Intestinal intraepithelial lymphocytes include precursors committed to the T cell receptor alpha beta lineage. Proc Natl Acad Sci U S A. 1998;95:9459–9464. doi: 10.1073/pnas.95.16.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pape KA, Catron DM, Itano AA, Jenkins MK. The humoral immune response is initiated in lymph nodes by B cells that acquire soluble antigen directly in the follicles. Immunity. 2007;26:491–502. doi: 10.1016/j.immuni.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Pope C, Kim SK, Marzo A, Masopust D, Williams K, Jiang J, Shen H, Lefrancois L. Organ-specific regulation of the CD8 T cell response to Listeria monocytogenes infection. J Immunol. 2001;166:3402–3409. doi: 10.4049/jimmunol.166.5.3402. [DOI] [PubMed] [Google Scholar]
- Rocha B, Guy-Grand D, Vassalli P. Extrathymic T cell differentiation. Curr Opin Immunol. 1995;7:235–242. doi: 10.1016/0952-7915(95)80008-5. [DOI] [PubMed] [Google Scholar]
- Rochman Y, Spolski R, Leonard WJ. New insights into the regulation of T cells by gamma(c) family cytokines. Nat Rev Immunol. 2009;9:480–490. doi: 10.1038/nri2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet C, Belz GT. Terminal differentiation of dendritic cells. Advances in immunology. 2013;120:185–210. doi: 10.1016/B978-0-12-417028-5.00007-7. [DOI] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol. 2013;13:145–149. doi: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- Spits H, Di Santo JP. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat Immunol. 2011;12:21–27. doi: 10.1038/ni.1962. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Duncan GS, Takimoto H, Mak TW. Abnormal development of intestinal intraepithelial lymphocytes and peripheral natural killer cells in mice lacking the IL-2 receptor beta chain. J Exp Med. 1997;185:499–505. doi: 10.1084/jem.185.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Ueno Y, Yajima T, Iwao Y, Tsuchiya M, Ishikawa H, Aiso S, Hibi T, Ishii H. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J Clin Invest. 1995;95:2945–2953. doi: 10.1172/JCI118002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber DA, Attinger A, Kemball CC, Wigal JL, Pohl J, Xiong Y, Reinherz EL, Cheroutre H, Kronenberg M, Jensen PE. Peptide-independent folding and CD8 alpha alpha binding by the nonclassical class I molecule, thymic leukemia antigen. J Immunol. 2002;169:5708–5714. doi: 10.4049/jimmunol.169.10.5708. [DOI] [PubMed] [Google Scholar]
- Weitkamp JH, Koyama T, Rock MT, Correa H, Goettel JA, Matta P, Oswald-Richter K, Rosen MJ, Engelhardt BG, Moore DJ, Polk DB. Necrotising enterocolitis is characterised by disrupted immune regulation and diminished mucosal regulatory (FOXP3)/effector (CD4, CD8) T cell ratios. Gut. 2013;62:73–82. doi: 10.1136/gutjnl-2011-301551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward J, Jenkinson E. Identification and characterization of lymphoid precursors in the murine intestinal epithelium. Eur J Immunol. 2001;31:3329–3338. doi: 10.1002/1521-4141(200111)31:11<3329::aid-immu3329>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wu W, Wong WM, Ward E, Thrasher AJ, Goldblatt D, Osman M, Digard P, Canaday DH, Gustafsson K. Human gamma delta T cells: a lymphoid lineage cell capable of professional phagocytosis. J Immunol. 2009;183:5622–5629. doi: 10.4049/jimmunol.0901772. [DOI] [PubMed] [Google Scholar]
- Yang Q, Saenz SA, Zlotoff DA, Artis D, Bhandoola A. Cutting edge: Natural helper cells derive from lymphoid progenitors. J Immunol. 2011;187:5505–5509. doi: 10.4049/jimmunol.1102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y, Mansouri A, Mori S, Sugawara S, Adachi S, Nishikawa S, Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–706. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1 related to Figure 1. Characterization of iCD8α cells present in the intestinal epithelium. (A) Analysis of CD16 and CD32, CD69 and CD103 surface markers of iCD8α cells present in the epithelium of the colon. Solid line represents iCD8α cells from the small intestine, dotted line iCD8α cells from the colon, and solid histogram represents isotype control staining. Data are from 3 mice. (B) Light microscopy analysis of FACS-enriched iCD8α cells, TCR−CD8α− cells, and conventional TCR+CD8α+ IEL stained with H&E (400X magnification). Two examples are shown for each cell type. (C) Immunohistochemical analysis of iCD8α cells present in the intestine of Rag2−/− mice (left, 200X and right, 400X magnification). Arrows indicate representative iCD8α cells and their intimate relationship with intestinal epithelial cells. Representative analysis of more than 3 mice analyzed independently. (D) Analysis of the presence of iCD8α cells in the indicated organs. (E) Analysis of the ontogeny of iCD8α cells in the intestinal epithelium of 14- and 21-day-old mice. Cells were gated as in Figure 1A. Data represent 3–4 mice from two independent experiments. (F) Immune cells associated with the epithelium of WT mice were isolated by mechanical disruption and gated for iCD8α cells as indicated in the top panels. Bottom panels indicate surface marker staining of iCD8α cells. Solid histograms represent isotype control staining. Data represent more than 5 mice. (G) Rag2−/− mice were reconstituted with total splenocytes from WT mice. After 4 weeks, recipient-derived iCD8α cells were analyzed for expression of the indicated surface markers and compared to cells from untreated Rag2−/− mice (right panels). Left panels indicate the expression level for the surface markers of iCD8α cells from WT mice. Solid histograms represent isotype control staining. Data represent 3 mice per group from two independent experiments.
Supplemental Figure 2 related to Figure 2. Role of Id2 in iCD8α cell development. Left, Id2 expression by iCD8α cells in Id2+/GFP mice, in comparison to TCR+ IEL. Center, iCD8α cell proportions and numbers in Id2GFP/GFP mice. Summary is represented by bar graphs. **P<0.01.
Supplemental Figure 3 related to Figure 3. Purity of iCD8α cells after FACS enrichment. Representative dot plot showing the purity of iCD8α cells post FACS-enrichment isolated from intestinal epithelium of Rag2−/− mice.
Supplemental Figure 4 related to Figure 4. iCD8α cells isolated from the colon behave similarly than cells from the small intestine, and their function is H2-T3-independent. (A) iCD8α cells from colon and small intestine were incubated in the presence of peptidoglycan for four hours. Decrease of intracellular OPN was determined by staining. Results represent 3–4 mice from two independent experiments. (B) Left, enriched iCD8α cells derived from Rag2−/− (H2-T3+) or H2-T3−/−Rag2−/− (TL−) mice were incubated in the presence of peptidoglycan and OPN reduction was measured as in Figure 4G. Right, enriched iCD8α cells derived from Rag2−/− mice were incubated in the presence of TL+ or TL− IEC (1:3 ratio) and treated as indicated above.
Supplemental Figure 5 related to Figure 6. Effects of iCD8α cells on innate immune responses to C. rodentium infection. C. rodentium-infected mice were sacrificed 6 days post treatment. (A) A portion of the distal colon was used to determine RegIIIβ and RegIIIγ expression by real-time PCR. (B) Lamina propria lymphoid cells from 4-days infected mice were isolated and cultured for 4 hrs in the presence of Golgi Stop and stained for the presence of IL-22+ NKp46+ ILC. Summary of the data is represented in the bar graph. Data are from 5–6 mice per group from two independent experiments.