Abstract
We describe here the design and initial implementation of the eMERGE-PGx project. eMERGE-PGx, a partnership of the eMERGE and PGRN consortia, has three objectives : 1) Deploy PGRNseq, a next-generation sequencing platform assessing sequence variation in 84 proposed pharmacogenes, in nearly 9,000 patients likely to be prescribed drugs of interest in a 1–3 year timeframe across several clinical sites; 2) Integrate well-established clinically-validated pharmacogenetic genotypes into the electronic health record with associated clinical decision support and assess process and clinical outcomes of implementation; and 3) Develop a repository of pharmacogenetic variants of unknown significance linked to a repository of EHR-based clinical phenotype data for ongoing pharmacogenomics discovery. We describe site-specific project implementation and anticipated products, including genetic variant and phenotype data repositories, novel variant association studies, clinical decision support modules, clinical and process outcomes, approaches to manage incidental findings, and patient and clinician education methods.
Keywords: pharmacogenetics, pharmacogenomics, next generation sequencing, study design, pre-emptive genotyping
Introduction
A widely-held vision arising from the Human Genome Project is to use information on genomic variation to guide preventive and therapeutic decision-making for individual patient treatment1–3. An area thought to be ready for evaluating the implementation of genomics-guided clinical decision-making is pharmacogenomics – the analysis of the genetic component of drug therapy response variability3. As is the case in many areas of genetic association research 4, there are few formal investigations into the real-world health practice, implementation, and clinical outcomes of genetically-tailored drug prescribing. Such investigations are particularly difficult due to logistic barriers in delivering variant information, along with genotype-specific clinical decision support (CDS), to prescribers.
Discovery efforts to identify novel pharmacogenetic variants have typically focused on a limited number of common variants in specific target “pharmacogenes”. Recent advances in DNA sequencing are permitting deep-coverage sequencing on large numbers of samples 5, resulting in the identification of large numbers of rare variants across the genome 6. Some of these pharmacogenetic rare variants will contribute to variable drug responses, but their detection and clinical effect assessment will be difficult both in the small cohorts that have adverse event data or more traditional larger cohorts, which typically have limited clinical outcome data.
The eMERGE-PGx project, described here, is a multi-center pilot of pharmacogenetic sequencing in clinical practice designed to address these issues and initiated through a collaboration between the Electronic Medical Records and Genomics (eMERGE) Network and the Pharmacogenomics Research Network (PGRN). Work in eMERGE-PGx will sequence 84 key pharmacogenes and examine the process for implementing preemptive genotyping for known pharmacogenetic drug-gene pairs at the 10 academic medical centers and health systems in the eMERGE-II Network 7 through three specific aims: 1) Deploy PGRNseq, a next-generation sequencing platform assessing sequence variation in the target pharmacogenes, in a patient population likely to be prescribed drugs of interest in a 1–3 year timeframe; 2) Integrate well-established CLIA-validated pharmacogenetic genotypes into the electronic health record (EHR) with associated clinical decision support (CDS) and assess process and clinical outcomes of this implementation; and 3) Develop a repository of pharmacogenetic variants of unknown significance linked to a repository of an EHR-based clinical phenotype data for ongoing pharmacogenomics discovery.
The eMERGE-PGx project is possible due to the unique and complementary strengths of the eMERGE Network and the Pharmacogenomics Research Network (PGRN). The 14 PGRN sites each advance a set of aims in a specific pharmacogenomics area 8 and PGRN includes a set of network-wide resources that enable sites’ specific aims. These resources include the PGRNseq platform, described below, to sequence 84 genes identified by PGRN investigators as likely important modulators of drug action. Another key activity of PGRN is the creation of multi-institutional consortia to study drug response across PGRN and other collaborating sites coordinated by PGRN and the Pharmacogenomics Knowledge Base (PharmGKB): examples include the international warfarin 9, 10, clopidogrel, and drug transporter consortia, and the Translational Pharmacogenetics Program, “real-world” pharmacogenetics implementation project11. The Clinical Pharmacogenetics Implementation Consortium (CPIC) is another PGRN-based effort that provides specific peer-reviewed guidelines on possible actions a clinician should consider when a drug is prescribed to a patient with relevant pharmacogenomic testing results12.
eMERGE is a multi-center network of health systems with biorepositories linked to EHRs 7, 13. The goals of research within the eMERGE Network are to identify novel genetic associations with disease using EHR data and to explore methods for integrating genotyping and sequencing results clinical practice. eMERGE provides expertise in defining and extracting clinical phenotypes from diverse EHRs 14–27, integrating complex genomic data generated across sites into unified datasets 28, incorporating large-scale genomics data into the EHR and pairing these data with relevant linked CDS 29, 30, and engaging with and educating patients and prescribers around issues associated with incorporating genetics into clinical care 31, 32.
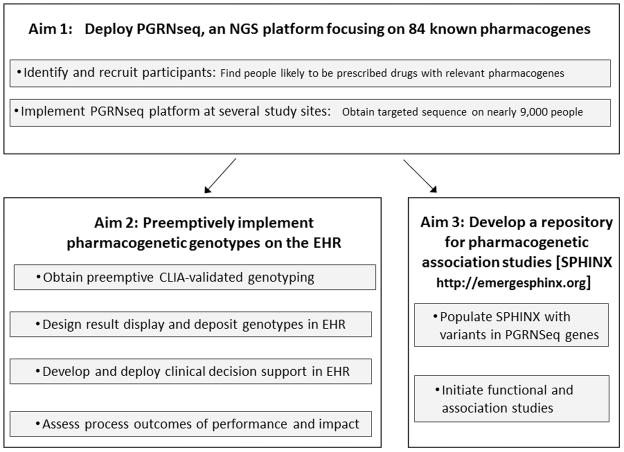
Figure 1 provides an overview of the design of the eMERGE-PGx study, driven by the three aims of the project detailed above. Where appropriate, we have indicated which methods and results sub-sections pertain to aim 1, aim 2, or aim 3. In the discussion section, we include anticipated lessons to be learned during this complex project. The implementation of eMERGE-PGx and the measureable outcomes vary, by necessity, across participating sites based on available patient populations, EHR systems, patient and prescriber preferences, and institutional policies. This variation allows for the critical evaluation of a real-world implementation of pharmacogenomics into clinical care. In this manuscript we seek to highlight the commonalities of the eMERGE-PGx project across sites, as well as site-specific differences that may influence outcomes.
Figure 1.
eMERGE PGx Project Design
Results--Anticipated Outcomes of the PGx Project
Generation of pharmacogene sequence data and validated pharmacogenetic genotypes for 9000 participants (Project Aims 1 and 2)
The 10 eMERGE-PGx sites will together recruit nearly 9000 individuals for this project. Individuals will have sequence data generated for 84 pharmacogenes using a novel platform (PGRNSeq). Participants will also have selected Clinical Laboratory Improvement Amendments (CLIA)-grade genotypes generated for deposit in EHR. Genotypes in the EHR will have associated CDS, helping providers to act on this genetic information. Details about sequencing, validation genotyping, sample and gene/drug pair selection, and genotype EHR integration with CDS are presented in the methods section of this manuscript.
Process and Clinical Outcomes (Project Aim 2)
eMERGE-PGx sites are collaborating to describe descriptive meta-data and define quantitative and qualitative outcomes across seven domains: recruitment, sequencing, genotype validation, EHR integration (including CDS), return of results, clinician and staff training, and patient education. The domains were selected to broadly compare pharmacogenomic implementation across sites and to define quantitative metrics that could be reported in aggregate. Key metrics include the number of eligible subjects genotyped and consented, PGRNseq validation performance using a common set of samples obtained from the Coriell Institute of Medical Research, CDS opportunities converted to genetically-tailored prescriptions, the reach of each implementation within the potential clinician and patient populations, and a detailed description of clinician training and patient education. Clinical outcomes related to genetically-tailored prescriptions such as stent thrombosis and myositis will be collected, as determined by individual institutional resources. Because of variability in targeted populations, released genetic variants, and enrollment selection criteria, clinical outcomes will be pooled among the subsets of sites who are implementing similar drug-gene interactions with comparable genotyping and study enrollment criteria.
Searchable Genomic Repositories (Project Aim 3)
A major contribution of recent sequencing efforts has been the creation of variant repositories, such as the Exome Variant Server 33. We will create such a resource for eMERGE-PGx, called SPHINX (Sequence, Phenotype, and pHarmacogenomics INtegration eXchange, http://emergesphinx.org). SPHINX will contain a secure, de-identified, web-accessible repository of genomic variants. Sequence variants determined from PGRNseq on all enrolled eMERGE-PGx participants will be available to the public via SPHINX in aggregate forms. eMERGE members will also have the ability to search a subset of clinical data important for clinical phenotyping for these participants including demographics, international classification of diseases diagnosis codes, current procedural technology codes, and medications mapped to RxNorm identifiers (http://www.nlm.nih.gov/research/umls/rxnorm/). We plan to map drug data from combinations of free text and electronic prescribing tools to structured vocabularies representing common drug ingredients using natural language processing tools such as the Medication Extraction (MedEx) 34 and clinical Text Analysis and Knowledge Extraction System (cTAKES) 35 systems. These clinical data will not adequately describe phenotypes for study, but will allow rapid hypothesis exploration. Once a potentially interesting population of individuals with an interesting set of variants is identified, each eMERGE site will have the potential – subject to local Institutional Review Board (IRB) approval – to access patients’ EHR for more extensive review of selected clinical records manually or via more rigorously defined phenotype algorithms 36.
Due to the risk of possible patient re-identification using demographic and clinical data, SPHINX will include important privacy safeguards. Genetic variation data will be presented in aggregate for the public interface, and cohort numbers on the public and private sides of SPHINX will be purposely obscured to protect individuals from re-identification.
Novel Pharmacogenetic Variant Gene Associations with Drug-Related Phenotypes (Project Aim 3)
The genotype-targeted preemptive programs implemented to date confine testing to a handful of common variants of known significance30, 37–39 in a small number of candidate pharmacogenes. By contrast, eMERGE-PGx may also identify novel variants in a larger set of 84 pharmacogenes and therefore provides a valuable discovery dataset. eMERGE, PGRN, and outside investigators will use data from the SPHINX repository described above, and, in some cases, with the cooperation of individual eMERGE-PGx sites, automated- or manually-abstracted data from EHR, to investigate phenotypic associations with novel variation uncovered by the PGRNseq platform.
Discussion
Lessons to be Learned from Consent and Education Procedures
The Consent, Education, Regulation and Consultation (CERC) working group has collaborated on a number of aspects of the eMERGE-PGx project, including engaging stakeholders about implementation32, developing model patient consent forms, and creating patient and physician/prescriber educational materials including a patient website. Initial focus on consent development for enrolling participants into the eMERGE-PGx study at the various sites has included determining the critical elements of consent, developing shortened consent forms, and monitoring and addressing individual site IRB responses to the study. A manuscript describing the development of consents for the eMERGE-PGx study and IRB responses at the various sites is in process. The workgroup has also helped with the development of a patient website (http://myresults.org) that contains patient educational materials and videos about the eMERGE-PGx study, as well a the eMERGE Network in general. Each site is populating the website with information about their eMERGE-PGx projects and specifically about the results patients will receive. The web site is available to the public and will provide easy access to information. Additionally, through collaboration with the eMERGE Electronic Health Record Integration (EHRI) working group, CERC Work Group members are evaluating and developing content to support CDS tools for physicians.
Lessons to be Learned from Next-Generation Sequencing Cross-Site Data Analysis and QC Pipelines
A significant lesson learned in genome-wide association studies (GWAS) that extends into next-generation sequencing (NGS) experiments is the importance of quality control (QC) pipelines and consideration of batch effects in sequencing results analysis. It has been observed that sequencing results can vary by capture reagent, sequencing platform, and bioinformatics filters, among other criteria. To ensure that eMERGE sites can combine the NGS data from this project, we have developed an NGS pipeline and quality control plan. First, all sites will be sequencing the identical set of Coriell Institute for Medical Research control samples, such that cross-site comparisons can be made in terms of concordance. [The Coriell Institute has a set of lymphoblastoid cell lines that are commonly used as controls for genotyping and sequencing experiments (http://www.coriell.org/)]. Second, the eMERGE Coordinating Center (eMERGE-CC) has developed a variant calling pipeline that will be used to process all of the NGS data. Sites will send variant call file (VCF) and BAM files (which are binary versions of sequence alignment files) to the eMERGE-CC, and the BAM files will be combined across all sites. In an effort to reduce batch effects, multi-sample calling using the Genome Analysis Toolkit (GATK) will be performed for all samples.
Two commercial sequencing platforms are being used; one site is sequencing on an Ion Torrent platform, while other sites are using Illumina platforms. We will be performing analyses based on the Coriell control samples to determine how these datasets can best be combined.
Lessons to be Learned from Approaches to Incidental Findings
The PGRNseq platform includes several genes associated with highly penetrant actionable disorders. These include genes suggested by the American College of Medical Genetics (ACMG) for return of incidental pathogenic variants (with, in some cases, consideration of returning mutations that are ‘expected’ to be pathogenic) found in clinical genomic testing: Long QT syndrome genes KCNH2 and SCN5A, malignant hyperthermia genes RYR1 and CACNA1S, a catecholaminergic polymorphic ventricular tachycardia gene RYR2, and a familial hypercholesterolemia gene LDLR 40. Sequencing these genes in eMERGE-PGx offers an opportunity to explore the phenotypes of enrolled patients carrying their variants, with potential for offering additional clinical follow-up for these incidental findings. Since eMERGE was developed as a genetic discovery network, the issue of returning genomic results is nonstandard across sites. Some sites have indicated that they are working with their IRBs to identify a process to return these incidental findings. We anticipate the publication of manuscripts summarizing approaches to return of incidental findings across the eMERGE-PGx network.
In conclusion, The eMERGE-PGx project will obtain sequence from 84 pharmacogenes using the PGRNseq platform in 9,000 individuals likely to be prescribed drugs of interest in a 1–3 year timeframe, will integrate a number of well-established CLIA-validated pharmacogenetic genotypes into the EHR with associated CDS in these same individuals, and will develop a repository of pharmacogenetic variants of unknown significance linked to a repository of clinical phenotypes for future pharmacogenetic discovery. Anticipated products include process outcomes of implementation, searchable genomic and repositories, novel pharmacogenetic variant disease associations, and lessons learned from cross-site sequencing, community engagement and consent activities, and return of incidental findings. The activities pursued as part of eMERGE-PGx will both further the implementation of pharmacogenetics into clinical care and provide an important tool for the discovery of new pharmacogenetic variants.
Methods—Design of eMERGE-PGx
Implementation of the PGRNseq platform (Project Aim 1)
Relevant portions of DNA from all participants in the eMERGE-PGx project will be sequenced using the PGRNseq custom capture reagent. An in-depth description of the development, implementation, and validation of the capture reagent is in preparation and thus a brief description is presented here. PGRNseq is a low-cost, high-throughput, next generation sequencing platform focused on the custom capture of 84 genes, selected collaboratively by the PGRN community, with associations to, or under investigation for, a wide variety of drug phenotypes. Supplementary Table 1 presents a list of genes targeted by the PGRNseq capture reagent. Sequence captured from each gene includes the complete coding regions plus 2 kilobases (kb) up- and 1 kb down-stream to assess variation within nearby regulatory regions – nearly 1 megabase of total DNA sequence. PGRNseq’s design also includes known variants present on other commercially available pharmacogenetic panel genotyping platforms, such as Affymetrix’s (Santa Clara, CA) DMET Plus panel and Illumina’s (San Diego, CA) VeraCode ADME panel, to facilitate meta-analyses with existing large datasets. PGRNseq was designed for multiplexed processing while still retaining the high coverage and sensitivity needed for effective novel genetic variation discovery and accurate genotyping of known variants. Batches of 24 or 48 samples are processed per flowcell lane, allowing for rapid throughput of many samples. (Flowcells, the site of the Illumina sequencing process, consist of 8 lanes). Preliminary testing of this platform using 24 samples / lane on cell lines derived from 32 diverse HapMap trios (two parent + 1 child trios with DNA extensively analyzed by the International HapMap Project http://hapmap.ncbi.nlm.nih.gov/) produced an average depth of coverage per sample of 496x, and genotypes derived from this PGRNseq data were 99.9% concordant with existing single nucleotide variant data on these samples from the 1000 Genomes project. Thus, overall, PGRNseq is a robust sequencing platform for the discovery and analysis of pharmacogenetic variation, both common and rare.
Individual eMERGE-PGx sites have varying sequencing protocols. Supplementary Table 2 details the locations and platforms used for PGRNseq sequencing for eMERGE-PGx. This design will permit comparison of sequencing quality control metrics across sequencing locations and recruitment sites. It will also inform the processes institutions may need to adopt to efficiently receive and process large amounts of sequence data from different sources.
Validation Genotyping (Project Aim 2)
In order to accomplish the second eMERGE-PGx objective of integrating a number of well-established, pharmacogenetic genotypes into the EHR with associated CDS, it will be necessary to generate CLIA-certified genotype results. Next generation sequencing assays are becoming more common in clinical genetics laboratories; however, the investment in test validation is significant, as are the ongoing needs for operational expertise 41. In eMERGE-PGx, two of the sites (Mount Sinai and Mayo Clinic42) are deploying selected portions of PGRNseq as a clinical test in their CLIA and CAP certified genetics/genomics laboratories, while using orthogonal platforms for other variants. Other sites are returning clinical genotype results only from orthogonal platforms (e.g. custom MASSARRAY (Sequenom), existing drug metabolism arrays, and Sanger sequencing) run in CLIA-certified laboratories on- and off-site. Generation of genotype data from CLIA-certified laboratories is further summarized in Supplementary Table 2.
Study Participants (Project Aim 1)
The eMERGE-PGx project involves a large (nearly 9000 subjects) cohort consented for returning genetic information to their EHR for the purpose of clinical care decision-making. The project was approved by the IRBs at each of the eMERGE-PGx sites. Institutions and potential study populations vary, and Table 1 provides comparative recruitment and enrollment information for the different eMERGE sites. An expanded version of Table 1 which includes more information about the characteristics of the recruiting sites, additional details about inclusion criteria, and the specific methods used to recruit patients at each site, is provided in Supplementary Table 3. Each site is recruiting subjects based on their likelihood of being prescribed given target drugs in the near-term (a 1–3 year timeframe). Recruitment is in progress at all sites and full enrollment is projected by September 2015.
Table 1.
Recruitment sites and patient enrollment for the eMERGE-PGx project
| Site | Participant Pool | Participant Selection Method | Target Enrollment |
|---|---|---|---|
| Boston Children’s Hospital (BCH) | Patients (inpatient and clinic) followed by the anticoagulation service | Participants 1 month of age or older who will potentially require warfarin | 250 |
|
| |||
| Children’s Hospital of Philadelphia (CHOP) | The entire hospital and outpatient clinics | Participants taking more than 3 medications at once, EHR records for serious adverse event (SAE), SAE noted during hospitalization | 1650 |
|
| |||
| Cincinnati Children’s Hospital Medical Center (CCHMC) | Pediatric general surgery, thoracic surgery, and orthopedic surgery clinics | 6–21 year olds being evaluated for pectus excavatum or scoliosis | 375 |
| Past participants in tonsillectomy studies | Participants 6–15 years old | 300 | |
|
| |||
| Geisinger Health System (GHS) | MyCode™ biobank participants | All males over the age of 50 who were not on one of more of the target medications at the time of ascertainment | 1100 |
|
| |||
| Group Health/University of Washington | Northwest Institute of Genetic Medicine (NWIGM) biorepository participants | Adults aged 50–65 years old at the time of entry to the repository indicating interest in return of clinically useful results. Preferential selection for individuals with certain diagnoses and medication history and all participants with Asian or African American descent. | 900 |
|
| |||
| Marshfield Clinic | Adults above the age of 50 with a primary care physician in the Marshfield Clinic. | Adults above the age of 50 with no prior use of simvastatin, warfarin or clopidogrel | 750 |
|
| |||
| Mayo Clinic | Mayo Clinic Biobank participants who are adult primary care patients | Predictive regression algorithm to estimate risk of receiving statin therapy | 1000 |
|
| |||
| Icahn School of Medicine at Mount Sinai | Patients with primary care providers at Mount Sinai Faculty Practice Associates—Primary Care Associates | Adults above the age of 50 with no prior use of simvastatin, warfarin, or clopidogrel | 750 |
|
| |||
| Northwestern University | Patients visiting the Northwestern Medical Group-General Internal Medicine Clinic | Predictive regression algorithm to estimate risk of receiving simvastatin, clopidogrel, or warfarin | 750 |
|
| |||
| Vanderbilt University | Vanderbilt internal medicine and cardiology clinics | Predictive regression algorithm to estimate risk of receiving simvastatin, clopidogrel, or warfarin | 1000 |
Several sites chose to identify the study population using a predictive algorithm incorporating site-specific data, such as that implemented in the Vanderbilt PREDICT program42, 43. Using historical EHR data, these sites identified demographic and clinical factors that predicted which patients were prescribed relevant drugs in the years preceding the eMERGE-PGx project. Then, using the most recently available data from EHR records and multivariable regression models, the investigators identified individuals in their clinical population most likely to be prescribed the relevant drugs as potential participants for the eMERGE-PGx study. Other sites used a similar, but simplified, approach of targeting patients they believed to be at high risk of being prescribed certain drugs based on demographic predictors such as diagnosis, age, and sex. At some sites these targeted approaches were implemented only among members of their biobank population (Geisinger Health System, Group Health Cooperative with University of Washington, and Mayo Clinic), while other sites implemented the approach in clinic populations. Other sites chose to use indication-based recruitment and recruited individuals assumed to be at high-risk for being prescribed certain drugs based on scheduled procedures (such as anticipated need for opioid pain medication after surgery).
In addition to patients, several sites are also enrolling prescribers into the eMERGE-PGx study to systematically capture their experiences with the implementation of pharmacogenetic variants into the EHR. Methods used to capture these experiences will include surveys, semi-structured interviews and focus groups.
Drug-Gene Pairs Implemented into Clinical Care (Project Aim 2)
The Clinical Pharmacogenetics Implementation Consortium (CPIC) 12 (http://www.pharmgkb.org/page/cpic), a shared project between PharmGKB and PGRN, publishes prescribing guidelines for using available genetic test results to make prescribing decisions. For eMERGE-PGx, each site engaged with local prescribers and institutional committees to choose a small number of drugs with CPIC guidelines for which pre-emptive pharmacogenetic genotyping could be obtained and results placed in the EHR with associated CDS to deploy at the time of drug order (hereafter referred to as drug-gene pairs). This study design addresses two primary barriers to the widespread implementation of pharmacogenetic genotyping in clinical practice, which are 1) lack of prescriber awareness or understanding of pharmacogenetics, and 2) time delay in receiving pharmacogenetic results at time of prescription 30, 37–39, 44. Table 2 lists the drugs with CPIC guidelines for which genotyping and associated CDS is being implemented at each of the eMERGE-PGx sites. The table also includes information about which prescribers at each site will see the CDS. Notably, six of the sites are implementing genotyping and CDS for the drug-gene pairs of clopidogrel/CYP2C19, warfarin/CYP2C9/VKORC1, and simvastatin/SLCO1B1. Some sites also chose to implement additional genotyping for drugs without current CPIC guidelines for this project (e.g., montelukast/SLCO2B145 or tacrolimus/CYP3A546). The chosen target drugs were used for defining the study population at each site, as discussed previously.
Table 2.
Implementation of well-established pharmacogenetic genotypes into the EHR, by eMERGE-PGx Site
| Site | CPIC Gene/Drug Pairs* | Prescribers seeing CDS | EHR vendor |
|---|---|---|---|
| Boston Children’s Hospital (BCH) |
TPMT/thiopurines CYP2C9/warfarin VKORC1/warfarin |
All prescribers including pharmacists | Cerner |
| Children’s Hospital of Philadelphia (CHOP) |
CYP2D6/codeine TPMT/thiopurines HLAB1502/carbamazepine Additional CPIC and Dutch Pharmacogenetics Working group recommendations |
All prescribing physicians | Epic |
| Cincinnati Children’s Hospital Medical Center (CCHMC) |
CYP2D6 /codeine CYP2C9/ warfarin VKORC1 / warfarin CYP2D6/ tricyclic antidepressants CYP2C19/ tricyclic antidepressants CYP2D6/ SSRIs TPMT/thiopurines |
All prescribers | Epic |
| Geisinger Health System (GHS) |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin SLCO1B1/simvastatin IL28B/interferon response |
All prescribing physicians | Epic |
| Group Health/University of Washington | HLAB1502/ carbamazepine | All prescribers | Epic |
| Marshfield Clinic |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin SLCO1B1/simvastatin |
All prescribing physicians | Internally Developed (Cattails) |
| Mayo Clinic |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin SLCO1B1/simvastatin CYP2D6 /codeine CYP2D6/tramadol CYP2D6/tamoxifen |
All prescribers | General Electric Centricity (Rochester Campus) Cerner (Arizona and Florida Campuses) |
| Icahn School of Medicine at Mount Sinai |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin SLCO1B1/simvastatin |
Primary care provider collaborators | Epic |
| Northwestern University |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin SLCO1B1/simvastatin |
Primary care provider collaborators | Epic outpatient |
| Vanderbilt University |
CYP2C19 / clopidogrel CYP2C9/ warfarin VKORC1 / warfarin TPMT/thiopurines |
All prescribing physicians | Internally Developed (StarChart) |
Some of the eMERGE-PGx sites are also implementing gene/drug pairs that do not have CPIC guidelines. These include CHOP: SLCO2B1 /montelukast, ABCB1 + CYP2C19 / ranitidine + omeprazole, CYP2D6 + ABCB1 + OPRM1 + COMT+ UGT2B7/ morphine, Marshfield: CYP4F2/ warfarin, Vanderbilt: CYP3A5/tacrolimus
Pediatric Sites (Project Aims 1 and 2)
eMERGE-PGx is recruiting a total of 2,575 patients from three pediatric sites. The pediatric focus presented additional design and implementation challenges. One challenge is that, to date, CPIC gene-drug guidelines focus primarily on medications typically prescribed in the adult population. One exception is the CYP2D6/codeine guideline. However, in February 2013, the FDA announced its intention to add a black box warning to the codeine label that strongly recommended avoiding codeine in all children following tonsillectomy. Two of the three pediatric sites are moving forward with pre-emptively integrating CYP2D6 genotypes into their EHR since, in addition to codeine, CYP2D6 metabolizes tramadol, hydrocodone and oxycodone, and the CPIC CYP2D6/codeine guideline also discusses these drugs. Another pediatric site will focus, in part, on the implementation of pre-emptive warfarin pharmacogenetics in children. This is an under-studied population for whom a validated dosing algorithm only recently became available47.
EHR Implementation and Clinical Decision Support (CDS) (Project Aim 2)
Each site is developing processes and workflow for genetic data integration into their institution’s EHR, and CDS is being developed locally based on the preferences of prescribers and the requirements of the EHR system in use. CDS required development of educational materials and automated systems to integrate validated genotype results into EHR. Many sites (CCHMC, CHOP, Geisinger, Group Health Cooperative / University of Washington, Marshfield, Mayo Clinic, Northwestern, and VUMC) are planning to place information for participants in electronic patient portals associated with the EHR. Table 2 includes information about which prescribers at each site will see the CDS. Supplementary Figure 1 shows examples of CDS alerts from several different eMERGE-PGx sites. Although the final design of decision support and educational materials vary by site, eMERGE-PGx sites are collaborating on educational materials and CDS design whenever possible.
Supplementary Material
Supplementary Figure 1. Clinical Decision Support examples from some sites A)Mayo Clinic
Supplementary Figure 1. Clinical Decision Support examples from some sites B) Northwestern Universit
Supplementary Figure 1. Clinical Decision Support examples from some sites C) Vanderbilt University
Study Highlights.
What is the current knowledge on the topic?
There are very few formal investigations into the real-world implementation and clinical outcomes of genetically-tailored drug prescribing. Additionally, little is known about the importance of rare variants in pharmacogenes.
What question does this study address?
eMERGE-PGx examines associations of rare variants in pharmacogenes with EHR-derived phenotypes in a large (~9,000) study sample. eMERGE-PGx also examines what happens when you implement large-scale genetically-tailored drug prescription across several large medical centers.
What will this study add to our knowledge?
Results from eMERGE-PGx will both inform the critical question of HOW best to implement genetically-tailored prescription in medical practice and will provide multiple new genetic targets for pharmacogenetic investigation.
How this might change clinical pharmacology and therapeutics?
eMERGE-PGx represents a first step toward a vision of incorporating large scale sequence information into the flow of routine healthcare.
Acknowledgments
The eMERGE Network was funded through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Harvard); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative and the University of Washington); U01HG006379 (Mayo Clinic).); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University); and U01HG006385 (Vanderbilt serving as the Coordinating Center). The development of the PGRN-Seq platform was supported by the Deep Resequencing Resources of the Pharmacogenomic Research Network: U01 HL069757, U19 GM61388, and U01 GM097119. The activities being performed at Johns Hopkins Center for Inherited Disease Research (CIDR) and Johns Hopkins DNA Diagnostic Lab are funded by grant U01HG004438. This work is also supported by grant (HL069757). One list of actionable genes was generated with funding from the NHGRI, 5U01HG006507. At Mayo Clinic this work was also supported in part by Mayo Clinic Center for Individualized Medicine and National Institutes of Health grants U19 GM61388 (The Pharmacogenomics Research Network), R01 GM28157, U01 HG005137, R01 CA138461, and R01 AG034676 (The Rochester Epidemiology Project). At Vanderbilt University, the work was also supported by the Pharmacogenomics of Arrhythmia Therapy PGRN site grant from the National Institute of General Medical Sciences and the National Heart Lung and Blood Institute (U19 HL65262), which also supports the site’s participation in the TPP and CPIC.
Abbreviations
- CDS
Clinical Decision Support
- CPIC
Clinical Pharmacogenomics Implementation Consortium
- EHR
Electronic Health Record
- eMERGE
Electronic Medical Records and Genomics
- PGRN
Pharmacogenomics Research Network
- CLIA
Clinical Laboratory Improvement Amendments
- IRB
Institutional Review Board
- ACMG
American College of Medical Genetics
Footnotes
Conflict of Interest
A.A.V.--Intellectual Property/Royalty Income: Cinncinnati Childrens’ Hospital and Medical Center holds a license agreement on a pharmacogenetics interpretation algorithm. I am an inventor, and I have received royalties from the company to which this has been licensed; E.B--Intellectual Property/Shareholder: I am a co-inventor of web-enabled technology for collecting and utilizing healthcare data, and I hold shares in a company that is licensing this technology from Mount Sinai School of Medicine; E.B.L. I receive royalties from UpToDate from authoring several sections.
Author Contributions
L.J.R.-T., S.C.S., A.S.G., D.M.R. and J.C.D wrote the manuscript
L.J.R.-T., S.C.S., A.S.G., B.A., M.A.B., S.J.B., A.B., M.H.B., D.S.C., J.J.C., D.R.C., K.F.D., C.J.G., O.G., D.S.K., K.A.L., S.L., S.M., A.R.M., J.A.P., V.P., J.P., C.L.P., J.F.P., C.A.P., J.R., L.V.R., M.D.R., S.S., S.A.S., M.S., A.V., A.A.V., W.A.W., E.B., R.L.C., C.G.C., J.L.H., J.B.H., B.K., I.A.H., I.J.K., G.P.J., E.B.L., C.A.M., and M.S.W., T.M., R.L., and S.V., D.A.N and S.E.S., D.M.R. and J.C.D. designed the research.
L.J.R.-T., S.C.S., A.S.G., B.A., M.A.B., S.J.B., A.B., M.H.B., D.S.C., J.J.C., D.R.C., K.F.D., C.J.G., O.G., D.S.K., K.A.L., S.L., S.M., A.R.M., J.A.P., V.P., J.P., C.L.P., J.F.P., C.A.P., J.R., L.V.R., M.D.R., S.S., S.A.S., M.S., A.V., A.A.V., W.A.W., E.B., R.L.C., C.G.C., J.L.H., J.B.H., B.K., I.A.H., I.J.K., G.P.J., E.B.L., C.A.M., and M.S.W., D.A.N and S.E.S., D.M.R. and J.C.D. performed the research
L.J.R.-T. and S.C.S. analyzed the data
A.S.G., D.A.N and S.E.S., contributed New Reagents / Analytic Tools
References
- 1.Green ED, Guyer MS National Human Genome Research I. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 2.Collins F. Opportunities and challenges for the NIH--an interview with Francis Collins. Interview by Robert Steinbrook. N Engl J Med. 2009;361:1321–3. doi: 10.1056/NEJMp0905046. [DOI] [PubMed] [Google Scholar]
- 3.Hamburg MA, Collins FS. The path to personalized medicine. N Engl J Med. 2010;363:301–4. doi: 10.1056/NEJMp1006304. [DOI] [PubMed] [Google Scholar]
- 4.McBride CM, Alford SH, Reid RJ, Larson EB, Baxevanis AD, Brody LC. Putting science over supposition in the arena of personalized genomics. Nat Genet. 2008;40:939–42. doi: 10.1038/ng0808-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER. Next-generation sequencing platforms. Annual review of analytical chemistry. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 6.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–9. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013 doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–45. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, et al. International Warfarin Pharmacogenetics C. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perera MA, Cavallari LH, Limdi NA, Gamazon ER, Konkashbaev A, Daneshjou R, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013 doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shuldiner AR, Relling MV, Peterson JF, Hicks JK, Freimuth RR, Sadee W, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin Pharmacol Ther. 2013;94:207–10. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4:13. doi: 10.1186/1755-8794-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc. 2012;19:212–8. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, et al. Electronic medical records for genetic research: results of the eMERGE consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crosslin DR, McDavid A, Weston N, Nelson SC, Zheng X, Hart E, et al. Genetic variants associated with the white blood cell count in 13,923 subjects in the eMERGE Network. Hum Genet. 2012;131:639–52. doi: 10.1007/s00439-011-1103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crosslin DR, McDavid A, Weston N, Zheng X, Hart E, de Andrade M, et al. Genetic variation associated with circulating monocyte count in the eMERGE Network. Hum Mol Genet. 2013;22:2119–27. doi: 10.1093/hmg/ddt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet. 2011;89:529–42. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shameer K, Denny JC, Ding K, Jouni H, Crosslin DR, de Andrade M, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet. 2013 doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeff JM, Ritchie MD, Denny JC, Kho AN, Ramirez AH, Crosslin D, et al. Generalization of Variants Identified by Genome-Wide Association Studies for Electrocardiographic Traits in African Americans. Ann Hum Genet. 2013 doi: 10.1111/ahg.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen-Torvik LJ, Pacheco JA, Wilke RA, Thompson WK, Ritchie MD, Kho AN, et al. High density GWAS for LDL cholesterol in African Americans using electronic medical records reveals a strong protective variant in APOE. Clin Transl Sci. 2012;5:394–9. doi: 10.1111/j.1752-8062.2012.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritchie MD, Denny JC, Zuvich RL, Crawford DC, Schildcrout JS, Bastarache L, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation. 2013;127:1377–85. doi: 10.1161/CIRCULATIONAHA.112.000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding K, de Andrade M, Manolio TA, Crawford DC, Rasmussen-Torvik LJ, Ritchie MD, et al. Genetic variants that confer resistance to malaria are associated with red blood cell traits in African-Americans: an electronic medical record-based genome-wide association study. G3. 2013;3:1061–8. doi: 10.1534/g3.113.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kullo IJ, Ding K, Shameer K, McCarty CA, Jarvik GP, Denny JC, et al. Complement Receptor 1 Gene Variants Are Associated with Erythrocyte Sedimentation Rate. Am J Hum Genet. 2011;89:131–8. doi: 10.1016/j.ajhg.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schildcrout JS, Basford MA, Pulley JM, Masys DR, Roden DM, Wang D, et al. An analytical approach to characterize morbidity profile dissimilarity between distinct cohorts using electronic medical records. Journal of biomedical informatics. 2010;43:914–23. doi: 10.1016/j.jbi.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton KM, Peissig PL, Kho AN, Bielinski SJ, Berg RL, Choudhary V, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc. 2013;20:e147–54. doi: 10.1136/amiajnl-2012-000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, et al. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. J Am Med Inform Assoc. 2012;19:225–34. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zuvich RL, Armstrong LL, Bielinski SJ, Bradford Y, Carlson CS, Crawford DC, et al. Pitfalls of merging GWAS data: lessons learned in the eMERGE network and quality control procedures to maintain high data quality. Genet Epidemiol. 2011;35:887–98. doi: 10.1002/gepi.20639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starren J, Williams MS, Bottinger EP. Crossing the omic chasm: a time for omic ancillary systems. JAMA. 2013;309:1237–8. doi: 10.1001/jama.2013.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther. 2012;92:87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clayton EW, Smith M, Fullerton SM, Burke W, McCarty CA, Koenig BA, et al. Confronting real time ethical, legal, and social issues in the Electronic Medical Records and Genomics (eMERGE) Consortium. Genet Med. 2010;12:616–20. doi: 10.1097/GIM.0b013e3181efdbd0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartzler A, McCarty CA, Rasmussen LV, Williams MS, Brilliant M, Bowton EA, et al. Stakeholder engagement: a key component of integrating genomic information into electronic health records. Genet Med. 2013;15:792–801. doi: 10.1038/gim.2013.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP) Seattle, WA: [Google Scholar]
- 34.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010;17:19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denny JC. Chapter 13: Mining electronic health records in the genomics era. PLoS computational biology. 2012;8:e1002823. doi: 10.1371/journal.pcbi.1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14:723–6. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farrugia G, Weinshilboum RM. Challenges in implementing genomic medicine: the mayo clinic center for individualized medicine. Clin Pharmacol Ther. 2013;94:204–6. doi: 10.1038/clpt.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schildcrout JS, Denny JC, Bowton E, Gregg W, Pulley JM, Basford MA, et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther. 2012;92:235–42. doi: 10.1038/clpt.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, et al. ACMG clinical laboratory standards for next-generation sequencing. Genet Med. 2013;15:733–47. doi: 10.1038/gim.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clinic proceedings. 2014;89:25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson JF, Bowton E, Field JR, Beller M, Mitchell J, Schildcrout J, et al. Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med. 2013 doi: 10.1038/gim.2013.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Driest S, Shi Y, Bowton E, Schildcrout J, Peterson J, Pulley J, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95:423–31. doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mougey EB, Feng H, Castro M, Irvin CG, Lima JJ. Absorption of montelukast is transporter mediated: a common variant of OATP2B1 is associated with reduced plasma concentrations and poor response. Pharmacogenetics and genomics. 2009;19:129–38. doi: 10.1097/FPC.0b013e32831bd98c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birdwell KA, Grady B, Choi L, Xu H, Bian A, Denny JC, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenetics and genomics. 2012;22:32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lala M, Burckart GJ, Takao CM, Pravica V, Momper JD, Gobburu JV. Genetics-based pediatric warfarin dosage regimen derived using pharmacometric bridging. The journal of pediatric pharmacology and therapeutics : JPPT : the official journal of PPAG. 2013;18:209–19. doi: 10.5863/1551-6776-18.3.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Clinical Decision Support examples from some sites A)Mayo Clinic
Supplementary Figure 1. Clinical Decision Support examples from some sites B) Northwestern Universit
Supplementary Figure 1. Clinical Decision Support examples from some sites C) Vanderbilt University