Abstract
Background
A subset of individuals present at autopsy with the pathological features of Alzheimer's disease (AD) having never manifest the clinical symptoms. We sought to identify genetic factors that modify the relationship between phosphorylated tau (PTau) and dilation of the lateral inferior ventricles.
Methods
We used data from 700 subjects enrolled in the AD Neuroimaging Initiative (ADNI). A genome-wide association study (GWAS) approach was used to identify PTau × single nucleotide polymorphism (SNP) interactions. Variance explained by these interactions was quantified using hierarchical linear regression.
Results
Five SNPxPTau interactions passed a Bonferroni correction, one of which (rs4728029, POT1, 2.6% of variance) was consistent across ADNI-1 and ADNI-2/GO subjects. This interaction also showed a trend level association with memory performance and levels of interleukin-6-receptor.
Conclusions
Our results suggest that rs4728029 modifies the relationship between PTau and both ventricular dilation and cognition, perhaps through an altered neuroinflammatory response.
Keywords: Alzheimer's Disease, Phosphorylated Tau, Ventricular Volume, MRI, CSF, Memory, GWAS, Genetic Interactions, Imaging Genetics, Quantitative Traits, Plasma
1. INTRODUCTION
The pathological cascade in Alzheimer's disease (AD) has been widely debated. Some suggest that tauopathies play a causal role and initiate the disease process when they arise early in life;1 whereas others posit that amyloid pathology plays a causal role arising through an independent disease process, perhaps after early tauopathies appear, and ultimately leading to an acceleration of the disease process.2,3 Emerging out of this literature is evidence that a subset of individuals present at autopsy with tauopathies and amyloid plaques having never shown clinical symptoms of AD during their lifetime.4-6 Some of this work has suggested that asymptomatic individuals show acute differences in brain activity,7 and longitudinal hypertrophy in hippocampal and cortical cell bodies that may act as a potential mechanism of resilience.6 Moreover, risk for AD pathology in asymptomatic individuals is much higher in those with a family history of AD than in those without a family history, suggesting that genetic factors may play an important role in the presence and response to AD pathology.8 Advances in biomarker detection have led to validated measures that can be used to approximate the pathological processes of AD in vivo including amyloid deposition imaged using positron emission tomography (PET) and levels of Aβ-42, of tau, and phosphorylated tau (PTau) in cerebrospinal fluid (CSF). PET and CSF biomarkers show a strong relationship to each other,9-11 and both predict risk for AD.11 The availability of genome wide association study (GWAS) data has led to the identification of a wide array of genetic risk factors for AD12,13 and associations with AD biomarkers.14-18 Yet, no study to date has leveraged the availability of these two rich data sources to investigate individual predictors of cognitive resilience seemingly present in asymptomatic individuals.
We sought to identify genetic variants that modify the relationship between biomarkers of tau pathology and a magnetic resonance imaging (MRI) measure of disease progression – lateral ventricle dilation. The lateral ventricles have shown a strong relationship to AD onset and progression,19,20 and measures of ventricular dilation have been successfully applied as quantitative endophenotypes in genetic interaction analyses previously.21 We approached this research by first characterizing the relationship between tau CSF measures and ventricular volume. Next we performed a tau-gene interaction analysis to test whether genetic variants modified the relationship between pathology and atrophy. Finally, in post-hoc analyses we tested whether observed tau-gene interactions were associated with cognitive performance or neuroinflammatory cytokine levels.
2. SUBJECTS AND METHODS
Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). The ADNI was launched in 2003 by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies and non-profit organizations, as a $60 million, 5-year public-private partnership. The primary goal of ADNI has been to test whether serial magnetic resonance imaging (MRI), PET, other biological markers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD. Determination of sensitive and specific markers of very early AD progression is intended to aid researchers and clinicians to develop new treatments and monitor their effectiveness, as well as lessen the time and cost of clinical trials.
The Principal Investigator of this initiative is Michael W. Weiner, MD, VA Medical Center and University of California – San Francisco. ADNI is the result of efforts of many co-investigators from a broad range of academic institutions and private corporations, and subjects have been recruited from over 50 sites across the U.S. and Canada. The initial goal of ADNI was to recruit 800 adults, ages 55 to 90, to participate in the research, approximately 200 cognitively normal older individuals to be followed for 3 years, 400 people with MCI to be followed for 3 years and 200 people with early AD to be followed for 2 years. For up-to-date information, see ww.adni-info.org.
2.1 Subjects
Participants were enrolled based on criteria outlined in the ADNI protocol (http://www.adni-info.org/Scientists/AboutADNI.aspx). Participants genotyped in both the ADNI-1 and ADNI-2/GO protocols were included. To avoid spurious genetic effects due to population stratification, only Caucasian participants were used in all analyses. Demographic data are presented in Table 1.
Table 1.
Demographic Information
| Baseline Clinical Diagnosisa | ||||
|---|---|---|---|---|
| Normal Control | Mild Cognitive Impairment | Alzheimer's Disease | ||
| ADNI-1 Dataset | Number of Patients | 102 | 180 | 92 |
| Number of APOE- ε4 Carriers | 24 | 99 | 64 | |
| Number of Females | 48 | 60 | 38 | |
| Mean Baseline Age (SD) | 75.75 (4.99) | 74.58 (7.61) | 75.01 (7.97) | |
| Mean Years of Education (SD) | 15.92 (2.71) | 15.76 (3.01) | 15.25 (3.22) | |
| Mean Number of Visits | 5.09 (1.42) | 5.17 (1.74) | 3.34 (0.89) | |
| Mean Interval in Days | 1320 (549) | 1032 (542) | 585 (252) | |
| Phosphorylated Tau pg/mL (SD) | 25.77 (15.23) | 35.11 (17.09) | 41.47 (18.48) | |
| ADNI-2/GO Dataset | Number of Patients | 95 | 208 | 23 |
| Number of APOE- ε4 Carriers | 22 | 90 | 15 | |
| Number of Females | 46 | 92 | 9 | |
| Mean Baseline Age (SD) | 74.66 (5.63) | 71.39 (7.32) | 74.96 (10.67) | |
| Mean Years of Education (SD) | 16.47 (2.56) | 15.96 (2.69) | 15.48 (2.76) | |
| Mean Number of Visits | 3.55 (1.03) | 3.94 (0.89) | 3.35 (0.93) | |
| Mean Interval in Days | 366 (150) | 470 (202) | 313 (143) | |
| Phosphorylated Tau pg/mL (SD) | 21.42 (7.94) | 24.76 (12.42) | 35.46 (12.86) | |
| Combined Dataset | Number of Patients | 197 | 388 | 115 |
| Number of APOE- ε4 Carriers | 45 | 189 | 79 | |
| Number of Females | 94 | 152 | 47 | |
| Mean Baseline Age (SD) | 75.22 (5.32) | 72.86 (7.62) | 75.00 (8.53) | |
| Mean Years of Education (SD) | 16.19 (2.65) | 15.87 (2.84) | 15.30 (3.13) | |
| Mean Number of Visits | 4.35 (1.46) | 4.51 (1.48) | 3.34 (0.90) | |
| Mean Interval in Days | 860 (628.06) | 731 (486) | 530 (258) | |
| Phosphorylated Tau pg/mL (SD) | 23.68 (12.43) | 29.56 (15.63) | 40.26 (17.61) | |
Normal Control subjects had a Mini-Mental Status Examination (MMSE) score between 24 and 30, a Clinical Dementia Rating (CDR) score of 0, and were not depressed (Geriatric Depression Scale score < 6).
Mild Cognitive Impairment subjects had a MMSE score between 24 and 30, objective memory impairment, subjective memory impairment, and a CDR score of 0.5.
Alzheimer's Disease subjects met clinical criteria for dementia, had an MMSE of between 20 and 26, and had CDR score of .5 or 1.
bSUVR - Standardized uptake value ratio for amyloid tracer
2.2 Genotyping
In ADNI-1, genotyping was performed using the Illumina Infinium Human-610-Quad BeadChip. In ADNI-2/GO, genotyping was performed on the Illumina OmniQuad array. After quality control (QC) procedures using PLINK,22 256,790 SNPs remained for data analysis (Appendix A).
2.3 Quantification of Ventricular Dilation
All volumetric data from 1.5 Tesla MRI scans in ADNI were used in our analyses.23,24 We used the volume of the left inferior lateral ventricle as our primary outcome measurement given its previous association with neurofibrillary tangle pathology,25 and included a measurement of intracranial volume (ICV) as a covariate in all volume analyses. Both were defined in Freesurfer.26-30 Slopes of change in left ventricular volume over time were calculated in SAS 9.3 (SAS Institute Inc., Cary, NC) using mixed model regression (PROC MIXED) to leverage the longitudinal data. In the mixed model regression, time was modeled based on days from baseline for each subject. This was then rescaled so that slopes would represent annual change (days from baseline/365.25). Details on the longitudinal data are presented in Table 1. For additional technical details see Appendix A.
2.4 Statistical Analyses: Relationship between Tau and Brain Volume
Biomarker quantification in ADNI was performed previously.11,31 To test the relationship between PTau and ventricular dilation we performed a univariate general linear model regression analysis using IBM SPSS v.20 (http://www-01.ibm.com/software/analytics/spss/) with slope of change in left ventricular volume set as the quantitative outcome measure. Predictors in the model included baseline age, baseline intracranial volume (ICV), gender, years of education, diagnosis at baseline, and PTau. In order to maximize our power to identify genetic effects we chose to exclude APOE genotype as a covariate. However, we also wanted to ensure that observed effects were not simply carrying signal related to APOE genotype, so we implemented an approach taken in previous research,32 in which we performed posthoc hierarchical linear regression including APOE genotype to calculate variance explained by observed genetic interactions above and beyond variance explained by APOE. This approach aimed to balance statistical power with interpretability.
2.5 Statistical Analyses: Gene – PTau Interaction Analysis
Genetic interaction analyses were performed in PLINK using the --linear command with slopes of ventricular volume set as the outcome variable. An additive model was used for all genotypes (0, 1, 2) with the same covariates entered above and the addition of the SNP predictor (Y = β0 + β1Baseline Age + β2BaselineICV + β3Gender + β4Education + β5Dx + β6PTau + β7SNP + β8SNPxPTau). Our term of interest was a PTau × SNP interaction term which we used to identify SNPs that modified the relationship between ventricular volume and CSF PTau. A correction for multiple comparisons using the Bonferroni procedure (256,790 total tests) was applied to the PTau × SNP interaction term. Finally, all significant interactions were stratified across the ADNI-1 and ADNI-2/GO data sources to verify the consistency of the observed effect.
2.6 Hierarchical Linear Regression to Identify Variance Explained
In order to place our observed SNP interaction effects in the context of known predictors of brain volume including APOE, we used hierarchical linear regression to calculate the change in R2 when adding in the SNP main effect, and after adding in the SNP × PTau interaction term. We report the variance explained by the final block in this model, which is the variance explained by the SNP × PTau interaction term. Population covariates were calculated using Structure.33 For additional details see Appendix A.
2.7 Post-hoc Statistical Analyses
Posthoc analyses were performed using cognitive34,35 and plasma based proteomic markers from ADNI (see Appendix A for technical details). Predictors included baseline age, gender, years of education, diagnostic group, APOE, PTau, SNP, and the SNP × PTau interaction term.
3. Results
3.1 Relationship between PTau and Brain Volume
As expected, we observed a relationship between levels of PTau in CSF and ventricular volume when controlling for known predictors (t = 3.08, p = 0.002). All predictors except gender and education were associated with ventricular volume. We chose to leave all of the covariates in our subsequent statistical models. The overall regression model explained 34% of variance in ventricular dilation.
3.2 PTau – Gene Interaction
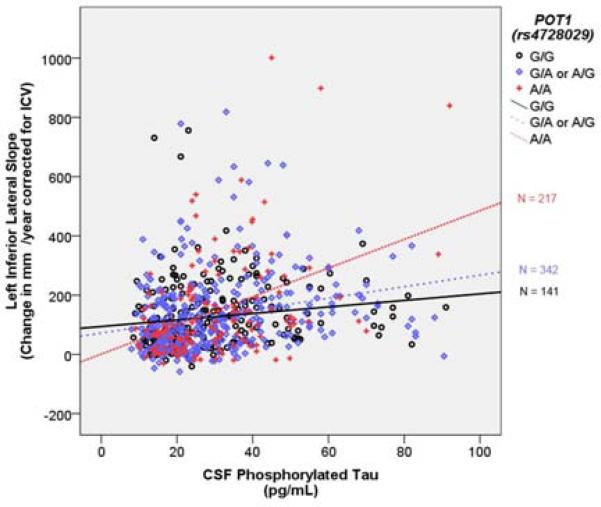
Results are presented in Table 2. Five SNP-PTau interactions remained significant after correcting for multiple comparisons. SNPS were annotated using the Illumina annotation file. Two SNPs (rs10973683, rs7859035) were annotated to the SHB gene, one SNP (rs10514128) was annotated to the TBCA gene, one SNP (rs2273704) was annotated to the EML1 gene, and one SNP (rs4728029) was annotated to the POT1 gene. When stratifying across the datasets, only the rs4728029-PTau interaction result remained significant in both ADNI-1 and ADNI-2/GO. Therefore we focused all post-hoc analyses on this SNP-PTau interaction. For the rs4728029 SNP, approximately 20% of participants were homozygous carriers of the A allele (N = 141) and 30% were homozygous carriers of the G allele (N = 217). This distribution was reflected in both the ADNI-1 and ADNI-2/GO cohorts. PTau was related to ventricular dilation, and this effect was strongly amplified in homozygous carriers of the rs4728029 A allele (Figure 1).
Table 2.
PTau-Gene Interaction Analysis
| SNP × SNP Interactions | |||||||
|---|---|---|---|---|---|---|---|
| Genes | ADNI-1 Dataset | ADNI2/GO Dataset | Combined Datasets | ||||
| t | p-value | t | p-value | t | p-value | FWE* | |
| POT1 (rs4728029) × PTau | 4.26 | 0.00003 | 2.33 | 0.020 | 5.36 | 1.11 × 10−7 | .045 |
| SHB (rs1097368) × PTau | 4.10 | 0.00005 | 1.40 | 0.164 | 5.44 | 7.26 × 10−8 | .019 |
| SHB (rs7859035) × PTau | 4.41 | 0.00001 | 1.33 | 0.184 | 5.44 | 7.60 × 10−8 | .019 |
| TBCA (rs10514128) × PTau | 4.25 | 0.00003 | 1.52 | 0.130 | 5.39 | 9.52 × 10−8 | .024 |
| EML1 (rs2273704) × PTau | 4.39 | 0.00001 | 1.23 | 0.220 | 2.28 | 1.74 × 10−7 | .029 |
FWE – p value when correcting for total number of comparisons using the bonferroni procedure.
Figure 1. POT1 (rs4728029) modifies the relationship between PTau and Lateral Inferior Ventricle Dilation.
The Y axis represents annual change in ventricular volume in mm3. The × axis represents CSF phosphorylated tau in pg/mL. Points and lines are color coded by rs4728029 genotype. The R2 linear for A/A carriers is 0.17, for A/G or G/A carriers it is 0.06 and for G/G carrier it is 0.02.
3.3 Hierarchical Linear Regression Result
Results are presented in Table 3. Our statistical model without APOE, rs4728029, or the rs4728029 × PTau interaction term explained 34% of variance in ventricular dilation. APOE genotype explained an additional 0.9% of variance. A weak, though significant, relationship between APOE and longitudinal change in volume is consistent with previous findings in which APOE was not related to volume loss in normal controls or MCI and only showed a moderate effect in AD.36 The main effect of rs4728029 explained an additional 0.1% of variance. Finally the rs4728029 × PTau interaction term explained an additional 2.5% of variance.
Table 3.
Post-hoc Hierarchical Linear Regression Result
| Change Statistics | ||||||||
|---|---|---|---|---|---|---|---|---|
| Model | R | R2 | Adj. R2 | R2 Change | F Change | df1 | df2 | Sig. F Change (P value) |
| 1a | 0.589 | 0.347 | 0.341 | 0.347 | 31.35 | 6 | 693 | 4.33 × 10−6 |
| 2b | 0.594 | 0.353 | 0.346 | 0.006 | 6.27 | 1 | 692 | 0.013 |
| 3c | 0.597 | 0.356 | 0.347 | 0.004 | 1.28 | 3 | 690 | 0.282 |
| 4d | 0.598 | 0.357 | 0.347 | 0.001 | 1.10 | 1 | 689 | 0.296 |
| 5e | 0.618 | 0.382 | 0.371 | 0.025 | 27.67 | 1 | 688 | 1.92 × 10−7 |
Predictors: Constant, Intracranial Volume, Phosphorylated Tau, Age, Education, Diagnosis, Gender
Predictors: Constant, Intracranial Volume, Phosphorylated Tau, Age, Education, Diagnosis, Gender, APOE
Predictors: Constant, Intracranial Volume, Phosphorylated Tau, Age, Education, Diagnosis, Gender, APOE, eigenvector1, eigenvector2, eigenvector3
Predictors: Constant, Intracranial Volume, Phosphorylated Tau, Age, Education, Diagnosis, Gender, APOE, eigenvector1, eigenvector2, eigenvector3, rs4728029
Predictors: Constant, Intracranial Volume, Phosphorylated Tau, Age, Education, Diagnosis, Gender, APOE, eigenvector1, eigenvector2, eigenvector3, rs4728029, rs4728029 × PTau
3.4 Post-hoc Statistical Analyses
3.4.1 Cognitive Performance
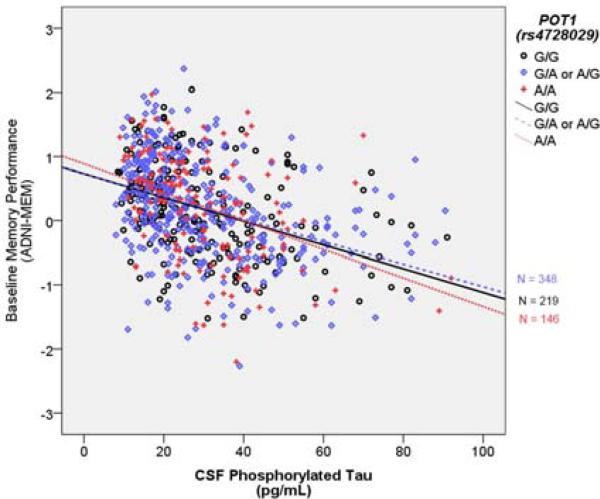
In the executive function analysis, the rs4728029 × PTau interaction term did not reach statistical significance. However, in the memory analysis the rs4728029 × PTau interaction term was statistically significant (t = −2.22, p = 0.026). This effect did not survive correction for multiple comparisons. PTau was related to poor memory performance, and the effect was amplified in homozygous carriers of the rs4728029 A allele (Figure 2).
Figure 2. POT1 (rs4728029) modifies relationship between PTau and Baseline Memory Performance.
The Y axis represents baseline memory performance. The × axis represents CSF phosphorylated tau in pg/mL. Points and lines are color coded by the rs4728029 genotype. The R2 linear for A/A carriers is 0.15, for A/G or G/A it is 0.141, and for G/G it is 0.167.
3.4.2 Neuroinflammatory Cytokine Analysis
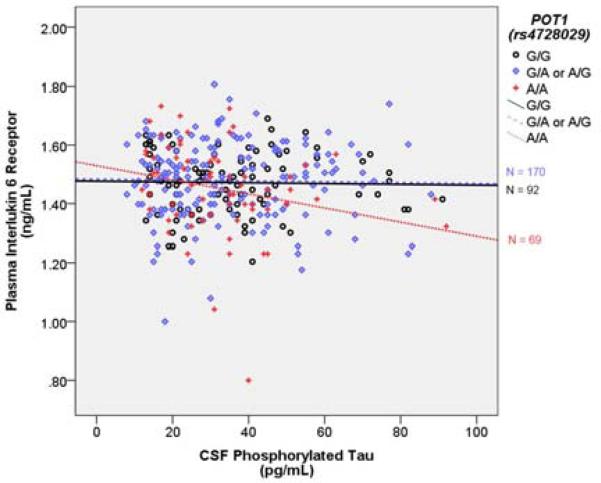
Proteomic data were only available from ADNI-1. There was no association between the rs4728029 × PTau interaction term and any of the neuroinflammatory cytokines. We also ran the model with rs4728029 recessively coded as A/A = 1 and all other genotypes = 0, and in this model, the interaction term approached nominal statistical significance (t = 1.950, p = 0.052). This effect did not survive a correction for multiple comparisons. PTau was related to lower levels of IL-6R, but only in A/A homozygotes (Figure 3).
Figure 3. POT1 (rs4728029) modifies relationship between PTau and Interleukin 6 Receptor.
The Y axis represents baseline plasma interlukin 6 receptor levels in ng/mL. The × axis represents CSF phosphorylated tau levels in pg/mL. Points and lines are color coded by the rs4728029 genotype. The R2 linear for A/A carriers is 0.062, for G/A or A/G carriers it is 0.0004, and for G/G carriers it is 0.0005.
4. DISCUSSION
The present project has replicated the relationship previously observed between CSF levels of phosphorylated tau and neurodegeneration.37,38 We have also identified a novel genetic-PTau interaction by which variation in a SNP annotated to POT1 modified the observed relationship between PTau and ventricular dilation. This interaction remained significant after correcting for multiple comparisons. Finally, we demonstrated that this SNP × PTau interaction was nominally associated with memory performance.
4.1 Genetic Modification of PTau-related Neurodegeneration
Across diagnostic categories, increased levels of PTau were related to increased ventricular dilation, although this effect was most pronounced in the MCI diagnostic group (the diagnosis by PTau interaction was not statistically significant, p = 0.16). The genetic modification of this effect highlighted a sub-group of individuals who showed the strongest relationship between PTau and neurodegeneration. This effect was not driven by a low frequency of homozygous minor allele carriers, as A/A allele carriers of rs4728029 made up 20% of the total sample (146 subjects) and the interaction effect remained present when stratifying across the two independent data sources. Importantly, although the rs4728029 SNP did not show a strong main effect on ventricular dilation, the PTau interaction explained 2.4% of variance above and beyond known predictors of neurodegeneration including the APOE- ε4 genotype. This highlights how the inclusion of genetic interaction effects, particularly when focusing on common variants, can improve prediction models and provide some suggestions as to the mechanisms of risk and resilience as outlined below.
The correlative nature of the current analyses precludes our ability to make definitive statements about the mechanisms of the observed interaction; however, some possible hints can be garnered from the rich data sources now available on SNP function. Rs4728029 is an intergenic SNP upstream from Protection of Telomeres 1 (POT1) and downstream from the metabotropic glutamate receptor 8 (GRM8). This SNP modifies three regulatory motifs and has the strongest effect on the CEBPB_known2 motif (Transfac M00201).39 In the case of this motif, the A allele of rs4728029 is associated with a strong decrease in regulatory action, suggesting that the A allele in the present analyses may be associated with lower levels of POT1. Previous research has demonstrated that POT1 expression is related to telomere length, with lower levels of POT1 expression showing a relationship to shorter telomeres,40 and the gene product of POT1 has demonstrated a similar relationship to telomere length.41 Interestingly, length of telomeres has also been associated with Alzheimer's disease status in previous research.42 Alzheimer's patients had shorter telomeres in T-cells than controls, and T-cell telomere length was inversely correlated with a serum marker of neuroinflammation. In posthoc analyses, we further investigated these possibilities by looking for similar POT1 modification of the relationship between Ptau and both cognitive performance and levels of neuroinflammatory cytokines in serum.
4.2 Posthoc Analyses: Cognitive Performance and Neuroinflammation
Beyond the increased variance explained in ventricular dilation in our primary analyses, our posthoc analyses suggested the POT1 × PTau interaction nominally predicts cognitive performance. The rs4728029 SNP modified the relationship between PTau and memory performance, though it did not have an effect on executive function performance. If such an effect is indeed driven by the ventricular dilation observed in our primary analysis, then it is not surprising that the effect is present in relation to memory rather than executive function, particularly given the crucial memory structures that surround the lateral inferior ventricles including the hippocampus. Moreover, the observed interaction was consistent with our primary finding as A/A allele carriers showed both more dilation in the inferior lateral ventricle in relation to PTau load and poorer memory performance at baseline in relation to PTau. These two factors are, of course, highly correlated (Pearson's r = −0.55), and for that reason should not be interpreted as independent signals as both are likely related to a single underlying factor. In both cases, PTau was related to dilation and memory performance in all genotype subgroups, but showed the strongest relationship in the A/A group. Given the relationship between POT1 and neuroinflammatory cytokines previously mentioned42 and the relationship between neuroinflammation and brain volume43 we chose to investigate whether our observed effect may be driven by such a neuroinflammatory response.
Previous research has demonstrated a relationship between telomere length and tumor necrosis factor alpha (TNF-α), Interleukin-6 and Interleukin-1.42,44 For that reason we chose to focus on the TNF-α and Interleukin cytokines available in our dataset. We were underpowered to look for such gene-biomarker interactions in relation to cytokines (only available in a sub-sample of ADNI-1 participants), so our results should be interpreted with caution. We were able to observe a trend gene × PTau interaction in line with our cognitive and brain volume findings, as A/A allele carriers of rs4728029 showed lower levels of the Interleukin-6 receptor (IL-6R) in relation to increased levels of PTau. IL-6R has previously been suggested to be an anti-inflammatory marker,45 and indeed, the association observed in this post-hoc analysis suggests in persons with lower levels of this anti-inflammatory marker (and possibly higher neuroinflammation), PTau leads to even higher levels of neurodegeneration. Soluble IL-6R appears to play a role in a large number of both pro- and anti-inflammatory activities mediated by IL-6,46 and these posthoc results must be interpreted within the many actions of the IL-6/IL-6R complex. The present results suggest that future work clarifying the relationship between PTau load, neuroinflammation, and neurodegeneration is warranted.
4.3 Previous GWAS of CSF markers and Resilience in ADNI
Previous GWAS analyses have looked for single marker predictors of Tau and PTau levels in CSF using data from ADNI. The earliest studies highlighted strong single marker effects in and around the APOE gene.18 Although we did not use PTau as our outcome, we did verify that APOE was strongly associated with PTau (p = 1.9 × 10−11). As highlighted in our hierarchical model, the effect of APOE on LILV slopes was more modest, particularly when including PTau in the prediction model (p = .013). This is consistent with the modest predictive power of APOE in relation to longitudinal change in bilateral hippocampal volume and ventricular volume reported previously.36,47 An additional GWAS reported a strong association between a SNP within APOE and a variety of baseline volume endophenotypes including the right hippocampus.48 This suggests future interaction analyses using a variety of smaller regions of interest may be warranted.
The most comprehensive GWAS of CSF biomarkers to date included data from ADNI and four other datasets,15 and reported a strong relationship between signals on chromosome 19 and biomarker levels. Interestingly, they also highlighted particular siginals within the APOE region that are related to Tau pathology independent of Aβ42 pathology or clinical status. In the present manuscript, the strong relationship between PTau and APOE, along with the moderate association between APOE and ventricular volume might explain why the strongest gene interaction effects identified do not cluster around the APOE signal. Moreover, this key difference highlights the strength of gene-biomarker interactions in identifying novel candidates for future analyses.
In addition, previous work by Cruchaga et al. demonstrated that genetic variation associated with CSF biomarker levels can meaningfully predict disease progression.14 Taken together, the results of the current manuscript and both Cruchaga et al. manuscripts suggest that genetic risk factors must be considered at all stages of the Alzheimer's disease cascade in order to fully elucidate the genetic etiology of LOAD. Genetic variants may increase susceptibility to the primary pathologies of LOAD, to neurodegeneration in the presence of a given pathology as presented in this manuscript, or even to cognitive decline in the presence of neurodegeneration (as reported most recently by Mukherejee et al.).49
4.4 Strengths and Weaknesses
The present manuscript presents a novel genetic interaction approach making use of MRI, CSF, and genotyping data. Combining the two independent data sources allowed us to maximize our power to detect genetic effects, and the stratified post-hoc analysis provides support for the consistency of the observed effect. The lack of consistency for some of the SNP interactions may be due to differences in the ADNI-1 and ADNI-2/GO protocols. For example, the longitudinal follow-up scans were slightly more frequent in ADNI-2/GO (approximately every 3 months v. approximately every 6 months in ADNI-1) but covered a shorter period of time. Certainly this longer follow-up interval in ADNI-1 could explain the cohort differences we observe, but other differences, such as the distribution of diagnostic categories in the two cohorts, cannot be ruled out.
In addition, it is possible that differences in CSF batches between ADNI-1 and ADNI-2/GO could be driving the differences in effect size between the two groups. In the current analysis, it is difficult to distinguish batch effects from group differences because we used the first CSF observation for each subject (thus the batches align roughly with ADNI-1 subjects v. ADNI-2/GO subjects). However, it should be noted that previous work has demonstrated the test-retest reliability of the biomarker measures from CSF in the ADNI dataset. 31 An additional independent sample with a comparable longitudinal follow-up interval to that of ADNI-1 could help clarify whether the cohort differences observed are simply due to longitudinal power, or differences due to batch effects, MRI follow-up interval, or sample characteristics.
Our posthoc analyses highlight a possible mechanism of the observed genetic interaction based on alterations in the neuroinflammatory response. It should be noted that these posthoc effects did not survive correction for multiple comparisons. The current project was not powered to look for all possible SNP effects across the entire proteomic panel, and the trend seen with IL-6R should be interpreted conservatively as a hint at a potential mechanism. Future functional work confirming the mechanism of the observed SNP interaction will be necessary to the underlying mechanism.
Future work replicating our findings in an independent sample with MRI, CSF, and genotype data is needed to confirm the observed SNP effect. To avoid possible confounding factors related to population substructure we chose to restrict all analyses to Caucasian individuals, and thus our results may not generalize to other ancestral populations. The statistical approach taken in the present manuscript provides a blue-print for future exploratory analyses aimed at identifying genetic modifiers of disease risk using other AD biomarkers such as amyloid load measured using PET and provides some possible targets for future functional analyses.
Research in Context.
Systematic Review
The authors performed a comprehensive review of existing work categorizing the relationship between phosphorylated tau (PTau) and brain volume, as well as genetic modifiers of this relationship. Previous research had been somewhat mixed with some studies finding only trend level associations between CSF tau/PTau and brain volume. No work to date has investigated genetic modifiers of this relationship beyond the APOE genotype.
Interpretation
Our results further clarify the relationship between CSF PTau and brain volume, identify a novel genetic interaction that explains substantial variance in brain atrophy, and identify a possible mechanism of the observed interaction related to a neuroinflammatory response.
Future Directions
Although subdividing our dataset into ADNI-1 and ADNI-2/GO suggested the interaction was consistent, a replication of this gene-biomarker interaction is warranted. Future functional analyses investigating the relationship between POT1 and the neuroinflammatory response is also necessary to confirm our suggested mechanism of action.
ACKNOWLEDGEMENTS
The authors report no conflicts of interest. This research was supported in part by the Vanderbilt NIMH Neurogenomics Training grant (T32 MH65215), the Vanderbilt Medical Scientist Training Program (T32 GM07347), the Recruitment for Genetic Aging Research (P30 AG036445), and the Pharmaceutical Research and Manufacturers of America Foundation Fellowship in Translational Medicine and Therapeutics. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Data collection and sharing for this project was funded by ADNI (National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Amorfix Life Sciences Ltd;AstraZeneca; Bayer HealthCare; BioClinica, Inc; Biogen Idec Inc; Bristol-Myers Squibb Company; Eisai Inc; Elan Pharmaceuticals Inc; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc; GE Healthcare; Innogenetics, N.V.; IXICO Ltd; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc; Merck & Co, Inc; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc; Servier; Synarc Inc; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation or the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for NeuroImaging at the University of California, Los Angeles. This research was also supported by NIH grants P30 AG010129 and K01 AG030514.
Appendix A
Genotyping
Quality control (QC) and statistical analyses were performed using PLINK software (version 1.07).22 During QC, we excluded single nucleotide polymorphisms (SNPs) with a genotyping efficiency < 90%, a minor allele frequency < 10%, or deviation from Hardy-Weinberg Equilibrium (p< 1e−6). This left 515,383 SNPs in ADNI-1 and 605,317 SNPs in ADNI-2/GO. Finally, we merged the two genotyping files for our analyses and again applied filters for genotyping efficiency < 90% and minor allele frequency < 10%. Subjects were excluded if they had a call rate < 98%, if there was a reported versus genetic sex inconsistency, or if relatedness to another sample was established (PI_HAT > 0.5).
Quantification of Ventricular Dilation
The MRI processing pipeline has been described elsewhere.23 Cortical reconstruction and volumetric segmentation were performed with the Freesurfer image analysis suite version 4.3 (http://surfer.nmr.mgh.harvard.edu/).26-30 Freesurfer processing in ADNI has been described in detail elsewhere.24 An early version of the longitudinal image processing framework was used to process the sequential scans.30 We excluded volumetric data from subjects who did not pass ventricle QC as outlined in the ADNI Freesurfer processing pipeline from the ADNI MRI core (http://adni.loni.ucla.edu/wp-content/uploads/2010/12/ADNI_UCSF_Freesurfer-Overview-and-QC.pdf). Past work has demonstrated that, when measured longitudinally, the left inferior lateral ventricle shows greater dilation than the right, both in AD patients and in controls.19 For that reason we chose to focus on the left ventricle to reduce the total number of comparisons, while allowing for maximal power in detecting genetic effects related to progression. Slopes of change in left ventricular volume over time were calculated in SAS 9.3 (SAS Institute Inc., Cary, NC) using mixed model regression (PROC MIXED) to leverage the longitudinal data available. In the mixed model regression, time was modeled based on days from baseline for each subject. This was then rescaled so that slopes would represent annual change (days from baseline/365.25). On average we had 4 MRI scans for each subject (3.66 for subjects in ADNI1 and 4.62 for subjects in ADNI2/ADNIGO).
CSF PTau Preprocessing
We compiled a dataset across the UPENN1 – UPENN5 data sources available for download on the ADNI site. For the present analyses we used the first measure of PTau for each subject, given its previous association with neurofibrillary tangle pathology.25 Two subjects had PTau values greater than three standard deviations beyond the mean levels in both the AD and MCI groups and were excluded from our analyses.
Hierarchical Linear Regression to Identify Variance Explained
In order to place our observed SNP interaction effects in the context of known predictors of brain volume, we used hierarchical linear regression to calculate the change in R2 when adding in the SNP main effect, and after adding in the SNP × PTau interaction term. For this model we also chose to include the APOE genotype as a predictor to determine whether our observed SNP effects explained variance beyond this known major genetic predictor. Although we restricted analyses to Caucasian individuals, we also included population structure covariates in this model (calculated using Structure: http://pritch.bsd.uchicago.edu/structure.html)33 to ensure there was no effect of ancestry contributing to the observed interaction.
Slopes of ventricular volume were again set as the quantitative outcome. Our first block included baseline age, gender, years of education, baseline ICV, baseline diagnostic category, and PTau. Our second block added in the APOE genotype (number of E4 alleles). Our third block added in the population structure eigenvectors. Our fourth block added in the SNP main effect term, and our final block added in the SNP × PTau interaction term. We report the variance explained by the final block in this model, which is the variance explained by the SNP × PTau interaction term.
Posthoc Cognitive Performance Analysis
Composite measures of memory and executive function were calculated previously in ADNI and are described elsewhere.34,35 We performed one cognitive analysis using memory as the outcome (ADNI-MEM) and one analysis using executive function as the outcome (ADNI-EF) to test whether the observed interaction effects were associated with cognitive performance. The outcome measure was either baseline ADNI-EF or baseline ADNI-MEM, and predictors included baseline age, gender, years of education, diagnostic group, APOE, PTau, SNP, and the SNP × PTau interaction term.
Posthoc Neuroinflammatory Cytokine Analysis
Plasma based proteomic markers have been previously calculated in ADNI and are described elsewhere (http://adni.loni.ucla.edu/wp-content/uploads/2010/11/BC_Plasma_Proteomics_Data_Primer.pdf). For posthoc analyses we chose to focus on all interleukin (IL) and tumor necrosis factor alpha (TNF-α) markers that passed QC leaving 7 markers for analysis: TNF-α, IL-3, IL-8, IL-13, IL-17, IL-18, and IL-6 receptor (IL-6R). Each of these was a quantitative outcome measure in regression models using the same predictors as the cognitive post-hoc analyses. We again corrected for multiple comparisons using the Bonferroni procedure.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braak H, Del Tredici K. The pathological process underlying Alzheimer's disease in individuals under thirty. Acta Neuropathologica. 2011;121:171–181. doi: 10.1007/s00401-010-0789-4. [DOI] [PubMed] [Google Scholar]
- 2.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Hypothetical model of dynamic biomarkers of the Alzheimer's pathological cascade. Lancet Neurology. 2010;9:119. doi: 10.1016/S1474-4422(09)70299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jack CR, Jr, Knopman DS, Jagust WJ, et al. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. The Lancet Neurology. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braak H, Braak E. Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiology of Aging. 1997;18:351–357. doi: 10.1016/s0197-4580(97)00056-0. [DOI] [PubMed] [Google Scholar]
- 5.Driscoll I, Troncoso J. Asymptomatic Alzheimers Disease: A Prodrome or a State of Resilience? Current Alzheimer Research. 2011;8:330–335. doi: 10.2174/156720511795745348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iacono D, O'Brien R, Resnick SM, et al. Neuronal hypertrophy in asymptomatic Alzheimer disease. Journal of neuropathology and experimental neurology. 2008;67:578. doi: 10.1097/NEN.0b013e3181772794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Codispoti KT, Beason-Held LL, Kraut MA, et al. Longitudinal brain activity changes in asymptomatic Alzheimer disease. Brain and Behavior. 2012;2:221–230. doi: 10.1002/brb3.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampert EJ, Roy Choudhury K, Hostage CA, et al. Prevalence of Alzheimer's Pathologic Endophenotypes in Asymptomatic and Mildly Impaired First-Degree Relatives. PloS one. 2013;8:e60747. doi: 10.1371/journal.pone.0060747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagan AM, Mintun MA, Mach RH, et al. Inverse relation between in vivo amyloid imaging load and cerebrospinal fluid Abeta42 in humans. Annals of neurology. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 10.Forsberg A, Engler H, Almkvist O, et al. PET imaging of amyloid deposition in patients with mild cognitive impairment. Neurobiology of Aging. 2008;29:1456–1465. doi: 10.1016/j.neurobiolaging.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 11.Jagust WJ, Landau SM, Shaw LM, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature genetics. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cruchaga C, Kauwe JS, Mayo K, et al. SNPs associated with cerebrospinal fluid phospho-tau levels influence rate of decline in Alzheimer's disease. PLoS genetics. 2010;6:e1001101. doi: 10.1371/journal.pgen.1001101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruchaga C, Kauwe J, Harari O, et al. GWAS of Cerebrospinal Fluid Tau Levels Identifies Risk Variants for Alzheimers Disease. Neuron. 78(2):256–268. doi: 10.1016/j.neuron.2013.02.026. 4-24-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kauwe JSK, Wang J, Mayo K, et al. Alzheimer's disease risk variants show association with cerebrospinal fluid amyloid beta. Neurogenetics. 2009;10:13–17. doi: 10.1007/s10048-008-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kauwe JSK, Cruchaga C, Karch CM, et al. Fine mapping of genetic variants in BIN1, CLU, CR1 and PICALM for association with cerebrospinal fluid biomarkers for Alzheimer's disease. PloS One. 2011;6:e15918. doi: 10.1371/journal.pone.0015918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim S, Swaminathan S, Shen L, et al. Genome-wide association study of CSF biomarkers Abeta-42, t-tau, and p-tau181p in the ADNI cohort. Neurology. 2011;76:69–79. doi: 10.1212/WNL.0b013e318204a397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thompson PM, Hayashi KM, de Zubicaray GI, et al. Mapping hippocampal and ventricular change in Alzheimer disease. Neuroimage. 2004;22:1754–1766. doi: 10.1016/j.neuroimage.2004.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Jack CR, Shiung MM, Gunter JL, et al. Comparison of different MRI brain atrophy rate measures with clinical disease progression in AD. Neurology. 2004;62:591–600. doi: 10.1212/01.wnl.0000110315.26026.ef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koran MI, Hohman TJ, Meda SA, et al. Genetic interactions within inositol-related pathways are associated with longitudinal changes in ventricle size. Journal of Alzheimer's Disease. 2013 doi: 10.3233/JAD-130989. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jack CR, Bernstein MA, Fox NC, et al. The Alzheimer's disease neuroimaging initiative (ADNI): MRI methods. Journal of Magnetic Resonance Imaging. 2008;27:685–691. doi: 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mormino EC, Kluth JT, Madison CM, et al. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132:1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buerger K, Ewers M, Pirttilä T, et al. CSF phosphorylated tau protein correlates with neocortical neurofibrillary pathology in Alzheimer's disease. Brain. 2006;129:3035–3041. doi: 10.1093/brain/awl269. [DOI] [PubMed] [Google Scholar]
- 26.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis: I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 27.Desikan RS, Segonne F, Fischl B, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 2006;31:968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 28.Fischl B, Sereno MI, Dale AM. Cortical Surface-Based Analysis: II: Inflation, Flattening, and a Surface-Based Coordinate System. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 29.Fischl B, Sereno MI, Tootell RB, et al. High-resolution intersubject averaging and a coordinate system for the cortical surface. Human brain mapping. 1999;8:272–284. doi: 10.1002/(SICI)1097-0193(1999)8:4<272::AID-HBM10>3.0.CO;2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reuter M, Schmansky NJ, Rosas HD, et al. Within-subject template estimation for unbiased longitudinal image analysis. Neuroimage. 2012;61(4):1402–18. doi: 10.1016/j.neuroimage.2012.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaw LM, Vanderstichele H, Knapik-Czajka M, et al. Qualification of the analytical and clinical performance of CSF biomarker analyses in ADNI. Acta Neuropathologica. 2011;121:597–609. doi: 10.1007/s00401-011-0808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramanan VK, Risacher SL, Nho K, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry. 2013 doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crane PK, Carle A, Gibbons LE, et al. Development and assessment of a composite score for memory in the Alzheimer's Disease Neuroimaging Initiative (ADNI). Brain Imaging and Behavior. 2012:1–15. doi: 10.1007/s11682-012-9186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gibbons LE, Carle AC, Mackin RS, et al. A composite score for executive functioning, validated in Alzheimer's Disease Neuroimaging Initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging and Behavior. 2012:1–11. doi: 10.1007/s11682-012-9176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuff N, Woerner N, Boreta L, et al. MRI of hippocampal volume loss in early Alzheimer's disease in relation to ApoE genotype and biomarkers. Brain. 2009;132:1067–1077. doi: 10.1093/brain/awp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Souza LC, Chupin M, Lamari F, et al. CSF tau markers are correlated with hippocampal volume in Alzheimer's disease. Neurobiology of Aging. 2012;33:1253–1257. doi: 10.1016/j.neurobiolaging.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 38.Fagan AM, Head D, Shah AR, et al. Decreased cerebrospinal fluid Abeta-42 correlates with brain atrophy in cognitively normal elderly. Annals of neurology. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Research. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo T, Oue N, Yoshida K, et al. Expression of POT1 is associated with tumor stage and telomere length in gastric carcinoma. Cancer research. 2004;64:523–529. doi: 10.1158/0008-5472.can-03-1196. [DOI] [PubMed] [Google Scholar]
- 41.Ning X, Yang S, Wang R, et al. POT1 deficiency alters telomere length and telomere-associated gene expression in human gastric cancer cells. European Journal of Cancer Prevention. 2010;19:345–351. doi: 10.1097/CEJ.0b013e32833b4812. [DOI] [PubMed] [Google Scholar]
- 42.Panossian LA, Porter VR, Valenzuela HF, et al. Telomere shortening in T cells correlates with Alzheimer's disease status. Neurobiology of Aging. 2003;24:77–84. doi: 10.1016/s0197-4580(02)00043-x. [DOI] [PubMed] [Google Scholar]
- 43.Jefferson AL, Massaro JM, Wolf PA, et al. Inflammatory biomarkers are associated with total brain volume The Framingham Heart Study. Neurology. 2007;68:1032–1038. doi: 10.1212/01.wnl.0000257815.20548.df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calvert PA, Liew TV, Gorenne I, et al. Leukocyte telomere length is associated with high-risk plaques on virtual histology intravascular ultrasound and increased proinflammatory activity. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31:2157–2164. doi: 10.1161/ATVBAHA.111.229237. [DOI] [PubMed] [Google Scholar]
- 45.Ferrucci L, Cherubini A, Bandinelli S, et al. Relationship of plasma polyunsaturated fatty acids to circulating inflammatory markers. Journal of Clinical Endocrinology & Metabolism. 2006;91:439–446. doi: 10.1210/jc.2005-1303. [DOI] [PubMed] [Google Scholar]
- 46.Jones SA, Horiuchi SANK, TOPLEY NICH, et al. The soluble interleukin 6 receptor: mechanisms of production and implications in disease. The FASEB Journal. 2001;15:43–58. doi: 10.1096/fj.99-1003rev. [DOI] [PubMed] [Google Scholar]
- 47.Nestor SM, Rupsingh R, Borrie M, et al. Ventricular enlargement as a possible measure of Alzheimer's disease progression validated using the Alzheimer's disease neuroimaging initiative database. Brain. 2008;131:2443–2454. doi: 10.1093/brain/awn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen L, Kim S, Risacher SL, et al. Whole genome association study of brain-wide imaging phenotypes for identifying quantitative trait loci in MCI and AD: a study of the ADNI cohort. Neuroimage. 2010;53:1051. doi: 10.1016/j.neuroimage.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mukherjee S, Kim S, Ramanan VK, et al. Gene-based GWAS and biological pathway analysis of the resilience of executive functioning. Brain Imaging and Behavior. 2013:1–9. doi: 10.1007/s11682-013-9259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]