Abstract
Calcium signaling is one of the most extensively employed signal transduction mechanisms in life. As life evolved into increasingly complex organisms, Ca2+ acquired more extensive and varied functions. Here, we compare genes encoding proteins that govern Ca2+ entry and exit across cells or organelles within organisms of early eukaryotic evolution into fungi, plants, and animals. Recent phylogenomics analyses reveal a complex Ca2+ signaling machinery in the apusozoan protist Thecamonas trahens, a putative unicellular progenitor of Opisthokonta. We compare T. trahens Ca2+ signaling to that in a marine bikont protist, Aurantiochytrium limacinum, and demonstrate the conservation of key Ca2+ signaling molecules in the basally diverging alga Cyanophora paradoxa. Particularly, our findings reveal the conservation of the CatSper channel complex in Au. limacinum and C. paradoxa, suggesting that the CatSper complex likely originated from an ancestral Ca2+ signaling machinery at the root of early eukaryotic evolution prior to the unikont/bikont split.
Keywords: animals, calcium channels, calcium signaling, CatSper, eukaryotes, evolution
Calcium ions (Ca2+) serve as a universal signal to modulate almost every aspect of cellular function in bacteria (Dominguez 2004), plants (Kudla et al. 2010), fungi (Zelter et al. 2004), and animals (Berridge et al. 2003; Clapham 2007). The core principles of Ca2+ signaling emerged very early in the life processes of bacteria (Case et al. 2007; Verkhratsky and Parpura 2014). The appearance of eukaryotes with intracellular organelles and the independent evolution of multicellular organisms from diverse ancestral unicellular lineages (Rokas 2008) prompted the refinement of versatile and complex Ca2+ signaling systems, which provide precise spatial and temporal control of Ca2+ concentration.
Although the basic principles of Ca2+ signaling appear to be universal in eukaryotes (Shemarova and Nesterov 2005), the components of the Ca2+ signaling machinery exhibit significant differences among distinct groups of organisms such as animals (Clapham 2007), plants (Nagata et al. 2004; Verret et al. 2010), and fungi (Zelter et al. 2004). In large skeletal and cardiac muscle cells, a highly distributed intracellular Ca2+ store, released by ryanodine receptor channels, was coupled directly or indirectly to plasma membrane voltage-gated Ca2+ (CaV) channels (Amador et al. 2013). Another specialized distribution system along the sperm flagella evolved via the sperm-specific CatSper Ca2+ channels that are critical for sperm cell hyperactivation (Lishko et al. 2012). It has been argued that in contrast to the complex Ca2+ signaling machinery in animals, plants and fungi have adopted more simplified Ca2+ signaling cascades to suit their physiological requirements (Nagata et al. 2004; Zelter et al. 2004). The origin and evolution of these distinct Ca2+ signaling machineries has remained a long-standing question—they might have evolved independently of one another, or they might share the same evolutionary origin in the last common unicellular ancestor with subsequent lineage-specific evolution after divergence.
Recent phylogenomics studies of close unicellular relatives of metazoans including two choanoflagellates Monosiga brevicollis and Salpingoeca rosetta and the filasterean Capsaspora owczarzaki have revealed the presence of ancestral signaling molecules previously thought to be restricted to animals (King et al. 2008; Sebe-Pedros et al. 2011; Fairclough et al. 2013; Suga et al. 2013). Monosiga brevicollis also has an extensive Ca2+ signaling system resembling that in animals (Cai 2008). Animals, fungi, and their unicellular relatives form the eukaryotic supergroup Opisthokonta (Stechmann and Cavalier-Smith 2002; Steenkamp et al. 2006; Ruiz-Trillo et al. 2008). The apusozoan protist, Thecamonas trahens, is a putative unicellular progenitor of Opisthokonta (Cavalier-Smith and Chao 2010). We recently identified a complex ancestral Ca2+ signaling network in T. trahens that thus predates the divergence of animals and fungi (Cai and Clapham 2012).
The eukaryotic root has been hypothesized to be placed between Unikonta, the eukaryotic supergroup composed of Opisthokonta and Amoebozoa, and Bikonta, the eukaryotic supergroup including Archaeplastida (plants and relatives) and Chromalveolata (fig. 1), and other groups (Stechmann and Cavalier-Smith 2002; Burki and Pawlowski 2006; Derelle and Lang 2012). Comparative genomics studies of several plant, algal, and other bikont species showed the conservation and divergence of many Ca2+ signaling molecules such as CaV channels, transient receptor potential (TRP) channels, and ligand-gated channels among animals, fungi, plants, and many unicellular organisms (Wheeler and Brownlee 2008; Verret et al. 2010; Chan et al. 2011). Thus, it has been demonstrated that many components of the Ca2+ signaling machineries in animals, plants, and fungi likely emerged from early eukaryotic lineages before their divergence.
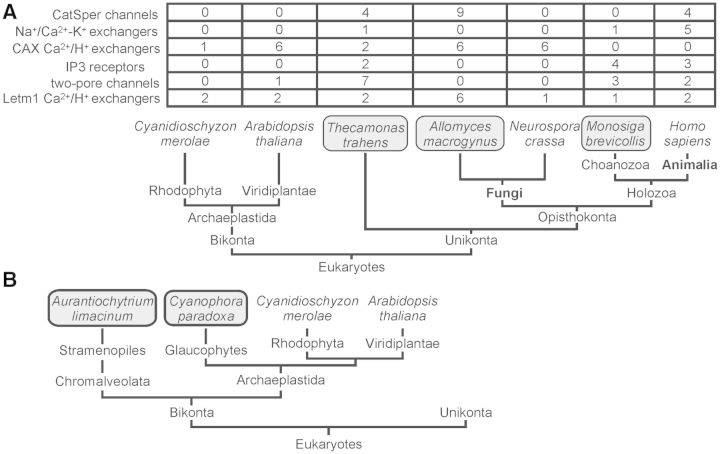
Fig. 1.
Distribution of Ca2+ signaling molecules in eukaryotes. (A) Comprehensive Ca2+ signaling network in the apusozoan protist Thecamonas trahens (Cai and Clapham 2012). Shown are representative classes of Ca2+ signaling molecules previously known to be animal specific, such as CatSper channels and K+-dependent Na+/Ca2+ exchangers, or fungal/plant specific, such as CAXs, or ubiquitous, such as mitochondrial Letm1 Ca/H exchangers (Tsai et al. 2014), from three recently analyzed genomes—T. trahens, the choanoflagellate Monosiga brevicollis (Cai 2008), and the basal fungus Allomyces macrogynus (Cai and Clapham 2012), as well as genomes of Homo sapiens, Neurospora crassa, Arabidopsis thaliana, and Cyanidioschyzon merolae (Wheeler and Brownlee 2008; Verret et al. 2010). (B) Schematic diagram illustrating the evolutionary history of the marine thraustochytrid protist Aurantiochytrium limacinum and the basally diverging alga Cyanophora paradoxa (Price et al. 2012) in Bikonta. Inferred from the Tree of Life project (http://www.tolweb.org/, last accessed July 11, 2014) and recent references on the eukaryotic tree (Burki and Pawlowski 2006; Derelle and Lang 2012). Letm1, leucine zipper-EF-hand containing transmembrane protein 1.
Several key types of Ca2+ signaling molecules conserved in T. trahens have not been extensively analyzed. For instance, CatSper channels and Na+/Ca2+ exchangers are present in T. trahens, the basal fungus Allomyces macrogynus and most animals but are absent in land plants such as Arabidopsis thaliana, Cyanidioschyzon merolae (a unicellular red alga), and most fungi (fig. 1A). To elucidate whether the ancestral Ca2+ signaling network shown in T. trahens is also preserved in bikonts, we examined related genomic databases of organisms in Bikonta to search for homologs of Ca2+ signaling molecules.
An Extensive Ca2+ Signaling Machinery in Aurantiochytrium limacinum
Aurantiochytrium limacinum is a common marine thraustochytrid protist within the class of labyrinthulomycetes, which is one of the earliest diverging lineages in the phylum of stramenopiles (Riisberg et al. 2009) (fig. 1B). We found that the genome of Au. limacinum encodes Ca2+ signaling machinery as comprehensive as that in T. trahens (fig. 2 and supplementary fig. S1, Supplementary Material online) including CatSper channels, CaV channels, ligand-gated channels, second messenger-gated channels, and TRP channels. Components of the animal Ca2+-release-activated Ca2+ channel complex—stromal interaction molecules (STIM) and Orai channels—are absent in Au. limacinum and T. trahens. Both Au. limacinum and T. trahens possess intracellular ion channels—inositol 1,4,5-trisphosphate receptors and two-pore channels but not ryanodine receptors. In addition, Au. limacinum and T. trahens contain a complete set of Ca2+ exchange systems—Ca2+ ATPases and three classes of the cation/Ca2+ exchangers (Cai and Lytton 2004)—the K+-independent Na+/Ca2+ exchangers, the K+-dependent Na+/Ca2+ exchangers, and the Ca2+/H+ exchangers (CAXs). Here, we focus our analysis on the early evolution of the CatSper channel complex. Description of Ca2+ signaling molecules other than the CatSper complex is discussed in Supplementary Material online.
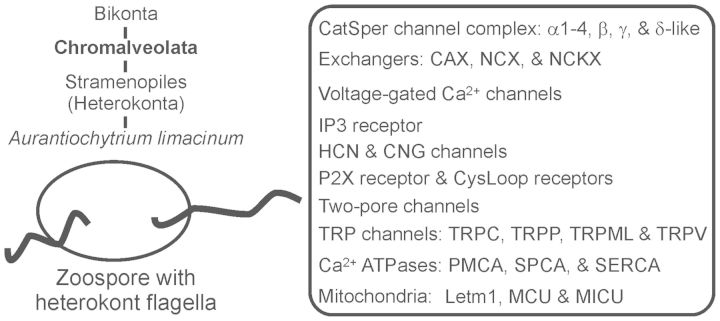
Fig. 2.
Ca2+ signaling machinery in Aurantiochytrium limacinum. The evolutionary relationship of A. limacinum is inferred from the Tree of Life project (http://www.tolweb.org/) and the JGI genome portal (http://genome.jgi-psf.org/Aurli1/Aurli1.home.html, last accessed July 11, 2014). Aurantiochytrium limacinum life stages include colonies of vegetative cells or zoospores with heterokont flagella (http://syst.bio.konan-u.ac.jp/labybase/Aurantiochytrium_limacinum_life_cycle.html, last accessed July 11, 2014). CatSper, sperm-associated cation channel; CNG, cyclic nucleotide-gated channel; CysLoop receptors, cysteine-loop ligand-gated receptor; IP3 receptor, inositol 1,4,5-trisphosphate receptor; Letm1, leucine zipper-EF-hand containing transmembrane protein 1; MCU, mitochondrial Ca2+ uniporter; MICU, mitochondrial EF hand Ca2+ uniporter regulator; NC(K)X, Na+/Ca2+ (K+-dependent) exchanger; P2X receptor, P2X purinergic receptor; PMCA, plasma membrane Ca2+ ATPase; SERCA, sarco/endoplasmic reticulum Ca2+ ATPase; SPCA, secretory pathway Ca2+ ATPase.
The CatSper Ca2+ Channel Complex
The typical “9 + 2” motile flagellum in human spermatozoa is conserved across species and likely evolved in the last common ancestor of all eukaryotic organisms (Mitchell 2007). Studies of flagellar axonemes have focused on sperm cells from unikont model organisms—marine invertebrates such as sea urchins and tunicates and also in bikont protists such as Chlamydomonas and Paramecium (Inaba 2011). Basic flagellar motility is controlled by the active sliding of paired outer doublet microtubules coupled with ATP hydrolysis by axonemal dyneins, which does not require the elevation of Ca2+ concentration or even an intact plasma membrane. In contrast, Ca2+ is necessary for complex flagellar activities such as capacitation, chemotaxis, and hyperactivated motility (Lishko et al. 2012).
As the most biochemically complex ion channel known to date, the CatSper complex is located in the principle piece of the sperm flagellum (fig. 3A). Ca2+ influx through CatSpers is essential for sperm cell hyperactivated motility and male fertility in mammals (Lishko et al. 2012). CatSpers are present in sea urchins and tunicates but have not been identified previously in any bikonts (Cai and Clapham 2008).
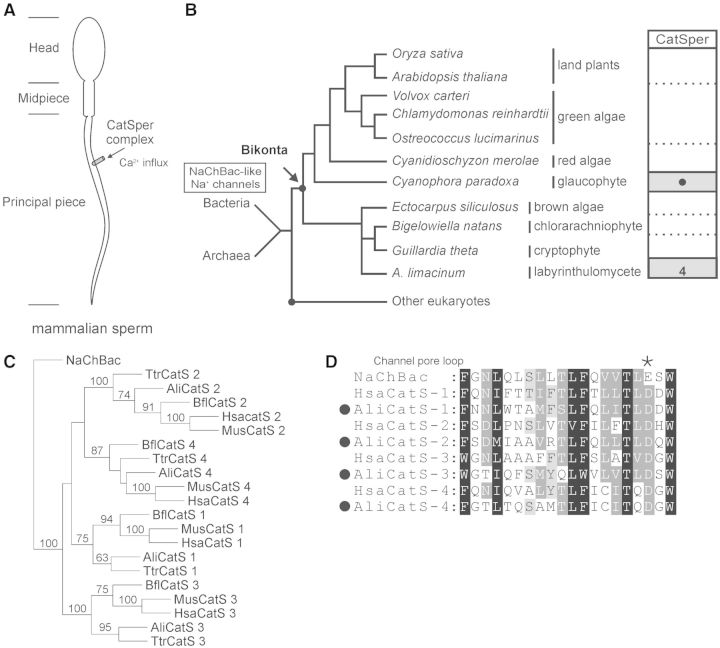
Fig. 3.
Identification of CatSper channels in Aurantiochytrium limacinum. (A) Schematic representation of mammalian spermatozoa. The CatSper channel complex is located at the principle piece that is critical for Ca2+-modulated sperm motility. (B) Distribution of CatSpers in select bikont species. Aurantiochytrium limacinum contains CatSper α subunits 1–4, whereas the exact number of CatSper α subunit in Cyanophora paradoxa is unknown due to incomplete genome assembly. (C) Phylogenetic relationship of CatSper α subunits. A maximum likelihood phylogenetic tree (LG + I + G + F model) showing the relationship of protist and animal homologs of CatSper α subunits 1–4 from the thraustochytrid protist A. limacinum (Ali), the amphioxus Branchiostoma floridae (Bfl), Homo sapiens (Hsa), Mus musculus (Mus), and the apusozoan protist Thecamonas trahens (Ttr). NaChBac, a prokaryotic voltage-gated Na+ channel isolated from Bacillus halodurans (Ren et al. 2001), was used as an outgroup. Bootstrap values above 60 are shown at the nodes. (D) Sequence alignment of the pore loop regions of NaChBac and CatSper α subunits from A. limacinum (Ali) and H. sapiens (Hsa). The asterisk symbol indicates the location of a key acidic residue important for ion selectivity. CatS, CatSper α subunit.
Except in the basal fungus A. macrogynus (Cai and Clapham 2012), all known CatSper channel complexes are composed of four pore-forming α subunits. Loss of either of the four α subunits by gene knockout results in male infertility and the complete loss of the whole protein complex (Qi et al. 2007). Similarly, numerous cases of lineage-specific simultaneous loss of all four CatSper α subunit and auxiliary subunit genes are present throughout metazoan evolution (Cai and Clapham 2008). The mammalian CatSper complex also contains at least three auxiliary subunits—CatSper-β, CatSper-γ, and CatSper-δ, all of which are sperm-specific transmembrane proteins (Lishko et al. 2012).
We found that Au. limacinum contains the same components of the CatSper channel complex in T. trahens; these include the pore-forming α subunits 1–4, auxiliary β and γ subunits, and a distantly related homolog of the δ subunit (figs. 2 and 3B and C, supplementary fig. S1, Supplementary Material online). The loop regions containing the key acidic residue aspartate are highly conserved in CatSper α subunits from Au. limacinum to humans (fig. 3D), suggesting the high Ca2+ selectivity observed in mammalian CatSpers is likely retained in Au. limacinum. The sequence regions outside the transmembrane segments and the pore loop of CatSpers are poorly conserved across species, which possibly indicates that modulation of complex flagellar motility differs in a species-specific manner.
The highly conserved composition of the CatSper complex between Unikonta and Bikonta suggests that CatSpers might play an important role in regulating flagellar activity in ancestral protists prior to the beginning of eukaryotic radiation, much earlier than previously thought (Cai and Clapham 2008). Alternatively, it remains a possibility that the presence of the CatSper complex in Au. limacinum might be caused by a horizontal gene transfer event from basal fungal species. In humans and in A. macrogynus, sperm cell motility can be regulated by Ca2+ influx in response to progesterone (Lishko et al. 2011; Strunker et al. 2011) or sex pheromones (Pommerville et al. 1990), respectively. Activation of the CatSper complex by environmental signals such as progesterone and pH changes induces Ca2+ influx in human sperm.
Thraustochytrid protists reproduce solely by means of biflagellate zoospores (fig. 2) (Raghukumar and Damare 2011). It is conceivable that in thraustochytrid protists such as Au. limacinum, modulation of cell motility by the CatSper complex prompts biflagellate zoospores to move toward environmental cues (Fan et al. 2002). The CatSper complex is absent in most bikonts and fungi. Although the basic “9 + 2” flagellar structure is highly conserved across bikonts and unikonts, the protein composition of flagellar axonemes and signaling cascades that regulate flagellar motility may have been modified to fit different physiological environments (Inaba 2011).
The CatSper channel pore-forming α subunits and NC(K)X and CAXs are believed to be closely related to their prokaryotic counterparts, the NaChBac Na+ channel (Clapham and Garbers 2005), and the YRBG exchanger (Philipson and Nicoll 2000), respectively. The identification of these genes in Au. limacinum and T. trahens supports a prokaryotic genesis of ancestral eukaryotic signaling systems (Shpakov and Pertseva 2008). It is speculated that gene duplication gave rise to four unique CatSper channel pore-forming α subunits and three classes of exchangers—K+-dependent Na+/Ca2+ exchangers, K+-independent Na+/Ca2+ exchangers, and CAXs. Indeed, two diatom genomes contain four and three copies of NaChBac-type channels, which are believed to have been acquired through horizontal gene transfer of an ancestral NaChBac-like channel from a prokaryote into the common ancestor of diatoms, followed by subsequent gene duplications (Verret et al. 2010). Notably, these diatom NaChBac-type channels exhibit sequence divergence that might render them more selective for Ca2+, a function adapted for the CatSper complex. Recent phylogenetic analyses of ion selectivity between animal and bacterial ion channels also suggest that NaChBac-type bacterial channels are closely related to CatSper channels (Liebeskind et al. 2013).
Conservation of Key Components of Ca2+ Signaling System in Cyanophora paradoxa
The glaucophyte Cyano. paradoxa is a basally diverging, photosynthetic freshwater alga that emerged before the divergence of green plants and red algae (Price et al. 2012). The land plant Ar. thaliana and the unicellular red alga C. merolae are known to retain very simplified Ca2+ signaling networks (Verret et al. 2010) (fig. 1A). We found conservation of several key Ca2+ signaling molecules in the Cyano. paradoxa genome including homologs of CatSper channels, CaV channels, and TRP channels, all of which are absent in Ar. thaliana and C. merolae (figs. 1A and 3B and supplementary fig. S1, Supplementary Material online). Cyanophora paradoxa lacks homologs of P2XRs, inositol 1,4,5-trisphosphate receptors, and NC(K)X exchangers, which are all present in Au. limacinum and T. trahens. The identification of CatSper homologs in two bikont groups—Archaeplastida (Cyano. paradoxa) and Chromalveolata (Au. limacinum)—further supports the ancestral evolution of the CatSper complex.
In summary, we have demonstrated that the bikont protist Au. limacinum possess an extensive Ca2+ signaling machinery comparable to that in the unikont protist T. trahens. Consistent with previous reports (Wheeler and Brownlee 2008; Verret et al. 2010), our findings also suggest that the extensive network of Ca2+ signaling molecules observed in animals originated in ancestral protists before the eukaryotic radiation, prior to the divergence of animals, fungi, and plants. The ancestral Ca2+ signaling machinery containing the CatSper channel complex is generally conserved in the lineage leading to animals and can also be occasionally identified in very few protists such as T. trahens and Au. limacinum. Although many ancestral Ca2+ signaling molecules were subsequently lost during the evolution of modern protists, plants and fungi, traces of relatively conserved Ca2+ signaling systems harboring CatSpers can still be found in basally divergent species, for instance, the basal chytridiomycete fungus A. macrogynus (Cai and Clapham 2012) and the basally diverging glaucophyte Cyano. paradoxa. Characterization of the function and regulation of these ancestral homologs will provide novel insight into the understanding of their physiological roles and the evolution of the ancestral Ca2+ signaling machinery.
Supplementary Material
Supplementary information and figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the Department of Energy Joint Genome Institute (http://genome.jgi.doe.gov) and investigators of the genome projects listed on the JGI genome portal and the C. paradoxa genome project (Price et al. 2012) for making data publicly available. X.C. also thanks Yanhong Zhang for excellent technical assistance and for critical reading of the manuscript.
References
- Amador FJ, Stathopulos PB, Enomoto M, Ikura M. Ryanodine receptor calcium release channels: lessons from structure-function studies. FEBS J. 2013;280:5456–5470. doi: 10.1111/febs.12194. [DOI] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Burki F, Pawlowski J. Monophyly of Rhizaria and multigene phylogeny of unicellular bikonts. Mol Biol Evol. 2006;23:1922–1930. doi: 10.1093/molbev/msl055. [DOI] [PubMed] [Google Scholar]
- Cai X. Unicellular Ca2+ signaling ‘toolkit’ at the origin of metazoa. Mol Biol Evol. 2008;25:1357–1361. doi: 10.1093/molbev/msn077. [DOI] [PubMed] [Google Scholar]
- Cai X, Clapham DE. Evolutionary genomics reveals lineage-specific gene loss and rapid evolution of a sperm-specific ion channel complex: CatSpers and CatSperβ. PLoS One. 2008;3:e3569. doi: 10.1371/journal.pone.0003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Clapham DE. Ancestral Ca2+ signaling machinery in early animal and fungal evolution. Mol Biol Evol. 2012;29:91–100. doi: 10.1093/molbev/msr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lytton J. The cation/Ca2+ exchanger superfamily: phylogenetic analysis and structural implications. Mol Biol Evol. 2004;21:1692–1703. doi: 10.1093/molbev/msh177. [DOI] [PubMed] [Google Scholar]
- Case RM, Eisner D, Gurney A, Jones O, Muallem S, Verkhratsky A. Evolution of calcium homeostasis: from birth of the first cell to an omnipresent signalling system. Cell Calcium. 2007;42:345–350. doi: 10.1016/j.ceca.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EE. Phylogeny and evolution of apusomonadida (protozoa: apusozoa): new genera and species. Protist. 2010;161:549–576. doi: 10.1016/j.protis.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Chan CX, Reyes-Prieto A, Bhattacharya D. Red and green algal origin of diatom membrane transporters: insights into environmental adaptation and cell evolution. PLoS One. 2011;6:e29138. doi: 10.1371/journal.pone.0029138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Clapham DE, Garbers DL. International union of pharmacology. L. nomenclature and structure-function relationships of CatSper and two-pore channels. Pharmacol Rev. 2005;57:451–454. doi: 10.1124/pr.57.4.7. [DOI] [PubMed] [Google Scholar]
- Derelle R, Lang BF. Rooting the eukaryotic tree with mitochondrial and bacterial proteins. Mol Biol Evol. 2012;29:1277–1289. doi: 10.1093/molbev/msr295. [DOI] [PubMed] [Google Scholar]
- Dominguez DC. Calcium signalling in bacteria. Mol Microbiol. 2004;54:291–297. doi: 10.1111/j.1365-2958.2004.04276.x. [DOI] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan KW, Vrijmoed LL, Jones EB. Zoospore chemotaxis of mangrove thraustochytrids from Hong Kong. Mycologia. 2002;94:569–578. doi: 10.1080/15572536.2003.11833185. [DOI] [PubMed] [Google Scholar]
- Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod. 2011;17:524–538. doi: 10.1093/molehr/gar034. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origins of metazoan multicellularity. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla J, Batistic O, Hashimoto K. Calcium signals: the lead currency of plant information processing. Plant Cell. 2010;22:541–563. doi: 10.1105/tpc.109.072686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebeskind BJ, Hillis DM, Zakon HH. Independent acquisition of sodium selectivity in bacterial and animal sodium channels. Curr Biol. 2013;23:R948–R949. doi: 10.1016/j.cub.2013.09.025. [DOI] [PubMed] [Google Scholar]
- Lishko PV, Botchkina IL, Kirichok Y. Progesterone activates the principal Ca2+ channel of human sperm. Nature. 2011;471:387–391. doi: 10.1038/nature09767. [DOI] [PubMed] [Google Scholar]
- Lishko PV, Kirichok Y, Ren D, Navarro B, Chung JJ, Clapham DE. The control of male fertility by spermatozoan ion channels. Annu Rev Physiol. 2012;74:453–475. doi: 10.1146/annurev-physiol-020911-153258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DR. The evolution of eukaryotic cilia and flagella as motile and sensory organelles. Adv Exp Med Biol. 2007;607:130–140. doi: 10.1007/978-0-387-74021-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Iizumi S, Satoh K, Ooka H, Kawai J, Carninci P, Hayashizaki Y, Otomo Y, Murakami K, Matsubara K, et al. Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Mol Biol Evol. 2004;21:1855–1870. doi: 10.1093/molbev/msh197. [DOI] [PubMed] [Google Scholar]
- Philipson KD, Nicoll DA. Sodium-calcium exchange: a molecular perspective. Annu Rev Physiol. 2000;62:111–133. doi: 10.1146/annurev.physiol.62.1.111. [DOI] [PubMed] [Google Scholar]
- Pommerville JC, Strickland JB, Harding KE. Pheromone interactions and ionic communication in gametes of aquatic fungus Allomyces macrogynus. J Chem Ecol. 1990;16:121–131. doi: 10.1007/BF01021274. [DOI] [PubMed] [Google Scholar]
- Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, et al. Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants. Science. 2012;335:843–847. doi: 10.1126/science.1213561. [DOI] [PubMed] [Google Scholar]
- Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, Kirichok Y, Ramsey IS, Quill TA, Clapham DE. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A. 2007;104:1219–1223. doi: 10.1073/pnas.0610286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghukumar S, Damare VS. Increasing evidence for the important role of Labyrinthulomycetes in marine ecosystems. Bot Mar. 2011;54:3–11. [Google Scholar]
- Ren D, Navarro B, Xu H, Yue L, Shi Q, Clapham DE. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. doi: 10.1126/science.1065635. [DOI] [PubMed] [Google Scholar]
- Riisberg I, Orr RJ, Kluge R, Shalchian-Tabrizi K, Bowers HA, Patil V, Edvardsen B, Jakobsen KS. Seven gene phylogeny of heterokonts. Protist. 2009;160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Rokas A. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet. 2008;42:235–251. doi: 10.1146/annurev.genet.42.110807.091513. [DOI] [PubMed] [Google Scholar]
- Ruiz-Trillo I, Roger AJ, Burger G, Gray MW, Lang BF. A phylogenomic investigation into the origin of metazoa. Mol Biol Evol. 2008;25:664–672. doi: 10.1093/molbev/msn006. [DOI] [PubMed] [Google Scholar]
- Sebe-Pedros A, de Mendoza A, Lang BF, Degnan BM, Ruiz-Trillo I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol. 2011;28:1241–1254. doi: 10.1093/molbev/msq309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemarova IV, Nesterov VP. [Evolution of Ca(2+) signaling mechanisms. Role of calcium ions in signal transduction in lower eukaryotes] Zh Evol Biokhim Fiziol. 2005;41:303–313. [PubMed] [Google Scholar]
- Shpakov AO, Pertseva MN. Signaling systems of lower eukaryotes and their evolution. Int Rev Cell Mol Biol. 2008;269:151–282. doi: 10.1016/S1937-6448(08)01004-6. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297:89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- Steenkamp ET, Wright J, Baldauf SL. The protistan origins of animals and fungi. Mol Biol Evol. 2006;23:93–106. doi: 10.1093/molbev/msj011. [DOI] [PubMed] [Google Scholar]
- Strunker T, Goodwin N, Brenker C, Kashikar ND, Weyand I, Seifert R, Kaupp UB. The CatSper channel mediates progesterone-induced Ca2+ influx in human sperm. Nature. 2011;471:382–386. doi: 10.1038/nature09769. [DOI] [PubMed] [Google Scholar]
- Suga H, Chen Z, de Mendoza A, Sebé-Pedrós A, Brown MW, Kramer E, Carr M, Kerner P, Vervoort M, Sánchez-Pons N, et al. The Capsaspora genome reveals a complex unicellular prehistory of animals. Nat Commun. 2013;4:2325. doi: 10.1038/ncomms3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MF, Jiang D, Zhao L, Clapham D, Miller C. Functional reconstitution of the mitochondrial Ca2+/H+ antiporter Letm1. J Gen Physiol. 2014;143:67–73. doi: 10.1085/jgp.201311096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkhratsky A, Parpura V. Calcium signalling and calcium channels: evolution and general principles. Eur J Pharmacol. 2014 doi: 10.1016/j.ejphar.2013.11.013. 739C:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verret F, Wheeler G, Taylor AR, Farnham G, Brownlee C. Calcium channels in photosynthetic eukaryotes: implications for evolution of calcium-based signalling. New Phytol. 2010;187:23–43. doi: 10.1111/j.1469-8137.2010.03271.x. [DOI] [PubMed] [Google Scholar]
- Wheeler GL, Brownlee C. Ca2+ signalling in plants and green algae–changing channels. Trends Plant Sci. 2008;13:506–514. doi: 10.1016/j.tplants.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Zelter A, Bencina M, Bowman BJ, Yarden O, Read ND. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet Biol. 2004;41:827–841. doi: 10.1016/j.fgb.2004.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.