Abstract
There is a need to develop novel targeted imaging and therapeutic agents that can aid in early diagnosis, detection of metastasis and treatment of melanoma. Alpha-3 integrin is overexpressed in 82% of metastatic melanomas in humans and may be a potential target for peptide ligands carrying therapeutic agents. Five melanoma cell lines were generated from canine primary oral and metastatic canine tumors, grown in mice, and validated with melanoma markers Melan A, S-100, Micropthalmia transcription factor (MITF), Tyrosinase, and MART-1. The melanoma cell lines were tested for binding affinity to previously published alpha-3 integrin-binding peptides containing the cdGXGXXc motif. Fluorescent conjugates of the alpha-3 integrin binding OA02 peptide were used to quantify receptor affinity in the cell lines, a specimen of canine primary oral melanoma, and melanoma xenografts. Alpha-3 integrin was expressed by all 5 canine melanoma cell lines. Four of the 5 lines as well as the primary canine tumor showed affinity to alpha-3 integrin binding peptides with the cdGXGXXc motif. Optical imaging of canine melanoma xenografts in nude mice indicates rapid, strong uptake of the optical tracer in the tumor with an average persistence of approximately 48 hours. Ex vivo images showed high tumor-to-background ratio, with tumor signals more than twice that of the kidney and other vital organs. We propose that integrin alpha-3 integrin binding ligands could potentially become useful probes for imaging and delivery of cytotoxic agents for the treatment of melanoma.
Keywords: melanoma, peptides, integrin, optical imaging, canine
1. Introduction
Melanoma in pet dogs has been established as a relevant model for human melanoma (Paoloni and Khanna, 2008; Porrello et al., 2006). The annual incidence rates of melanoma in people is 12.6/100,000, compared to 25/100,000 in dogs (Vail and MacEwen, 2000). Although canine melanoma is not typically attributed to sun exposure as in humans, the tumor biology and therapeutic response of malignant phenotypes are similar to that in human melanoma. 85% of canine cutaneous melanomas are benign, while greater than 90% of melanomas arising from the oral cavity or mucocutaneous junction have a malignant biological behavior (Bolon et al., 1990). Oral melanoma may be melanotic or amelanotic in both canines and humans, and a major difference in survival rates occur between each phenotype (58% vs. 20% in humans).
Canine melanoma has become recognized as a cancer model that has concrete applications to translational research. Melanoma responded to a novel tyrosine kinase inhibitor (London et al., 2003) in a clinical trial, which had inhibitory activity toward several receptor tyrosine kinases common to human and canine cancers, including C-KIT and vascular endothelial growth factor receptor (VEGFR). A DNA-based melanoma vaccine has also been developed to treat canine melanoma, providing safety and efficacy data for similar therapy in humans (Bergman et al., 2006).
Overexpressed or differentially expressed tumor cell surface receptors make good targets for specific drug delivery. Inhibitors of mitogen activated protein kinase (MAPK) pathway have been investigated as activators of cell response, and c-Kit and CRAF have also been identified as potential targets (Smalley et al., 2009) for melanoma. Other strategies for tumor therapy include targeting cell surface integrins with antibodies, peptides, small molecules and peptidomimetics that can facilitate carriage of nanoparticle drugs or radiopharmaceuticals, delivered directly to the tumor while sparing normal cells. Human malignant melanoma has been shown to express ICAM-1, alpha-v-beta-3, and alpha-3 beta-1 integrins, which are significantly associated with tumor thickness and progression (Melchiori et al., 1995; Moretti et al., 2008; Natali et al., 1997; Natali et al., 1993). Alpha-3 integrin has also been shown to be overexpressed in 82% of metastatic melanoma lesions isolated from human patients (Natali et al., 1997). Further, alpha-3 integrin expression has been associated with increased cellular motility in cell culture as well as increasing the invasiveness and metastatic potential of melanoma cells in a nude mouse model (Johnson, 1999; Yoshinaga et al., 1993). These properties make the alpha-3 integrin a potential therapeutic target.
Research into targeted therapy using small molecules has provided new hope for improving therapeutic outcomes in cancer patients. High-affinity peptides have been developed to bind to the alpha-3 integrin cell surface receptor of ovarian cancer cells (Aina et al., 2005b). Given the similarities of canine and human melanoma in potential over expression of alpha-3 integrin, previously identified peptides and focused alpha-3 integrin One-Bead-One-Compound (OBOC) libraries (Aina et al., 2007) were tested for their ability to selectively bind to canine melanoma cells. The cdG-HCit-GPQc (“OA02”) peptide, when conjugated to near-infrared dyes or copper 64, has been shown to be a useful optical and PET imaging agent of alpha-3 integrin expressing ES-2 ovarian cancer xenografts in nude mice (Aina et al., 2007; Aina et al., 2002). This non-invasive imaging strategy can be used to determine ligand affinity to canine melanoma xenografts in vivo, and to evaluate the potential for diagnostic imaging and drug delivery in canine patients.
2. Materials and methods
2.1 Canine melanoma cell lines
Canine melanoma cell lines were generated from primary oral tumors (UCDK9M3, UCDK9M4), skin metastasis from a primary oral tumor (UCDK9M1), and lymph node metastasis from primary oral tumors (UCDK9M2, UCDK9M5). Immediately after biopsy the tissue was placed in enough Hank’s pen/strep and fungizone so that the tumor was completely covered. Using a scalpel blade the tumor was then minced in a petri dish.
The minced pieces were then transferred to a T25 flask with 12ml of enzyme solution, consisting of 2.3 U/ml Dispase (Gibco, Invitrogen, Carlsbad, CA) and 200U/ml Collagenase III (Gibco III, Invitrogen, Carlsbad, CA) in RPMI-1640 media supplemented with 10% heat-inactivated FBS (GIBCO, Invitrogen, Carlsbad, CA). A magnetic stir bar was placed in the flask and put on submersible stirrer in a 37°C water bath. The tissue was allowed to digest with stirrer on low setting for up to one hour. When most of tissue had dissolved it was then strained through a mesh sieve.
The dissociated cells were then added to a 50ml conical tube and spun down at 1500rpm for 5min. The supernatant was discarded and the pellet resuspend in 10mls of RPMI-10 media. 5 × 105 cells were then added to a T75 culture flask along with media consisting of 90% DMEM medium (High Glucose- GIBCO no. 11995, Invitrogen, Carlsbad, CA) supplemented with 10% heat-inactivated FBS (GIBCO, Invitrogen, Carlsbad, CA), 1% non-essential amino acids (MEM, Invitrogen, Carlsbad, CA), 50 U/mL penicillin and 50 mg/mL streptomycin (Penicillin-streptomycin, Invitrogen, Carlsbad, Calif.). Cells were detached by rinsing twice with Ca and Mg-free DPBS (Invitrogen, Carlsbad, Calif.), then treating with 0.05% Trypsin (Trypsin, Invitrogen, Carlsbad, Calif.), 0.53 mM Na-EDTA for 3–5 minutes, at room temperature or at 37°C. Cells were passaged a minimum of 20 times. Cells were then maintained in the DMEM growth medium listed above.
2.2 Test for binding of UCDK9M melanoma cell lines to peptide beads
Eight known alpha-3 integrin-binding peptides, one alpha-4 beta-1 targeting peptidomimetic (LLP2A), and one negative control peptide (cdG-Bta-GPYc), were tested for binding to the UCDK9M1-5 cell lines (Courtesy of Dr. Kit Lam’s laboratory; Table 1)(Aina et al., 2005b; Peng et al., 2008). Single compound peptide beads were prepared using the OBOC method described elsewhere (Lam et al., 1997). Seven of the nine alpha-3 integrin peptides had 100% surface concentration “substitution”, (cdG-Cha-G-Hcit-Qc, cdG-Chg-G-Hyp-Nc, cdGIGPQc, cdGLGQ-Bta-c, cdGMG-Hser-Nle-c, cdG-Tyr(3NO2)-GI-Pra-c, cdG-Bta-GPYc) and two had 10% surface substitution (cdG-Phe(4NO2)-GP-Cha-c, cdG-Hcit-GPQc). 50 µL sample of beads from each peptide were added to individual 2 mL fritted columns, washed in sterile PBS, water, 70% ethanol, and tissue culture medium. A 10 µL aliquot from each of the washed peptides was transferred into 5 separate multi-well tissue culture dishes. All cell lines were detached as described, counted, and approximately 106 cells/mL from each cell line were incubated with the peptide beads in each single multi-well dish. Plates were observed for cell binding at 30 minutes, 3 hours and 24 hours under a light microscope.
Table 1.
Results of screening canine melanoma cell lines UCDK9M1-5 with known alpha-3 integrin, alpha-4 integrin, and negative control peptides. Affinity of the cells to the beads is rated on a semi-quantitative visual scale of 1–4, with 1+ representing occasional cells bound to the beads, and 4+ representing cells bound to all beads in the field of view. The assay was evaluated at 30 minutes, 3 hours, and 24 hours after combining the cells and the beads.
| Peptide | Group | Substitution % | Time point | UCDK9M1 | UCDK9M2 | UCDK9M3 | UCDK9M4 | UCDK9M5 |
|---|---|---|---|---|---|---|---|---|
| cdG- Bta - GPYc |
negative | 100 | 30 min | − | − | +/− | +/− | +/− |
| 3 hrs | − | − | − | − | − | |||
| 24 hrs | − | − | − | − | − | |||
| cdG- Cha -G- Hcit-Qc |
alpha-3 | 100 | 30 min | ++++ | − | +/− | ++ | ++ |
| 3 hrs | ++++ | − | + | ++ | +++ | |||
| 24 hrs | ++++ | ++ | +++ | +++ | ++++ | |||
| cdG- Chg -G- Hyp-Nc |
alpha-3 | 100 | 30 min | ++++ | − | +++ | +++ | ++ |
| 3 hrs | ++++ | − | +++ | +++ | +++ | |||
| 24 hrs | ++++ | ++ | +++ | +++ | ++++ | |||
| cdG- Hcit- GPQc |
alpha-3 | 10 | 30 min | +++ | − | +/− | +/− | +/− |
| 3 hrs | +++ | − | − | − | + | |||
| 24 hrs | ++++ | +/− | +++ | +++ | + | |||
| cdGIGP Qc |
alpha-3 | 100 | 30 min | ++++ | − | + | ++ | ++ |
| 3 hrs | ++++ | − | +/− | + | ++ | |||
| 24 hrs | ++++ | +/− | +++ | +++ | ++++ | |||
| cdGLG Q-Bta-c |
alpha-3 | 100 | 30 min | ++++ | − | ++ | ++++ | ++ |
| 3 hrs | ++++ | − | +++ | +++ | +++ | |||
| 24 hrs | ++++ | +++ | +++ | +++ | ++++ | |||
| cdGMG- Hser- Nle-c |
alpha-3 | 100 | 30 min | ++++ | − | + | +++ | ++ |
| 3 hrs | +++ | − | ++ | +++ | +++ | |||
| 24 hrs | ++++ | +/− | +++ | +++ | ++++ | |||
| cdG- Phe(4N O2) -GP- Cha-c |
alpha-3 | 10 | 30 min | + | − | − | − | +/− |
| 3 hrs | + | − | − | + | +/− | |||
| 24 hrs | ++++ | − | ++ | +++ | + |
2.3 Screening of alpha-3 integrin OBOC libraries with UCDK9M melanoma cell lines
In order to identify novel peptide sequences that bind with specificity to canine melanomas, UCDK9M1 and UCDK9M2 cell lines were screened with 50 µL each of the cXGXGXXc 20% and 100% substitution libraries using the OBOC library screening method described elsewhere (Aina et al., 2005b). The beads and cells were incubated in complete RPMI 1640 medium in 100×50 mm plates in a humidified CO2 incubator. The cells and beads were agitated at 20 rpm for 4 hours and then incubated overnight without agitation. After 24 hours, positive beads were identified, isolated, stripped and cleaned. These beads were then re-bound to a normal human melanocytes cell line (NHEM, Cambrex BioScience, Walkersville, MD) which were maintained according tomanufacturer’s instructions. Beads that did not bind to normal melanocytes were then isolated and processed for sequencing.
2.4 Western Blot Analysis of whole cell lysates of UCDK9M cells for alpha-3 integrin protein
Western Blot analysis was performed to confirm the expression of alpha-3 integrin on UCDK9M1-5 cell lines using MAB2290 (Chemicon, Temecula, CA), a mouse monoclonal antibody to the alpha-3 integrin A subunit. The procedure has been described (Chen et al., 1995). Briefly, cell lysates were made with 2× SDA sample buffer and heated for 5 minutes. Proteins were then resolved in an SDS-PAGE gel and transferred onto nitrocellulose membranes. Membranes were then subjected to blocking, washing, antibody incubation, and detection by enhanced chemiluminescence (ChemiDoc-It Imaging System, UVP, Upland, CA).
2.5 Fluorescent staining of UCDK9M monolayer cells with “OA02”-biotin-SA-Alexa-488
UCDK9M1 and UCDK9M2 cells were grown as a monolayer on glass chamber slides in a humidified CO2 incubator. When cells were 70% confluent, slides were detached from the chambers, fixed with 90% methanol in water at −80°C overnight and then washed several times in PBS. Slides were blocked with 1mL each of avidin D and biotin (“Avidin-Biotin” blocking kit; Vector Labs, Burlingame, Calif.). A 10 µM aliquot of “OA02”-biotin peptide in 1mL media was added to the slides and incubated for 1 hour at room temperature in a humidified chamber. Subsequently slides were washed and incubated with 1:1000 dilution of Streptavidin-Alexa-488 (Invitrogen, Carlsbad, Calif.) in PBS for 15 minutes, washed and mounted with 4',6-diamidino-2-phenylindole (DAPI) counterstain and mounting medium (Vector Labs, Burlingame, Calif.). Slides were then observed under a fluorescent microscope camera (Zeiss, North Maple Grove, Minn.).
2.6 Fluorescent staining of canine primary mucosal amelanotic melanoma tumor biopsy
Several 4 um sections from a frozen canine primary oral melanoma were prepared. The tissue was fixed in acetone, dried and stored −4°C until used. Slides were warmed at room temp for 30 minutes, fixed in ice-cold acetone for 5 minutes, air dried for 30 minutes, and washed in PBS (containing calcium and magnesium) twice for 2 minutes. Slides were then blocked with 1%BSA/PBS (1ml/slide) for 30 minutes, rinsed in PBS for 5 minutes, blocked with avidin/ biotin block for 15 minutes each (rinsing for 2 minutes after each block). 10uM OA02-biotin in PBS was added (1ml/slide to positive control, PBS only to negative control) and incubated overnight at −4°C in a humidified chamber. Subsequently slides were rinsed twice in PBS and the secondary antibody Streptavidin-Alexa 488 (Invitrogen, Carlsbad, Calif.) was added at 1:500 dilution,1ml/slide for 30 minutes. Slides were rinsed in PBS, mounted with slow-fade mounting medium, and observed under a fluorescent microscope.
2.7 Generating Canine melanoma xenografts in nude mice
The following protocol was approved by the institution’s animal care and use committee. Six 4–6 week old athymic nude (Nu/Nu) mice were purchased from Harlan Sprague Dawley (Indianapolis, Indiana). Mice were housed in the animal facility and fed ad lib with rodent pellets and water. Melanoma cells were suspended in 100 µL incomplete 1– 2.5×106 cells subcutaneously on the shoulder and hip, or right and left flank, in up to three injection sites, while under mild inhalation anesthesia (Halothane, Sigma Aldrich, MO). Tumor latency varied for each cell line, however, imaging experiments commenced when tumors were approximately 10 mm3 in diameter. All procedures were conducted under an approved protocol according to guidelines specified by the National Institute of Health Guide for Animal Use and Care.
2.8 Immunohistochemistry with melanoma-associated antigens
Immunohistochemistry was performed on canine melanoma cell pellets and tumor xenograft sections of UCDK9M1, UCDK9M2, and UCDK9M5 cells using S100, GP100, MITF, Mart 1, Tyrosinase, and Melan A. For the cell pellets cells were expanded until they reached 70% confluency in six T185 flasks. Cell monolayers were detached by scraping the flasks with a cell scraper, rather than by trypsinization. Cells were centrifuged, washed in PBS, and the resultant cell pellet was gently transferred to the tip of a “tea bag” pouch. The cell pellet was then fixed in formalin and embedded in paraffin blocks. 4 um sections were cut from these blocks and mounted on “plus” coated slides and allowed to dry overnight. For the xenografts, 4 um sections were cut from paraffin embedded tissue blocks.
Sections were deparaffinized twice in xylene for 10 minutes each time, and rehydrated in a graded ethanol series to 70% ethanol. Slides were soaked 30 minutes in 0.3% hydrogen peroxidase in methanol to inactivate endogenous peroxidase, followed by a triple rinse in phosphate buffered saline (PBS). Slides were steamed in citrate buffer (Dako - Carpinteria, CA) in a steamer for 30 minutes, allowed to cool for 20 minutes, and triple rinsed in PBS. Serum-free protein block (Dako - Carpinteria, CA) was added to each slide for 10 minutes in moist chamber at room temperature. After draining off the protein block, the tissue sections were incubated with the following antibodies: S100α(Sigma, St. Louis, MO), diluted 1:750, Melanoma Ab-2 (gp100) (Neomarkers, Fremont, CA), diluted 1:25, Microphthalmia (Neomarkers, Fremont, CA), diluted 1:100, and Melan A (Novocastra, Newcastle, U.K.), diluted 1:100.
Each antibody was diluted with PBS and incubated overnight at 4°C in a moist chamber. After approximately 18 hrs, the slides were triple rinsed in PBS and incubated with the biotinylated secondary antibody (Goat anti-mouse IgG) (BioCare – Walnut Creek, CA) for 10 minutes in a moist chamber at room temperature. Slides were again triple rinsed with PBS and incubated with strepavidin-HRP (BioCare – Walnut Creek, CA) for 10 minutes in a moist chamber at room temperature. The slides were then soaked twice in PBS for 5 minutes. The chromagen Aminoethyl Carbazole (AEC) (Zymed – South San Francisco, CA) was added to each slide and monitored for staining for 1–3 minutes. Slides were rinsed in tap water, then in distilled water and counterstained in Meyers Hematoxylin (Sigma – St. Louis, MO) for 1 – 2 minutes. Slides were rinsed again in distilled water and coated with Cover Care aqueous mounting media (BioCare – Walnut Creek, CA) on a plate warmer for approximately ½ hour. Negative controls were obtained using purified mouse myeloma IgG1 (Zymed Laboratories - S. San Francisco, CA) in place of the primary antibody on each cell line.
2.9 In vivo Imaging of UCDK9M melanoma xenografts with “OA02”-Cy5.5
This procedure has been described in detail elsewhere (Aina et al., 2005a). Briefly, xenograft bearing mice were anesthetized intra-peritoneally using pentobarbitol sodium (Nembutal®, Western Medical Supply, Los Angeles, CA). All mice were imaged with 50µg of “OA02”-Cy5.5 (one UCDK9M2 mouse with cdG-Cha-G-Hcit-Qc-biotin-Streptavidin-Cy5.5 dissolved in 100 µL sterile saline solution) injected intravenously. Optical fluorescence imaging was performed serially at 0, 2, 24, 48, 96, and 168 hours on an IS2000MM image station (Kodak, Rochester, NY). After imaging was complete, the mice were sacrificed and tumors, organs, muscle and skin tissues were excised for ex vivo imaging. Operator-defined regions of interest (ROI) were drawn around each organ and mean optical intensity was obtained. To correct for background, signal from the blood (heart) was subtracted from signals of all other organs. A tumor-time contrast profile was created using the organ to tumor ratio (n=5) (GraphPad Prizm, La Jolla, CA).
3. Results
3.1 Test for binding of UCDK9M melanoma cell lines to peptide beads
UCDK9M1 bound very strongly to all the alpha-3 integrin peptides, but not to the negative control or the alpha-4 beta-1peptidomimetic LLP2A. With regards to the alpha-3 integrin peptides, binding was moderate to strong for lines UCDK9M3 and UCDK9M4. UCDK9M5 also had strong binding for alpha-3 integrin peptides except for the cdG-Phe(4NO2)-GP-Cha-c, and cdG-Tyr(3NO2)-GI-Pra-c peptides. Line UCDK9M2 on the other hand showed negative to very weak binding to all the alpha-3 integrin peptides except the cdGLGQ-Bta-c peptide. Results are summarized in Table 1.
3.2 Screening of alpha-3 integrin OBOC libraries with UCDK9M melanoma cell lines
Eleven beads were isolated from screening cXGXGXXc 20% substitution (high stringency) library against UCDK9M1 cell line. To eliminate those binding to non-neoplastic cells, these beads were re-screened with normal melanocytes. Two beads were selected for sequencing and further evaluation. The sequences of the two “UCDK9M1 specific” beads were cdG-Cha-G-I-Nle-c, and c(L/D) GLG-Phe(3-Cl)-Tc. These sequences matched originally published motifs for alpha-3 integrin binding ligands (Aina et al., 2007). Since high affinity beads were identified from the 20% substitution library, we saw no further need for re-screening this cell line with the lower stringency 100% substitution cXGXGXXc library.
No beads were isolated from screening the UCDK9M2 cell line with the cXGXGXXc 20% substitution (high stringency) library. We subsequently screened this cell line with the 100% substitution (low stringency) cXGXGXXc library and retested the isolated beads with normal melanocytes. Four beads with relatively good affinity to UCDK9M2 cells and no affinity to normal melanocytes were isolated and sequenced. The sequences of the peptides were c-Phe(3Cl)-G-Phe(3Cl)G-Chg-Nal1-c, cHG-Bpa-G (M/Q)-HoPhe-c, cDGRGA-Phe(4Me)-c, and cNGRGAYc. These peptides shared very little similarity with the initial eight cdGXGXXc peptide motifs identified as ligands to alpha-3 integrin.
3.3 Western Blot Analysis of whole cell lysates of UCDK9M cells for alpha-3 integrin protein
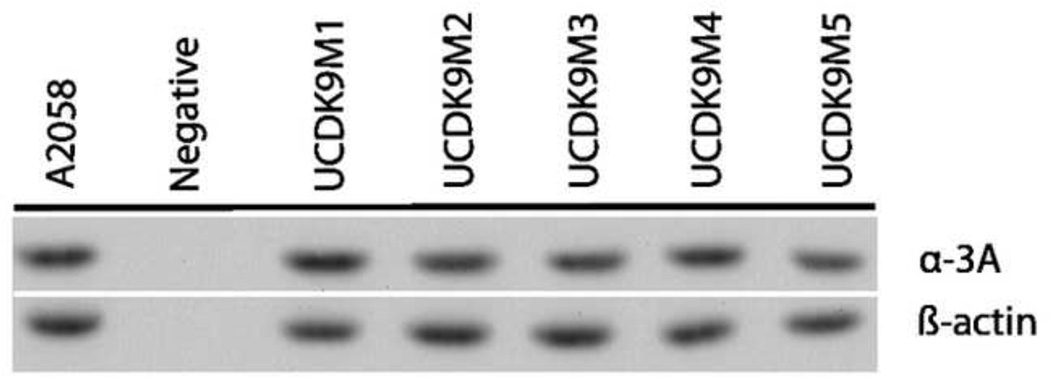
Western Blot analysis of each of the canine cell lines showed uniform expression of the alpha-3 A integrin subunit (Figure 1).
Figure 1.
Western Blot analysis of canine melanoma cell line expression of the alpha3 integrin A subunit. The cell lines were uniformly positive. The human melanoma cell line A2058 was used as a positive control. β-actin was used as a positive loading control.
3.4 Fluorescent staining of UCDK9M monolayer cells with “OA02”-biotin-SA-Alexa-488
A strong fluorescence signal was seen on the surface of the cells when UCDK9M1 were labeled with OA02-biotin-Streptavidin-Alexa-488. In some cells it appeared that the nucleus was also labeled but on closer examination of the DAPI slide these “bright spots” were overlapping cells. The UCDK9M2 cell line showed similar staining characteristics with the probe (data not shown).
3.5 Fluorescent staining of canine primary mucosal amelanotic melanoma tumor biopsy
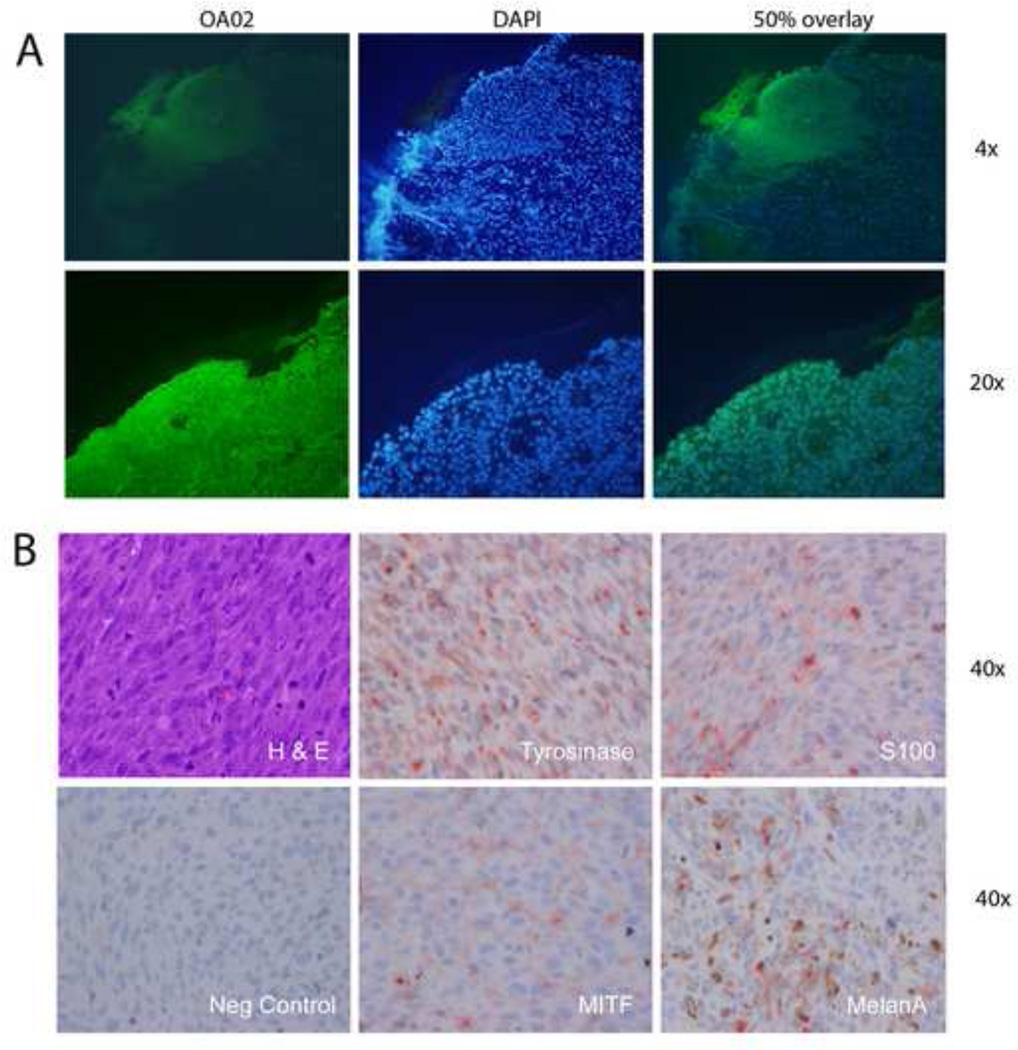
One section of frozen canine melanoma tumor tissue was stained with fluorescent-labeled α3-integrin peptide “OA02”-Streptavidin-Alexa 488. There was strong staining of the tumor cells on the oral mucosal surface and mild background staining of the lamina propria (Figure 2A),. The corresponding DAPI image showed that densely packed nuclei correlated with the tumor mass which corresponded to the peptide-labeled cells.
Figure 2.
Cell surface expression of primary canine melanoma and cell lines. A. Canine tumor tissue specimen labeled with OA02-Streptavidin-Alexa 488. The tumor cells on the mucosal surface showed strong staining. The DAPI image shows the densely packed tumor nuclei which correspond with the stained tumor in the overlaid image. Higher magnification of the tissue shows similar affinity to the ligand. B. The sample was positive for Tyrosinase, S100, MITF, and MelanA.
3.6 Immunohistochemistry with melanoma-associated antigens
UCDK9M1 was positive for S100, on both the cell and xenograft samples, positive for GP100 on the cell samples, and positive for Melan A, Mart 1 and Tyrosinase on the xenograft samples. UCDK9M2 was positive for MITF, S100, and Melan A on both samples, and positive for Tyrosinase on the xenograft sample only (Figure 2B). UCDK9M5 was positive for S100 on both samples, GP100 on the cell sample, and MITF, Melan A and Tyrosinase on the xenograft sample.
Canine melanoma xenografts in nude mice
Cell lines UCDK9M1, UCDK9M2, UCDK9M3 and UCDK9M5 grew subcutaneously, but with varying latencies. Time to tumor growth of about 10 mm3 varied from 2 weeks (in lines UCDK9M5 and UCDK9M1), to 8 weeks (UCDK9M2) and 12 weeks (UCDK9M3). Line UCDK9M4 did not grow subcutaneously after multiple attempts. Although all the cell lines grew as non-pigmented (amelanotic) cells in vitro, line UCDK9M2 grew as pigmented (melanotic) tumor xenografts in nude mice.
3.7 In vivo Imaging of melanoma xenografts with fluorescent labeled peptides
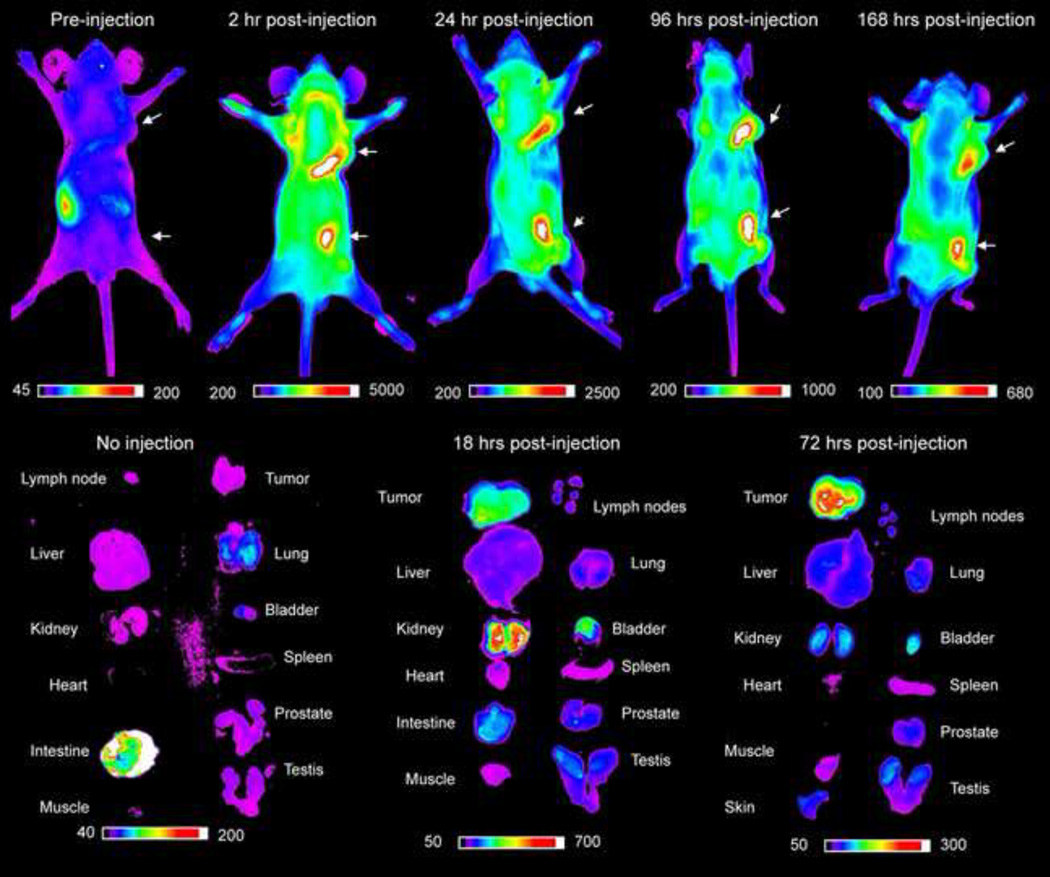
Six mice were imaged with “OA02”-Cy 5.5, and one mouse was imaged with cdG-Cha-G-Hcit-Qc-biotin-Streptavidin-Cy5.5. The tumors included 2 mice with UCDK9M1 xenografts, 1 mouse with UCDK9M2 (melanotic) xenografts, 1 mouse with UCDK9M3 xenografts, and 2 mice with UCDK9M5 xenografts. There were 12 amelanotic tumors and 3 melanotic tumors in total. In vivo imaging data showed tumor uptake of “OA02”-Cy5.5 tracer in 11/12 amelanotic tumors. On pre-injection images, a background signal of ~200 arbitrary units (A.U.) was seen in the gastrointestinal tract and can be attributed autoflorescence of alfalfa in the diet (Figure 3). As early as 10 minutes post injection a mild signal could be seen in the tumor (data not shown). The highest signal intensity was seen at 2 hours post injection (~5000 A.U.). At 24 hours post injection, there was still a strong fluorescence signal in the tumor even though the mean intensity was approximately half of the value at 2 hours. Between 48–168 hours, a much slower rate of decrease in mean intensity was observed, but mean intensity values never returned to background levels throughout the duration of the study. None of the 3 melanotic tumors showed any uptake with “OA02”-Cy5.5. However in vivo imaging of the same UCDK9M2 xenograft-bearing mouse with cdG-Cha-G-Hcit-Qc-biotin-Streptavidin-Cy5.5 tracer, showed a signal coming from the amelanotic edges of a single tumor that had both melanotic and amelanotic regions.
Figure 3.
In vivo imaging of canine melanoma xenograft tumors in nude mice with OA02-Cy5.5. The color scales represent fluorescent intensity detected with optical imaging, with the greatest fluorescence to the right of the scale. There is a high uptake of peptide in the tumor tissue in whole body images and ex-vivo images. The tumor remains strongly fluorescent up to 168 hours after injection.
Ex vivo images showed that without tracer injection, no signal was seen in the organs except for the intestine. The tracer was eliminated from the blood as seen by the lack of signal in the heart at 18 hours post injection. The probe was cleared primarily through the kidneys and the bladder with significant accumulation in the tumor. There was high tumor: background ratio at 72 hours (Figures 3 and 4). The cdG-Cha-G-Hcit-Qc-biotin-Streptavidin-Cy5.5 tracer had affinity to regions of amelanotic growth arising from the edges of a melanotic tumor.
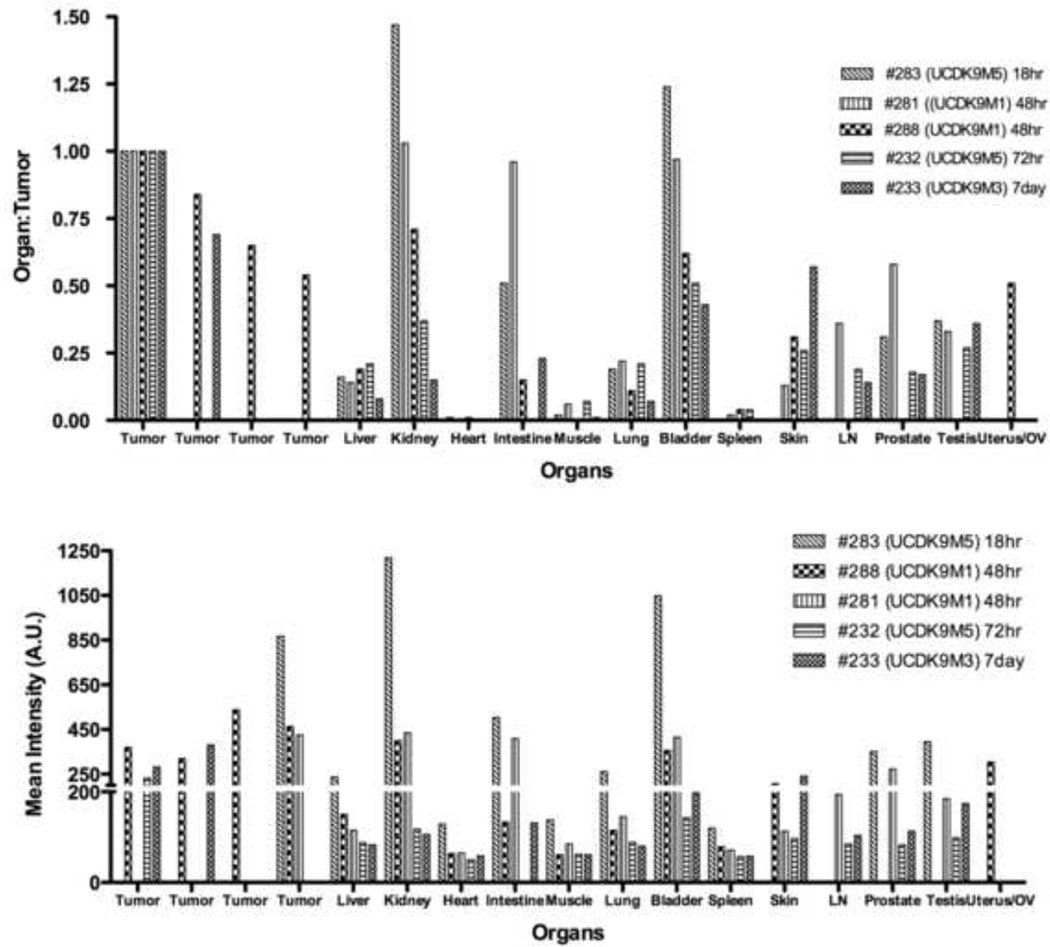
Figure 4.
Biodistribution of OA02-Cy 5.5 in tissues. Mice were sacrificed at 18 hours, 48 hours, 72 hours, and 7 days, and the fluorescent intensity for each organ and tissue was measured to display affinity of the peptide to tissues over time. Results of representative amelanotic tumors are displayed as organ to tumor ratio, and as optical intensity in semiquantitative arbitrary units (AU). The kidney and bladder had a high tumor:organ ratio and is the assumed route of excretion of the tracer.
4. Discussion
Alpha-3 integrin is present in canine melanoma cell lines as determined by Western Blot analysis. These tumor cell surface integrins can bind the “OA02” peptide and other alpha-3 integrin peptides from the cdGXGXXc library in vitro using bound bead assays, and in vivo using fluorescent-labeled optical imaging.
Although there was some difference in the early binding of “OA02” to different UCDK9M cell lines in vivo and in vitro, the expression of alpha-3 integrin receptor was similar across the cell lines. Our previous work suggests that our ligands bind specifically to the activated form of the integrin, since adding integrin activating cations (Ca2+ or Mn2+) did not qualitatively or quantitatively increase the cell binding to peptide beads.
The amino acids and their positions are important in specificity of the ligands’ binding to alpha-3 integrin receptor. The negative control peptide, cdG-Bta-GPYc, although differing from other binding peptides by as few as two amino acids, did not bind to any of the cell lines. The library screening results show that UCDK9M1 bound with high affinity to the high stringency cXGXGXXc 20% library, and the peptide sequence data obtained was equivelent to previously published sequences. This strongly supports the presence of similar alpha-3 integrin receptor conformation on the cell membrane.
None of the cell lines bound to the alpha-4 beta-1 integrin-specific peptidomimetic ligand, LLP2A, despite reports of increased gene expression and cell surface expression of alpha-4 integrin in human melanoma cell lines (Hoek et al., 2004; Klemke et al., 2007). This lack of affinity may be due to a species difference, post-translational protein expression, or conformational status of the receptor. Because of the uniformly negative binding of LLP2A to the canine melanoma cell lines, we did not pursue further investigations.
The cell line UCDK9M2 had the least affinity to all of the peptide beads, showing binding only after 24 hours. This again may indicate a difference in receptor concentration, conformation or post-translational modification, or a slower recovery/turnover of the receptor when the cells are detached from the growth substrate prior to bead binding experiments. In contrast, UCDK9M1, which had strong binding to most peptides at all time points examined. Phenotypically, UCDK9M1 showed amelanotic growth both in vivo and in vitro while UCDK9M2 was melanotic in vivo and amelanotic in vitro. Both UCDK9M1 and UCDK9M2 showed similar strong fluorescent membrane labeling pattern when stained on glass slides. One UCDK9M2 xenograft grew primarily as a melanotic tumor but phenotypically changed to amelanotic along the periphery (possibly the rapidly growing / leading edge of the mass). This area showed no affinity to “OA02”-Cy5.5, but moderate affinity to cdG-Cha-G-Hcit-Qc, which correlated with its affinity for this peptide in in vitro screening tests. The melanotic regions of the tumor showed no affinity to either peptide. The apparent affinity differences between the two testing methods could be explained by the permeablization and fixing of the cells, and use of a high concentration of peptide for the slide assay. The other three cell lines had slightly lesser binding properties. Although alpha-3 integrin receptor is present in each line, the peptide affinity is somewhat variable.
We had predicted that since UCDK9M2 (melanotic) did not bind with high affinity to the already identified alpha-3 integrin ligands, that, it may have a preference for a different type of peptide sequence, possibly with affinity to an integrin other than alpha-3. This proved to be the case. The four ligands identified from the low stringency cXGXGXXc 100% library had no similarity with previously identified peptide binding motifs for alpha-3 integrin. Interestingly, two of the identified peptides, cDGRGA-Phe(4Me)c and cNGRGAYc, showed strong consensus N/D in X2, R in X4, A in X6 and F/Y in X7. The -NGR- peptide has been published as a ligand for aminopeptidase N, and-DGR- peptide as a ligand for alpha-5 beta-1 (Koivunen et al., 1993; Pasqualini et al., 2000). However, NGRGAY and DGRGA(F) seem to be novel emerging sequences for the melanotic melanoma phenotype. A query of the Basic Local Alignment Search Tool-P (BLASTp, NCBI), showed that DGRGA(F) shared sequence similarities with the following proteins: Seprase (Fibroblast activation protein alpha / Integral membrane serine protease /170 kDa melanoma membrane-bound gelatinase); vascular endothelial growth factor receptor 2 precursor (VEGFR-2/Protein-tyrosine kinase receptor Flk-1); Frizzled-7 precursor (Fz-7); Dipeptidyl aminopeptidase-like protein 6 ; ADAMTS-1 precursor (A disintegrin and metalloproteinase with thrombospondin motifs 1); and FRAS1-related extracellular matrix protein 2 precursor (ECM3 homolog). These cell surface proteins may become additional targets for ligands binding to melanotic melanoma cells.
90% of the amelanotic UCDK9M tumors were imaged successfully in vivo. The high tumor:background ratio and specificity to tumor tissue with long retention times confirms that “OA02”-Cy 5.5 can be a novel tool for imaging canine oral amelanotic melanomas that express alpha-3 integrin. Alpha-3 integrin plays a critical role in migration, invasion, and metastasis, and so is an important marker of malignant cells. Pre-injection in vivo images showed autofluorescence artifact in the gastrointestinal tract that has been attributed alfalfa in the mouse diet. The intensity of the post-injection signal in other organs and in the tumor was sufficient to overcome this background autofluorescence signal.
Non-invasive imaging plays an important role in diagnosis, staging and monitoring of neoplasia in general. Specific tumor targeting agents combined with imaging tracers such as F-18 could aid in early detection of primary tumors and metastasis without the use of more invasive procedures such as surgery. Additional applications include surgical planning with intra-operative visualization of tumor margins, as well as targeted chemotherapy, immunotherapy, and radiotherapy. In its current form, the “OA02” peptide has high uptake in the urinary system through which it is excreted. Strategies exist to optimize the biological properties of the peptide and peptidomimetic, such as developing water-soluble benzimidazole analogs (Carpenter et al., 2007), to improve solubility and decrease kidney uptake.
5. Conclusions
Canine oral melanoma expresses the alpha-3 integrin and can bind to the OA02 peptide which has been shown to target the alpha-3 integrin in human models of cancer. Further, dogs with naturally occurring melanoma are a promising model for translational, targeted imaging and therapeutic strategies using alpha-3 integrin binding ligands.
Acknowledgments
Eddie Sanchez, Ray Liu, Ninhuan Yao, Yan Wang for chemistry support. Changying Shi and Fang Ye for help performing the immunohistochemistry. This study was supported in part by the UCD Cancer Center Core Support Grant NIH CA093373 and by the Toni Wiebe memorial fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aina OH, Liu R, Sutcliffe JL, Marik J, Pan C-X, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- Aina OH, Marik J, Gandour-Edwards R, Lam KS. Near-infrared optical imaging of ovarian cancer xenografts with novel alpha 3-integrin binding peptide "OA02". Mol Imaging. 2005a;4:439–447. doi: 10.2310/7290.2005.05169. [DOI] [PubMed] [Google Scholar]
- Aina OH, Marik J, Liu R, Lau DH, Lam KS. Identification of novel targeting peptides for human ovarian cancer cells using "one-bead one-compound" combinatorial libraries. Mol Cancer Ther. 2005b;4:806–813. doi: 10.1158/1535-7163.MCT-05-0029. [DOI] [PubMed] [Google Scholar]
- Aina OH, Sroka TC, Chen M-L, Lam KS. Therapeutic cancer targeting peptides. Biopolymers. 2002;66:184–199. doi: 10.1002/bip.10257. [DOI] [PubMed] [Google Scholar]
- Bergman PJ, Camps-Palau MA, McKnight JA, Leibman NF, Craft DM, Leung C, Liao J, Riviere I, Sadelain M, Hohenhaus AE, Gregor P, Houghton AN, Perales MA, Wolchok JD. Development of a xenogeneic DNA vaccine program for canine malignant melanoma at the Animal Medical Center. Vaccine. 2006;24:4582–4585. doi: 10.1016/j.vaccine.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Bolon B, Calderwood Mays MB, Hall BJ. Characteristics of canine melanomas and comparison of histology and DNA ploidy to their biologic behavior. Vet Pathol. 1990;27:96–102. doi: 10.1177/030098589002700204. [DOI] [PubMed] [Google Scholar]
- Carpenter RD, Andrei M, Lau EY, Lightstone FC, Liu R, Lam KS, Kurth MJ. Highly potent, water soluble benzimidazole antagonist for activated alpha 4 beta 1 integrin. J Med Chem. 2007;50:5863–5867. doi: 10.1021/jm070790o. [DOI] [PubMed] [Google Scholar]
- Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, Kluger HM, Berger AJ, Cheng E, Trombetta ES, Wu T, Niinobe M, Yoshikawa K, Hannigan GE, Halaban R. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Johnson JP. Cell adhesion molecules in the development and progression of malignant melanoma. Cancer Metastasis Rev. 1999;18:345–357. doi: 10.1023/a:1006304806799. [DOI] [PubMed] [Google Scholar]
- Klemke M, Weschenfelder T, Konstandin MH, Samstag Y. High affinity interaction of integrin alpha4beta1 (VLA-4) and vascular cell adhesion molecule 1 (VCAM-1) enhances migration of human melanoma cells across activated endothelial cell layers. J Cell Physiol. 2007;212:368–374. doi: 10.1002/jcp.21029. [DOI] [PubMed] [Google Scholar]
- Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- Lam KS, Lebl M, Krchnak V. The "One-Bead-One-Compound" Combinatorial Library Method. Chem Rev. 1997;97:411–448. doi: 10.1021/cr9600114. [DOI] [PubMed] [Google Scholar]
- London CA, Hannah AL, Zadovoskaya R, Chien MB, Kollias-Baker C, Rosenberg M, Downing S, Post G, Boucher J, Shenoy N, Mendel DB, McMahon G, Cherrington JM. Phase I dose-escalating study of SU11654, a small molecule receptor tyrosine kinase inhibitor, in dogs with spontaneous malignancies. Clin Cancer Res. 2003;9:2755–2768. [PubMed] [Google Scholar]
- Melchiori A, Mortarini R, Carlone S, Marchisio PC, Anichini A, Noonan DM, Albini A. The alpha 3 beta 1 integrin is involved in melanoma cell migration and invasion. Exp Cell Res. 1995;219:233–242. doi: 10.1006/excr.1995.1223. [DOI] [PubMed] [Google Scholar]
- Moretti RM, Montagnani Marelli M, Mai S, Limonta P. Gonadotropin-releasing hormone agonists suppress melanoma cell motility and invasiveness through the inhibition of alpha3 integrin and MMP-2 expression and activity. Int J Oncol. 2008;33:405–413. [PubMed] [Google Scholar]
- Natali PG, Hamby CV, Felding-Habermann B, Liang B, Nicotra MR, Di Filippo F, Giannarelli D, Temponi M, Ferrone S. Clinical significance of alpha(v)beta3 integrin and intercellular adhesion molecule-1 expression in cutaneous malignant melanoma lesions. Cancer Research. 1997;57:1554–1560. [PubMed] [Google Scholar]
- Natali PG, Nicotra MR, Bartolazzi A, Cavaliere R, Bigotti A. Integrin expression in cutaneous malignant melanoma: association of the alpha 3/beta 1 heterodimer with tumor progression. Int J Cancer. 1993;54:68–72. doi: 10.1002/ijc.2910540112. [DOI] [PubMed] [Google Scholar]
- Paoloni M, Khanna C. Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer. 2008;8:147–156. doi: 10.1038/nrc2273. [DOI] [PubMed] [Google Scholar]
- Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- Peng L, Liu R, Andrei M, Xiao W, Lam KS. In vivo optical imaging of human lymphoma xenograft using a library-derived peptidomimetic against alpha4beta1 integrin. Mol Cancer Ther. 2008;7:432–437. doi: 10.1158/1535-7163.MCT-07-0575. [DOI] [PubMed] [Google Scholar]
- Porrello A, Cardelli P, Spugnini EP. Oncology of companion animals as a model for humans. an overview of tumor histotypes. J Exp Clin Cancer Res. 2006;25:97–105. [PubMed] [Google Scholar]
- Smalley KS, Nathanson KL, Flaherty KT. Genetic subgrouping of melanoma reveals new opportunities for targeted therapy. Cancer Res. 2009;69:3241–3244. doi: 10.1158/0008-5472.CAN-08-4305. [DOI] [PubMed] [Google Scholar]
- Vail DM, MacEwen EG. Spontaneously occurring tumors of companion animals as models for human cancer. Cancer Invest. 2000;18:781–792. doi: 10.3109/07357900009012210. [DOI] [PubMed] [Google Scholar]
- Yoshinaga IG, Vink J, Dekker SK, Mihm MC, Jr, Byers HR. Role of alpha 3 beta 1 and alpha 2 beta 1 integrins in melanoma cell migration. Melanoma Res. 1993;3:435–441. doi: 10.1097/00008390-199311000-00006. [DOI] [PubMed] [Google Scholar]