Abstract
Ritonavir, an HIV protease inhibitor, is successfully used for the prevention and treatment of HIV infections. Ritonavir pharmacokinetics are complicated by inhibition, induction and pharmacogenetics of cytochrome P450 (CYP) enzymes mediating its clearance. This investigation revealed that CYP2J2, along with CYP3A4/5 and CYP2D6, efficiently metabolizes ritonavir, and to a CYP2J2-specific (minor) metabolite. Chemical inhibition of ritonavir metabolism, clearance, KI/kinact and abundance of CYP2J2 in liver microsomes were evaluated and then applied to an in vitro-in vivo static scaling model to estimate the contribution of each isozyme, as a function of CYP abundance, activity, and genotype. Disposition of the CYP2J2- specific metabolite was also evaluated in vivo. In plasma, metabolite abundance was well above previously reported levels with circulating concentrations measured at 2 μM for the main hydroxylisopropyl metabolite. Ritonavir and metabolite plasma profiles were simulated using Simcyp®. A modest (2-6%) contribution of CYP2J2 to ritonavir clearance is predicted which increases to more than 20% in subjects carrying CYP2D6 poor metabolizer polymorphisms and CYP3A4 irreversible inhibition. These results indicate that minor drug metabolizing enzymes could become quantitatively important in RTV clearance if main metabolic pathways are impeded.
Keywords: CYP2J2, CYP2D6, CYP3A4, time dependent inhibition, ritonavir, in vivo, Simcyp
1. Introduction
Ritonavir (RTV) has been successfully used in combination with a range of antiretroviral drugs to treat HIV infections, prevent the emergence of resistance, and is widely used as a protease inhibitor pharmacoenhancer [1]. RTV is used as a ‘booster’ because it is a strong inhibitor of CYP3A4-mediated intestinal and hepatic drug clearance, which leads to increased bioavailability of the co-administered CYP3A4 substrate drugs [2, 3]. The RTV metabolic pathway is complex due to multiple metabolites formed [4] and the ability to inhibit and auto-induce its own metabolism [5]. Several isozymes including CYP3A5 and CYP2D6 [6, 7] also mediate RTV metabolism but seem less susceptible to inhibition [7, 8]. A significantly compromised hepatic activity of CYP3A4 (∼90%) [9] through RTV mechanism-based inhibition [6] seems to implicate a metabolic shift towards CYP2D6-dependent clearance. Particularly in chronic dosing of RTV, CYP3A4 activity is greatly reduced as evidenced in vivo using midazolam as a CYP3A4 activity probe [10]. RTV inhibition of CYP2D6 was also observed in vivo [11] and CYP2D6 is known for its extensive genetic polymorphisms (such as inactive variants CYP2D6*4 and CYP2D6*41) which require dose adjustments [12]. Therefore, a combination of CYP3A4 and CYP2D6 inhibition and/or poor CYP2D6 metabolizer status (CYP2D6 *2,*4 or *41) can augment the contribution of other minor CYP enzymes to the clearance of RTV.
CYP2J2 is particularly known for its major role converting arachidonic acid to epoxyeicosatrienoic acids (EETs) in the heart and various extrahepatic tissues. EETs regulate a range of physiologically important functions, such as vasodilation, angiogenesis and inflammation [13]. CYP2J2's function in drug metabolism is, however, less established. CYP2J2 is known to metabolize a number of antihistamine drugs including terfenadine, astemizole and ebastine [14-18], but also a broad range of common medications and potential substrates in vitro [19] such as amiodarone and tamoxifen. Pharmacogenetic variants of CYP2J2 are mostly rare and their prevalence in the population depends on the ethnic group studied. One variant that seems common among various ethnicities is CYP2J2*7 which results in reduced protein expression and therefore activity [20]. Although several studies show that CYP2J2 is expressed in low abundance in the liver and intestine, it is still anticipated to contribute to first pass metabolism [21]. In fact, the contribution of CYP2J2 to intestinal ebastine hydroxylation is projected to be as high as 70% [14]. So far, no clinical study has addressed the contribution of CYP2J2 to drug metabolism in vivo.
Investigating various CYP-isozymes and their contribution to RTV metabolism in this study revealed that CYP2J2 was efficient at oxidizing RTV. To characterize CYP2J2 contribution to RTV clearance, we used in vitro experiments, plasma samples from prior completed clinical studies, and simulations to estimate the contribution of CYP2J2, CYP3A4 and CYP2D6 to RTV metabolism. We initially identified CYP2J2 specific metabolites in vitro, then used selective chemical inhibitors of CYP isozyme-mediated RTV metabolism in human liver microsomes to estimate individual CYP contribution. Next, we measured RTV-clearance and competitive/time-dependent isozyme inhibition in vitro to determine CYP contribution through in vitro-in vivo scaling. Finally, we profiled RTV and metabolites in plasma samples to trace CYP2J2 activity in vivo and conclusively simulated the pharmacokinetics of RTV and metabolites based on the above data to determine CYP-contribution to hepatic clearance using Simcyp®.
2. Materials and Methods
2.1 Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise stated and used without further purification. Human liver samples were obtained from the University of Washington School of Pharmacy Human Tissue Bank (Seattle, WA). Selected livers for this study were Caucasian, of mixed gender (equal ratio) and an age range of 7-70 (average 43) [21, 22].
2.2. In vitro assays with Supersomes® and human liver microsomes
2.2.1. Metabolite formation experiments
Reactions were conducted at 10 μM RTV and 20 pmol P450 mL-1 (Supersomes®, BD Biosciences, San Jose, CA) in potassium phosphate buffer (100 mM, pH 7.4). After a 5 min preincubation at 37 °C, NADPH was added (1 mM final concentration) and the incubation allowed to proceed for 30 min. The reaction was quenched by extracting the mixture three times with ice-cold ethyl acetate. The organic phase was combined, dried under nitrogen, and reconstituted in 50 μL acetonitrile:water (1:1). Calibration standards were prepared using similar assay conditions with heat inactivated Supersomes®.
2.2.2. RTV-depletion experiments
CYP-Supersomes® (40 pmol mL-1) were incubated under assay conditions described in 2.2.1 using 1 μM RTV. The reaction was initiated by adding NADPH (1 mM final concentration) and allowed to proceed for 30 min. 100 μL aliquots were removed at 0.25, 1, 2, 4, 6, 10, 15, 20, 30 min and quenched as described in 2.2.1. Intrinsic clearance (Clint) was calculated using the half-life (t1/2) derived from the first order decay Clint = 0.693 / t1/2,in vivo × mL incubation / pmol recombinant CYP as described previously [23].
For the determination of the apparent Km, as described in Obach and Reed Hagen [24], RTV-depletion was measured over a 30 min time period at concentrations from 0.01 to 30 μM. Reactions were quenched with an equal volume of acetonitrile containing midazolam (0.03 μM) as internal standard. Heat-denatured microsomes and incubation mixtures deprived of NADPH served as negative controls.
2.3. Inhibition of RTV metabolism in supersomes and HLMs
2.3.1. Competitive inhibition experiments
CYP2D6 and CYP2J2 recombinant protein (40 pmol P450 mL-1) or individual human liver microsomes (0.4 mg microsomal protein mL-1) were incubated using the procedure mentioned under in vitro assays (2.2.1), using a 5 min incubation time. Reactions were quenched as described in 2.2.2.
2.3.2. Time dependent inhibition
Experiments were conducted in HLMs (0.4 mg microsomal protein mL-1) over 15 min with troleandomycin (10 μM) and NADPH (1 mM) in the preincubation step, which was then diluted 10-fold into a reaction vial to measure RTV turnover containing RTV (1 μM) and additional NADPH (1 mM). These RTV turnover reactions were quenched by adding ice-cold ethyl acetate 2 mL containing 0.03 μM midazolam as internal standard and processed as described in 2.2.1.
2.3.3. Mechanism-based inhibition
Inhibition was measured in Supersomes. CYP2J2 Supersomes (40 pmol P450 mL-1) were incubated for up to 8 min with terfenadine concentration (2 μM) and RTV (inhibitor) concentrations ranging from 0.1 to 10 μM. CYP2D6 Supersomes were incubated up to 30 min with dextromethorphan (20 μM) and RTV-concentrations ranging from 1 to 100 μM. Reactions were terminated as mentioned above under in vitro assays (2.2.2).
Assay was performed under linear conditions with regards to protein and time, which were established beforehand.
2.3.4. Protein binding was measured as described in Barre et al. [25]
2.4. Substrate and metabolite quantification and identification
2.4.1.Dextromethorphan to dextrorphan and terfenadine to hydroxy terfenadine metabolism
Metabolites and parent were quantified on a Sciex API4000 LC/MS/MS (Applied Biosystems) connected to a Shimazu LC system (LC-10AD, SCL-10A) equipped with a CTC-PAL autosampler (LEAP Technologies, Carrboro, NC). 10 μL of supernatant was injected on an Agilent Zorbax XDB C8-column (2.1 μm, 5 cm) column. Both compound-metabolite pairs were separated with a mobile phase that consisted of aqueous phase A: 10 mM ammonium acetate (pH 5.5) and organic phase B: 10 mM ammonium acetate in methanol and analyzed using the following gradient: mobile phase B: 0 -1 min, 30% ; 1-2 min, 30 to 70% , 2 - 4 min 70 to 100% , 4-6.5 min 100% , 6.5-6.6 min 100-30%. The column was re-equilibrated at initial conditions for 1.4 min. The flow rate was 0.3 ml/min. MS/MS-parameters: ion spray 5,500 V, temperature 450 °C, collision gas 6 L/min, ion gas 15 L/min, curtain gas 10 L/min. The following MRM ion transitions were used: dextromethorphan N-demethyl metabolite (Santa Cruz Biotechnology, Santa Cruz, CA) (258>157; DP 60; CE 50), parent (272>171; DP 60; CE 50) and internal standard dextromethorphan-d3 (Santa Cruz Biotechnology, Santa Cruz, CA) (261>157; DP 60; CE 50).
For terfenadine, the following transitions were used: terfenadine (472.2 > 436.1; declustering potential, DP, 80; collision energy, CE, 37), hydroxy terfenadine (488.3 > 452.2; DP 90; CE 40), terfenadine acid (502.4 > 466.3; DP 100; CE 40) and midazolam (326.0 >291.2; DP 50; CE 30). The dwell time for each ion was 50 ms. The flow rate was 0.3 ml/min. MS/MS-parameters: ion spray 5,500 V, temperature 450 °C, collision gas 6 L/min, ion gas 15 L/min, curtain gas 10 L/min.
2.4.2. RTV metabolite detection
Products of RTV-metabolism were separated on a Waters UPLC Acquity BEH C18 (1.7 um 2.1 × 100 mm) column using a Waters Acquity Ultra Performance separation module. Flow rate was 0.35 mL min-1 using solvent A: 10 mM ammonium acetate pH, 4.5, and solvent B: 10 mM ammonium acetate. Solvent B was increased from 30% to 90% from 0.25 to 6.25 min, then held at 90% for 0.75 min, and then returned to 30%. Compounds were detected in positive electrospray ionization and multiple reaction mode (Quattro Premier XE-quadrupole MS, Waters, Milford, Massachusetts), with extractor 5 V, desolvation temperature 350 °C, source temperature 120 °C, desolvation gas (nitrogen) 800 L hr-1. Parent, metabolites and internal standard were detected using the following conditions: RTV 721>426 (cone voltage CV 35V, collision energy CE 20V), midazolam 326>291.2 (CV 42, CE 27), R5 580>296 (CV 35, CE 25), R1 582>426 (CV 35, CE 20), R9 634 > 296 and R7 707 > 282 (CV 35, CE 25), R2 737 > 59 and R4 737 > 114 (CV 35, CE 35), R6 737 > 426, R2/R3 737 > 426, and R4/R6 737 > 442 (CV 35, CE 20). RTV-metabolites were quantified by UV in in vitro incubations using the identical extinction coefficient measured from a ritonavir calibration curve. The concentration was then translated into a metabolite specific UV/MS-response factor by measuring peak areas of identical diluted samples on the mass spectrometer. [26]
Identity of metabolites was confirmed using a time of flight mass spectrometry in MSe and MS/MS mode (Waters Micromass High-Definition MS System, Quadrupole/ Triwave™/Orthogonal Acceleration Time-of-Flight Tandem Hybrid Mass Spectrometer QToa -TOF MS/IMS/MS) and the chromatographic conditions as mentioned under RTV-metabolite detection.
2.5. Quantification of CYP2J2 in human liver microsomes
CY2J2 protein content was determined in 40 human liver microsomes. 50 μg of microsomes were loaded onto a NuPAGE 4-12% Bis-Tris SDS PAGE gel electrophoresis performed using manufacturer's conditions (Invitrogen, CA). The SDS gel was transferred onto a PVDF membrane for 1h and the CYP2J2 protein was detected using a CYP2J2 specific primary antibody (ABNOVA, Taipei, Taiwan), a goat-antimouse IRDye 800CW as a secondary antibody and visualized on an Odyssey imaging system (LICOR-Bioscience, Lincoln, NE). CYP2J2 content was calibrated using commercially available Supersomes® in the range of 0.00315 to 0.065 pmol/μL.
2.6. Plasma samples and in vivo plasma profile
All studies were approved by the University of Washington Institutional Review Board. Plasma samples were obtained from previous studies [9-11] designed to determine the inhibition of CYP3A4 and other isozymes by protease inhibitors. Plasma samples were extracted three times with ice-cold ethyl acetate (2 × sample volume) containing dextromethorphan-d3 as internal standard. The organic phase was combined and dried under nitrogen. Samples were reconstituted in 100 μL acetonitrile:water 1:1. Calibration standards were performed with human plasma spiked with ritonavir (0.1 - 20 μM).
2.6.1. Concentrations-time profile study
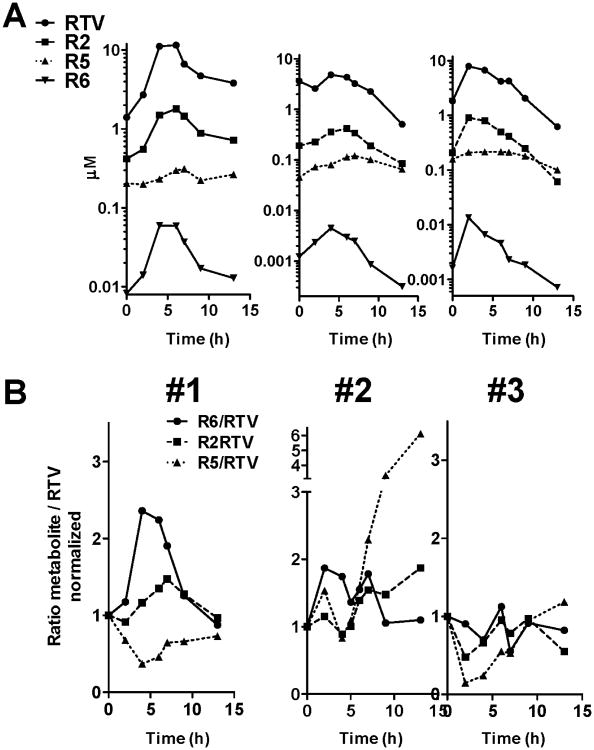
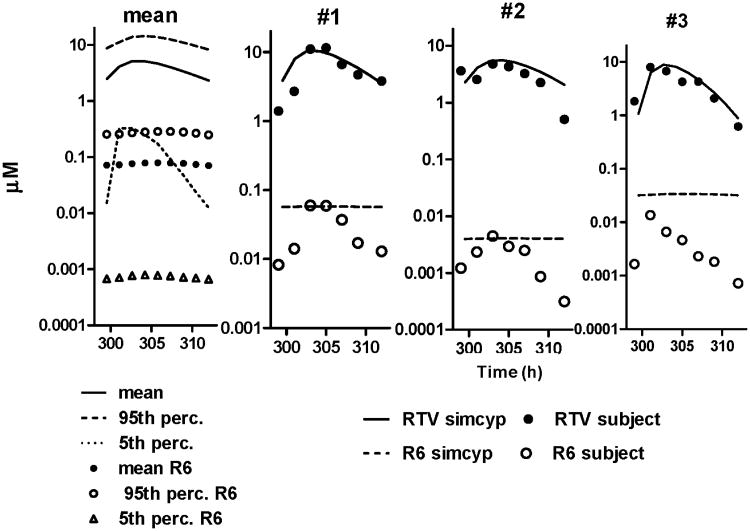
In “study 1” (as identified in the previous publication for consistency [9]), the effect of ritonavir on digoxin dosing was assessed by using a simultaneous dosing design. Blood from three subjects representing low, medium and high RTV-plasma levels after a 14 day RTV treatment (400 mg bid, at steady state) was drawn over a period of 24 hours. Banked plasma samples from “study 1” were analyzed in this present study to follow RTV and metabolite formation over time (figure 5).
Figure 5.
Plasma concentration profile of RTV and metabolites in three subjects with high (#1), low (#2), and medium (#3) RTV Cmax-values. Blood was sampled for 12 hours after the last RTV dose. A: Measured concentrations, B: Metabolite/RTV ratio for R2, R5 and R6.
2.6.2 RTV metabolite formation vs. dextromethorphan metabolism
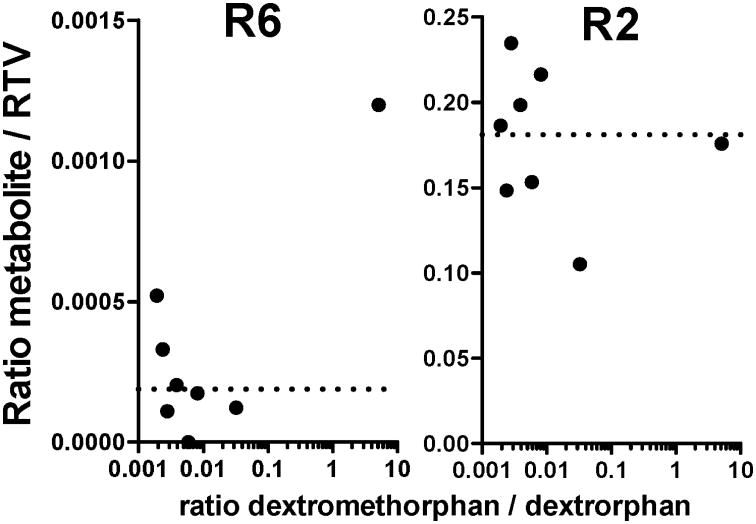
From “study 2” (as identified in the previous publication for consistency [9], in which the effect of ritonavir on digoxin dosing was assessed by using a staggered dosing design (treatment 14 days RTV, escalating dose to 400 mg b.i.d.), a total of eight subjects of the RTV arm were chosen for the assessment of RTV parent compound and metabolite concentrations. In addition, the assessment of CYP2D6-activity by measuring the urinary excretion of dextromethorphan and dextrorphan was performed. Banked plasma samples from “study 2” were used in the current study to assess differences in the R6/RTV formation ratio in relation to dextromethorpham metabolism (figure 6).
Figure 6.
Influence of CYP2D6 activity on ritonavir metabolism. Results are shown as the R6/RTV and R2/ RTV plasma concentration ratio at Cmax time point. CYP2D6 metabolic activity was determined by the ratio of total urinary dextromethorphan over dextrorphan. Each data point represents a single subject (N=8). The dotted line shows the average value for all samples measured.
2.7. Prediction model (static and dynamic using Simcyp®)
2.7.1 Static modeling
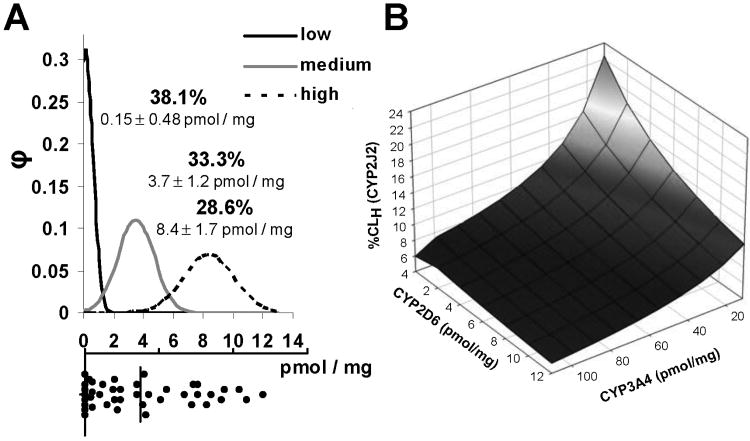
In the static model, hepatic clearance was scaled using 54.7 mg CYP per g of liver, 21.43 g liver per kg body weight, and 78 kg body weight, a well-stirred model, fu 0.02, hepatic blood flow of 0.927 L min-1, based on 0.0216 L/min/kg and 0.52 (1-Hct). CYP2J2 contribution to hepatic clearance was expressed as percent of total hepatic clearance (figure 4B).
Figure 4.
A: Distribution of CYP2J2 protein in human liver microsomes (n=42). Protein content was measured by Western blot as detailed in the methods. The histogram at the bottom portion of A shows the distribution of measured CYP2J2 content (vertical line depicts the average). The top portion of A shows the Gaussian distribution used to group livers into low, medium and high expressors (with the percentage of total and the mean CYP2J2 content in HLMs ± SD). B: Scaling the contribution (percentage) of CYP2J2 to hepatic clearance of RTV (%CLH CYP2J2) at different CYP2D6 and CYP3A4 contents (with CYP2J2 set at maximum of 12 pmol/mg, CYP3A5 content was correlated to CYP3A4 content).
2.7.2 Dynamic modeling
Simcyp® was used to predict ritonavir and metabolite profiles and account for a 14 day dosing scheme (bid), time dependent and competitive inhibition, and intestinal metabolism. The last 12 h following a 400 mg twice-daily dosing scheme, similar to the in vivo study, was used to estimate a CYP2J2 contribution to the RTV hepatic clearance. Pharmacokinetic parameters were obtained from previous publications (Vd = 0.41 L kg-1, fu= 0.01, t1/2 = 3.5 h; [27] and experimental in vitro clearance values (including fu) were used. A first order absorption model was used in a minimal Physiologically Based Pharmacokinetic (PBPK) model with enzyme kinetics driven clearance. The formation clearance of the CYP2J2 specific metabolite R6 was assessed from measured parent compound depletion Km with CYP2J2 and using the measured formation rate as Vmax, assuming saturation kinetics (measuring Km for R6 was not determined due to expected low levels of formation and limits of detection). Metabolite clearance was estimated from an average of in vivo metabolite profiles of R6 from each subject using four to five subsequent time points and assuming a similar volume of distribution as RTV. The plasma concentration time profile was then predicted for parent compound RTV and metabolite R6 in a virtual population of 500 subjects obtained from a Simcyp healthy population with manually adjusted CYP2J2 content according to measured results reported in figure 4A. Intestinal CYP2J2 content was set at 1-3 pmol/mg [17, 21]. Two modeling approaches were used: a) inhibition determined by microsomes and supersomes, and b) inhibition from sandwich-cultured hepatocytes. For the latter, kinact and KI for CYP3A5 and CYP2J2 were derived by linear scaling of kinact/KI values from microsomes of CYP2J2 or CYP3A5-enzyme over CYP3A4 at a concentration of 1 μM to accommodate overprediction of inhibition [11]. Kinetic input values are identical to table 1. fu,supersomes was measured with 0.49 for CYP2J2, 0.66 for CYP3A4 and 0.29 for CYP2D6. Simcyp parameters other than ones included above were kept as default (for reprint of input parameters please contact authors).
Table 1.
Kinetic data for RTV-metabolism and time dependent inhibition of CYPs by RTV.
| Metabolism | Inhibition | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Km (μM) |
Clint (μL pmol-1 min-1) |
IC50 (μM) |
kinact (μM min-1) |
KI (μM) |
kinact/KI (min-1 nM) |
slope at 1 μM (μM min-1) |
|
| CYP2J2 | ∼ 0.016 (dep.) | 2.0 ± 0.7 (dep.) | 0.98 (0.6-1.6) 5 | 0.042 ± 0.003 | 0.51 ± 0.07 | 82 | 0.00444 |
| CYP3A4 | 0.068 (R2)1 | 3.7 ± 0.5 (dep.) | 0.032 | 0.322 | 0.12 | 3200 | 0.172 |
| CYP3A5 | 0.047 (R2)1 | 1.4 ± 0.03 (dep.) | 0.032 | 0.082 | 0.122 | 672 | 0.0364 |
| CYP2D6 | 1.6 (R2)1 | 11.6 ± 1.1 (dep.) | 11.9 (8.7-16.3) 5 | n.d.3 | n.d.3 | - | |
3. Results
3.1 Identification of CYP2J2 -specific metabolites
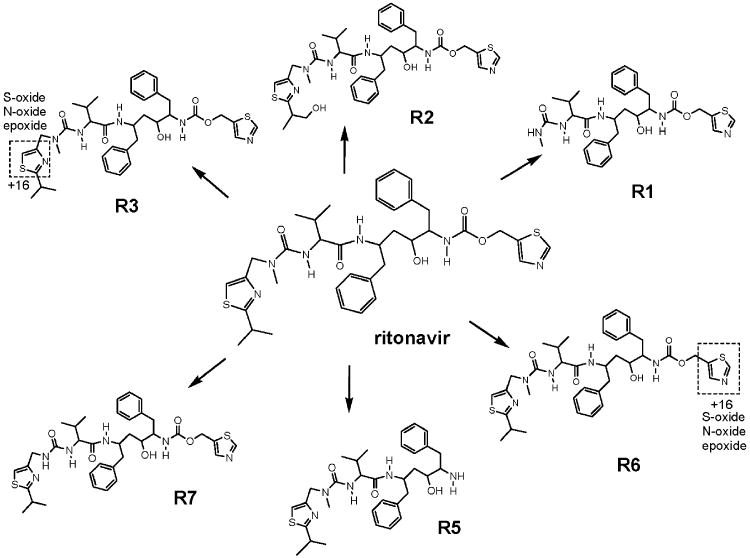
Metabolites were identified in incubations using CYP2J2 Supersomes®. Hydroxylation at the isopropyl group of RTV (R2, figure 1) was the major metabolite formed primarily by CYP2D6 (the only metabolite from this CYP) with the highest turnover number (figure 2A). In addition to R2, dealkylations were favored by CYP3A4 (R1, R5, R7). CYP2J2 generated not only R2 but also specifically R6 and R3 resulting from oxidation of the thiazole rings on the “east” and “west” side of the molecule respectively, and traces of R7 (demethylation) and R5 (dealkylation) in a 30 min incubation. In shorter 5 min incubations, R3 and R6 were the predominant metabolites formed by CYP2J2 (data not shown). Metabolites in in vivo samples were identified by matching retention times and mass transitions with metabolites observed in vitro since authentic standards were not available.
Figure 1.
Scheme depicting the primary metabolites of ritonavir, which were measured in this study. For R3 and R6, possible structural modifications with an increase of +16 in the molecular weight are depicted.
Figure 2.
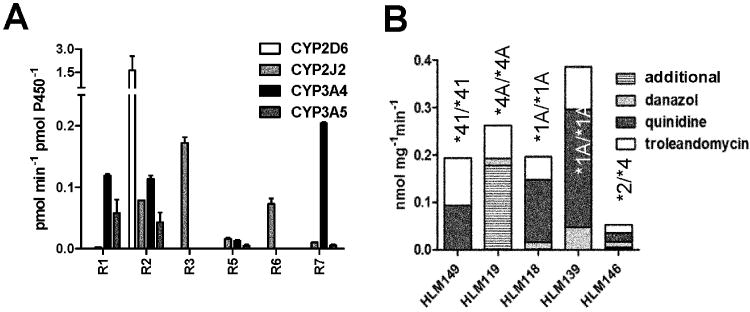
A: Comparison of RTV-metabolite formation in Supersomes® of CYP3A4, CYP3A5, CYP2D6, and CYP2J2 (mean, error bars indicating SD). B: R2 formation rate in human liver microsomes of selected donors and approximate contribution to R2 formation by CYP isoform, measured through inhibition by selective probes. Isoform-selective inhibitors of CYP3A4, CYP2D6 and CYP2J2 were troleandomycin, quinidine and danazol, respectively. The CYP2D6 genotype for each liver is shown above the column. For CYP3A contents see table 2.
3.2. Inhibition of R2 formation in individual human liver microsomes
Five individual HLMs, selected according to their CYP2D6 genotype and relatively high CYP2J2 content, were used to investigate the inhibition of RTV metabolism (figure 2B, for enzyme abundance see table 2). Homozygous CYP2D6 PM with a reasonable high CYP2J2 content (>5 pmol/mg) were scarce in the available liver samples, thus the best genotypical representative microsomal samples were selected as examples in this evaluation. R2 was observed in at least 10-fold excess over any other metabolite formed and was used to measure the % inhibition relative to control. In HLMs 118 and 139, carrying the CYP2D6*1/*1 genotype, the CYP2J2-selective inhibitor danazol reduced R2 formation by up to 15%. In HLM 146 (with low CYP3A4 content and carrying a CYP2D6 PM genotype CYP2D6*2/*4) danazol inhibition of CYP2J2 reduced R2 formation by 25%. Inhibition of CYP2J2 activity was insignificant in HLM 119, also a CYP2D6 PM but with high CYP3A4 content. No inhibitory effect was observed in HLM 149 (CYP2D6*41/*41 PM genotype with high CYP3A4 content). Quinidine (CYP2D6 inhibitor) reduced R2 formation up to 70% only in CYP2D6 moderate metabolizers (carrying the allele CYP2D6*1 and *2), but expectedly much less, or not at all, in HLMs carrying CYP2D6 PM genotypes (alleles *4 and *41). The inhibition of R2 by troleandomycin (TAO), used as the CYP3A4 inhibitor, was about 20-30% and moderately increased to 50% in HLMs carrying CYP2D6*41/*41. In recombinant CYP2J2, R2-formation was inhibited by 95%, using danazol and by 20% using TAO, whereas quinidine had no significant effect (data not shown).
Table 2.
HLM characteristics used for activity and inhibition assay in Figure 2.
| HLM | CYP2J2 | CYP3A4 pmol mg-1 | CYP3A5 | CYP2D6 genotype |
|---|---|---|---|---|
| 149 | 11.4 | 151.1 | 3.3 | *41/*41 |
| 119 | 9.7 | 106 | 101 | *4A/*4A |
| 118 | 7.2 | 20.1 | 2.1 | *1A/*1A |
| 139 | 6.8 | 67.2 | 2.4 | *1A/*1A |
| 146 | 7.6 | 5.0 | 0.6 | *2/*4 |
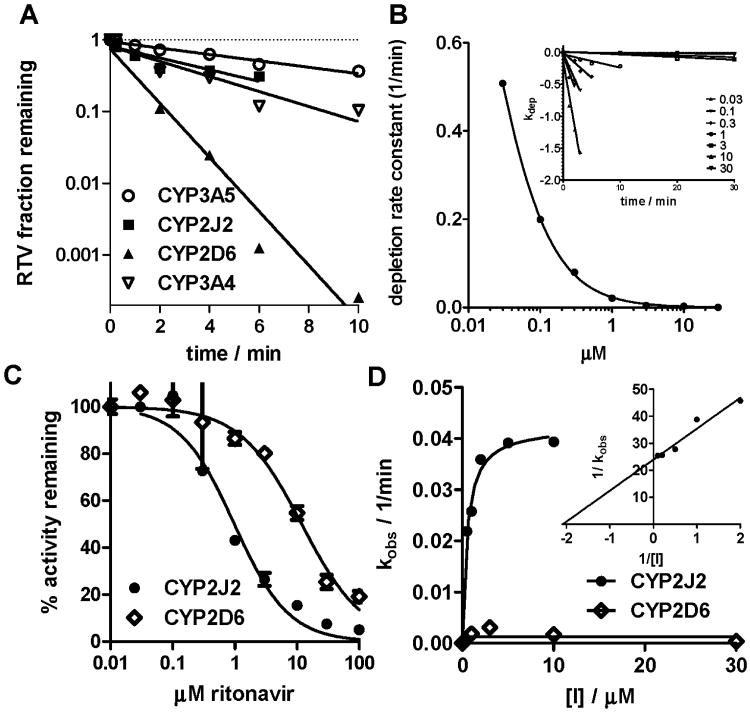
3.3 Evaluation of RTV in vitro clearance in Supersomes®
Ritonavir metabolism by CYP2J2 was compared to that by isozymes known previously to play a role in RTV clearance (CYP2D6, CYP3A4, and CYP3A5) [6]. CYP2D6 demonstrated the greatest metabolic clearance measured through substrate depletion at 1 μM RTV compared to CYP3A4, CYP2J2, and CYP3A5 (table 1, figure 3A). Clearance was measured by the depletion method because of limitations of Vmax/km approach for this system (Km in nanomolar range, products sequentially metabolized and thus depleting the primary product, substrate depletion due to significant turnover needed to generate product.
Figure 3.
A: Depletion of RTV in CYP-supersomes at 1 uM RTV concentration; B: depletion experiment to determine Km in CYP2J2 supersomes; C: Competitive inhibition of CYP activity by RTV; D: time dependent inhibition of CYP activity by RTV. Each experiment was performed with a minimum of n=2, standard errors are reported in table 1.
The often conventionally used concentration of 1 μM for substrate depletion experiments is lower than RTV concentration in vivo, but less sensitive to complications due to isozyme inhibition and worked well in later predictions (see below). RTV metabolic clearance using CYP2J2 was almost as high as CYP3A4, when depletion was measured over 6 min. The short incubation time was necessary, as the stability of CYP2J2-supersomes® decreased over longer incubation periods and therefore underprediction of CYP2J2 dependent clearance is possible. The Km for RTV measured through substrate depletion is expected to be in the low nM region (figure 3B), but insufficient detection levels of the analytical assay complicated precise determination.
3.4 Competitive and time dependent inhibition by RTV
RTV was a stronger competitive inhibitor of CYP2J2-mediated terfenadine metabolism in Supersomes® compared to CYP2D6-mediated dextromethorphan metabolism (figure 3C), but weaker compared to CYP3A4 (table 1). Weak time dependent inhibition of CYP2J2-mediated terfenadine metabolism by RTV in Supersomes® was observed, (figure 3D), in comparison to the extraordinary strong inhibition of CYP3A as reported in the literature (testosterone was used as a substrate for CYP3A4 and CYP3A5 inhibition, [28]). No significant time dependent inhibition was observed with CYP2D6.
3.5 CYP2J2-protein content in human liver microsomes
Average CYP2J2 protein content was approximately 4 pmol mg-1 (median 2.4, ranging from undetectable to 12 pmol mg-1) in 40 HLM samples (figure 4A). To account for the variability in CYP2J2 protein expression in subsequent modeling, HLMs were grouped into low, medium, and high CYP2J2 expressers, with each group accounting for about one third of the total number of HLMs.
3.6 In vitro to in vivo scaling of RTV hepatic clearance
Modeling the impact of CYP2J2 on hepatic ritonavir clearance through scaling of measured in vitro clearance reflected changing CYP3A4 and CYP2D6 activities (CYP3A4 inhibition or CYP2D6 PM). The static model predicted a 2-6% contribution of CYP2J2 to hepatic RTV clearance in individuals with normal CYP2D6 activity and high CYP3A4 activity (figure 4B). Assuming 90% reduction in CYP3A4 activity (at steady state) and inactive CYP2D6 (e.g. as in CYP2D6 poor metabolizers), the contribution of CYP2J2 is predicted to increase to 20%.
3.7 Plasma concentrations-time profile of RTV and RTV metabolites
Ritonavir and metabolite concentrations were determined at steady-state in three subjects (figure 5, upper panel A). RTV-concentrations were consistent with previously reported values [29]. Peak RTV plasma concentration (Cmax) occurred after 2-4 h, and are shown for three subjects with high (#1), low (#2), and medium (#3) Cmax-values. R2 and R5 were the predominant metabolites found in plasma, with R2 observed in the micromolar range. The concentration of R6 was higher in subject #1 than the other two subjects. The R6/RTV concentration ratio (depicted in figure 5, lower panel B) was used as an indicator of CYP2J2 contribution to RTV metabolism. The ratio increased only in subject #1 (high RTV Cmax) to about 2.5-times relative to the first time point. Ratios of R2 or R5 over RTV (predominant metabolites) did not vary with time.
In study two (figure 6), one subject with low CYP2D6 activity, evidenced by a high dextromethorphan (parent compound) to dextrorphan (metabolite) ratio had a 6-fold higher R6/ RTV ratio compared to the other subjects (figure 6, left). However, the second highest R6/RTV ratio was achieved by a subject with the lowest dextromethorphan over dextrorphan ratio. In contrast, the R2/RTV ratio did not increase over the mean value (figure 6, right). This subject also had the highest RTV plasma concentration within the chosen sample set (11.6 μM).
3.8 Simcyp® population pharmacokinetic simulation
Plasma concentration-time profiles of RTV and metabolites were simulated using Simcyp®. About 7% of the population (n=500) were CYP2D6 PM and the estimated average in vivo metabolite clearances for R6 (ClR6) was 2.8 L/h. Initial modeling using KI and kinact obtained from microsomes failed to simulate a reasonable mean RTV and R6 profile particularly for extreme scenarios of CYP2D6 poor metabolism. RTV AUC was over predicted (table 3, bottom rows, labeled HLM/CYP-approach). In contrast, the prediction was more successful using inhibition data derived using SCHH in incubations [11]. Consequently, HLM/CYP inhibition data were linearly scaled using the kinact/KI ratio. Using the scaled SCHH data, the predicted AUC and Cmax were within the range of the in vivo AUC-profiles and literature data [29]. From the simulated population, examples with similar PK profile (RTV, R6, R6/RTV ratio) were extracted to cover the in vivo profiles and AUC of subject #1 to #3 (figure 7, table 3). Whereas RTV plasma concentration-time profile of prediction to in vivo results matched well, some discrepancies in the Cmax of R6 in subject #3 with the predicted concentration-time profile were observed. An approximate 95%-inhibition of hepatic CYP3A4 in these chosen data was simulated in Simcyp, whereas hepatic CYP2J2 was predicted to be inhibited by about 14%. Overall contribution of CYP2J2 to hepatic RTV clearance within the population was predicted to be 6% on average, and exceeded 20% in about 4% of all in silico data sets, with a maximum value of 87% found (Cmax,RTV = 24 μM, AUC 322 μM h-1), which were mostly CYP2D6 PM and CYP2J2 EM. Two scenarios for the potential CYP2J2 contribution are described in table 3. CYP2J2 contribution can be 22% or increase as high 72% in CYP2D6 PM. The high contribution (72%) is predicted, if CYP2J2 is highly expressed and CYP3A4 content is low, this contribution decreases to 22% with a five-fold decrease in CYP2J2 expression but CYP3A4 protein remains constant.
Table 3.
Comparison of simulation data with in vivo samples from subjects.
| RTV | R6 | Cmax ratio R6/RTV (10-3) |
calc. % Clhep (CYP2D6/3A4/ 2J2/3A5) |
|||
|---|---|---|---|---|---|---|
| AUC (μMh) | Cmax (μM) | AUC (μMh) | Cmax (nM) | |||
| Simulation data (SCHH approach) | ||||||
| Mean (5th/95th percentile) | 80 (12-237) | 7.9 (1.4-21) | 2.8 (0.02-8.7) | 23 (1.7-710) | 29 (1.2-33) | 79/14/4/2 |
|
| ||||||
| Subjects with measured concentration profiles and matched simcyp simulation (SCHH approach) | ||||||
| Subject #1 (repr. sim) | 83 (70) 1 | 1.5 (11.1) | 0.41 (0.46) | 60 (33) | 5.2 (3) | 45/53/1/0 |
|
| ||||||
| Subject #2 (repr. sim) | 40 (46) | 4.8 (5.1) | 0.028 (0.03) | 4.4 (2.2) | 0.92 (0.43) | 87/10/0/2 |
|
| ||||||
| Subject #3 (repr. sim) | 54 (55) | 8 (7) | 0.062 (0.053) | 14 (3.8) | 1.8 (0.5) | 93/7/0.1/0 |
|
| ||||||
| CYP2D6 PM1 | 146 | 14 | 1.7 | 130 | 9.3 | 0/78/22/0 |
| CYP2D6 PM1 | 200 | 17 | 6.5 | 520 | 31 | 0/28/72/0 |
|
| ||||||
| Simulation data, examples (HLMs/CYP approach) | ||||||
| CYP2D6 EM | 46.3 | 5.14 | 0.449 | 34.5 | 6.72 | 97/3/0/0 |
| CYP2D6 PM | 527 | 101 | 0.227 | 16.3 | 0.16 | 0/73/27/0 |
representative Simcyp® simulation for CYP2D6 PM scenario; repr. sim. - means representative simulation.
Figure 7.
Simulation of plasma concentration of RTV and R6 over time (graphical depiction of table 3 to demonstrate the matching profiles). Results are displayed as the population mean value, and the 95th and 5th percentiles (over complete Simcyp population, left), and representative simulations for the three individual subjects (from figure 6, from left to right).
Discussion
This investigation revealed that in vitro recombinant CYP2J2 cleared RTV with about 50% efficiency compared to CYP3A4. In individual HLMs, CYP2J2 contribution to R2-formation was highest in carriers of CYP2D6 poor metabolizer genotypes and low CYP3A protein content. RTV was a weak competitive and time dependent inhibitor of CYP2J2 compared to CYP3A4, whereas no time dependent inhibition was observed with CYP2D6. CYP2J2 expression in human liver microsomes was variable and ranged from undetectable up to 12 pmol mg-1, well within levels of other important drug metabolizing isozymes such as CYP2D6 or CYP2B6. In vitro-in vivo scaling predicted a modest 2-6% contribution of CYP2J2 to metabolic clearance, increasing to over 20% when CYP2D6 PM status is accounted for in the simulations. In vivo, RTV-metabolites were readily measured in plasma of subjects from previous studies (dominant metabolite R2) with varying profiles with respect to RTV concentration. The ratio of R6 (CYP2J2 specific metabolite) to RTV was highest in carriers of CYP2D6 PM genotypes. In vivo plasma concentration-time profiles matched well in Simcyp® simulations and predicted an overall low contribution to Clhep for CYP2J2, but a contribution of >20% in CYP2D6 PM, with the residual carried by CYP3A. These extreme cases within a healthy population matched findings of IVIVE. Similar but weaker trends were found in in vivo studies indicating an increased contribution of CYP2J2 with higher RTV exposure. These data highlight the impact of a minor CYP-enzyme, CYP2J2, in the metabolism of RTV under specific conditions such as the lack of active CYP2D6.
The metabolism of RTV is complex both in vitro and in vivo generating the main metabolite (R2 hydroxylation of isopropyl group) and at least six measurable primary metabolites [4, 29]. Human metabolite identification in vivo was published for an oral single 600 mg dose solution, primarily in urine and bile. In feces, R2 was reported as dominant metabolite of almost equal radioactivity compared to parent compound followed by R5, about 1/10 of abundance, whereas R3 or R6 were not detectable in urine or feces [29]. In plasma, low metabolite abundance for the major metabolite R2 was reported (Norvir FDA labeling). In this study, measured R2-concentrations were as high as 2 μM using a multiple dosing scheme at lower (400 mg) dose indicating that metabolites were actually readily observed in plasma. The difference in the ratio of R2 to RTV parent compound was only 5-fold, demonstrating the accumulating effect of a continuous dose and potentially through enterohepatic circulation. Measurable CYP2J2 specific metabolites (R3 and R6) in vitro and available banked subject plasma samples from previous investigations [9-11] presented a unique opportunity to investigate the contribution of CYP2J2 to RTV metabolism in vivo. Although R3 seems more abundant in vitro, R6 was chosen to trace CYP2J2 metabolism as the latter was more robustly detected in plasma samples. Parent compound and metabolite degradation in frozen samples over time appears to be low as RTV concentrations matched well with previously determined levels.
With respect to metabolic clearance in vitro, RTV was depleted by CYP2J2 with similar efficiency and exhibited similar metabolic kinetics to CYP3A4 but was less affected by time dependent inhibition. This is not surprising since both CYP2J2 and CYP3A4 are proposed to have similar active site structure [19] and CYP2J2 often metabolizes CYP3A4 substrates at different sites of the molecule [19]. Although Supersomes® do not reflect cellular enzyme to coenzyme-cosubstrate compositions, they demonstrated the only feasible way to clearly distinguish between CYP2J2 and CYP3A4 in vitro clearance. The previous observation that RTV is a strong competitive inhibitor of CYP2J2-mediated terfenadine metabolism [30] already suggested that it could also serve as a substrate for CYP2J2 as RTV obviously bound well into the CYP2J2 active site. Both CYP2J2 and CYP3A4 seem to be high affinity enzymes for RTV - CYP3A4 being the high abundance isozyme but having low capacity (due to auto-inhibition). CYP2D6 seems to have substantially higher capacity, but lower affinity, judged by the clearance values and turnover numbers at saturating concentrations.
The grouping of CYP2J2 hepatic expression into low/medium/high content was justified due to a more pronounced variation, in mostly Caucasian subjects, compared to previously published data [14, 17] and a high number of livers with low CYP2J2 protein below the limit of detection. Thus, although similar median values were found, the grouping represented a more accurate distribution for the higher or extremely low values and would improve addressing extremes in a population modeling study. There is currently no adequate explanation for CYP2J2 protein variability. CYP2J2 genetic variants are rare and the regulation of CYP2J2 is not well characterized [20]. Investigating the variation of intestinal CYP2J2 in a similar fashion would also enrich the later simulation approach especially with regards to the first pass metabolism of ritonavir, but we were restricted to the reported median value in the literature [21] due to the lack of sufficient intestinal samples that are required for this approach.
The static in vitro to in vivo scaling demonstrated a modest CYP2J2-contribution to RTV-metabolism (2-7%) neglecting the effect of CYP3A4-inhibition, CYP2D6-pharmacogenetics and intestinal metabolism. However, CYP2J2-contribution increases to 20% when these conditions are taken into consideration, a situation achieved during chronic administration of RTV to CYP2D6 PM (frequency ∼ 10% in mostly European population [31]). Using CYP-specific inhibitors in incubations with human liver microsomes confirmed a possible maximal 20% contribution to R2 formation implicating a higher CYP2J2 contribution at low CYP3A4 content in CYP2D6 PMs. Overall the IVIVE approach and inhibition experiments provided good evidence for the possible extremes of CYP content/inhibition for CYP3A4 and CYP2D6. Specifically, the inhibition of R2 formation in HLM 119 carrying the CYP2D6*4/*4 genotype suggests contribution of additional enzymes to its formation. It is important to note that the sample size of HLMs tested was limited due to the low number of livers with CYP2D6 PM genotypes and therefore inadequate to develop a correlation between CYP2J2 contribution to RTV clearance and CYP2D6 genotype. Despite the limitations, the HLM data provides potential insight into probable scenarios without controlling for interindividual variation.
The initial simulation using kinact and KI values determined in HLMs and Supersomes® over- predicted the RTV exposure in CYP2D6 PM to a great extent. Based on the previous investigation [11], we chose to simulate the pharmacokinetic profiles using the inhibition data measured in SCHH and scaled the kinact and KI values according to the HLM/CYP inhibition using a linear model. Direct measurement of time dependent inhibition for CYP2J2 in a complex model such as HLM or SCCH was not possible due to the lack of a specific CYP2J2 probe in TDI experiments. Inhibition of dextromethorphan clearance by RTV was previously reported [11], but no TDI was observed experimentally in our study, thus we incorporated RTV only as a competitive inhibitor of CYP2D6. A possible long lasting inhibition of CYP2D6 caused by RTV-metabolites or an incomplete understanding of dextromethorphan clearance necessitates further investigation. This can include inhibition studies in intestinal or liver microsomes using dextromethorphan and RTV metabolites as precipitants or potential transporters involvement. Ritonavir, however, is a poor substrate for ABC transporters [32]. It has been reported to induce P-gp under certain clinical conditions [11], and inhibit P-gp in acute but less in chronic administration [33]. Thus, the impact of transporters in this simulation is likely limited.
With regards to pharmacokinetic modeling, some model related simplifications were necessary, but justifiable, as any sophisticated PK model would require many input parameters (volume of distribution, plasma protein binding and renal clearance of primary/secondary oxidative metabolism). However, using the above described reduction and assuming that R6 and RTV would possibly be eliminated in a similar fashion, predicted the in vivo metabolite exposure (Caverage) well. We also decided not to include CYP3A4 auto-induction [5], as initial simulations demonstrated no significant effect to counterbalance inhibition. This effect was previously confirmed in in vivo induction studies [9]. In addition, the calculation of AUC in the time interval of 1 to 12 h after a multiple dosing scheme applied the principle of superimposable plasma concentration time profiles. Despite these simplifications, AUC, Cmax concentrations and RTV concentration-profiles were predicted adequately demonstrating the feasibility of the chosen inhibition model.
In concentration time profiles of existing plasma samples, the Cmax and AUC of RTV [34, 35] were similar as previously reported [29]. Metabolite profiles showed formation rate limited clearance except for R5 indicating a lower clearance. The chosen concentration-time profiles from the simulations estimated a CYP2J2 contribution to RTV metabolism up to 2%, indicating that all three subjects were not CYP2D6 PM and subject #2 had very low CYP2J2 content.
Interestingly, a high CYP2J2 contribution of up to 70% in outliers in the upper 95th percentile can increase AUC and Cmax by two fold; an increase considered significant in drug interaction studies e.g. with didanosine [5]. From this simulation, a high CYP2J2 contribution is contingent on three factors: CYP2D6 genotype, CYP2J2 content, and also a higher RTV exposure likely impacting CYP3A4 inhibition more strongly. According to the simulation with 500 subjects, about 4% would have these characteristics. Thus, as hypothesized, when CYP2D6 activity is compromised and RTV AUC is high CYP2J2 could become a quantitatively important contributor to hepatic clearance due to slightly stronger inhibition of CYP3A. Such effect could be more pronounced if intestinal variability of CYP2J2 is also considered.
We believe that this study is an interesting example highlighting increased importance of minor pathways due to compromised main clearance pathways. It is also an interesting demonstration that inhibition data from SCCH might lead to more conclusive simulations than data generated by microsomal incubations. A similar study reported an increased contribution of CYP2C8 to imatinib metabolism due to autoinhibition of CYP3A4 [36].
In conclusion, this investigation shows that the contribution of a minor CYP-enzyme to drug metabolism could increase by the inhibition or genotypic inactivation of the major clearance pathways. In the case of RTV, the contribution of CYP2J2 seems to become a secondary metabolic pathway after CYP3A4 inhibition and CYP2D6 poor metabolic activity. As the metabolism of RTV is indeed complex, further investigation in a larger patient population is warranted to confirm the importance of CYP2J2 to clearance in vivo.
Acknowledgments
This work was supported by the Plein Project Grant of the University of Washington, Seattle, WA and from National Institutes of Health, Heart Blood and Lung Institute – RO1-HL096706 (Totah), GM032165 and R01-DA14211, R01-DA02931 and K24-DA00417 (Kharasch).
Non-standard abbreviations
- AUC
area under the plasma concentration-time curve
- CYP
cytochrome P450 monooxygenase
- EM
extensive metabolizer
- IM
intermediate metabolizer
- HLM
human liver microsomes
- PM
poor metabolizer
- RTV
ritonavir
- SCHH
sandwich culture human hepatocytes
- TDI
time dependent inhibition
- MRM
Multiple Reaction Monitoring
- Clhep
hepatic clearance (intrinsic)
- SD
standard deviation
- IVIVE
in vitro-in vivo extrapolation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Rüdiger Kaspera, Email: kaspera@uw.edu.
Brian J. Kirby, Email: brian.kirby@gilead.com.
Tariku Sahele, Email: tarikucs@gmail.com.
Ann C. Collier, Email: acollier@u.washington.edu.
Evan D. Kharasch, Email: kharasch@wustl.edu.
Jashvant D. Unadkat, Email: jash@uw.edu.
References
- 1.Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA. 2012;308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 2.Mathias AA, West S, Hui J, Kearney BP. Dose-response of ritonavir on hepatic CYP3A activity and elvitegravir oral exposure. Clin Pharmacol Ther. 2009;85:64–70. doi: 10.1038/clpt.2008.168. [DOI] [PubMed] [Google Scholar]
- 3.Zeldin RK, Petruschke RA. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-infected patients. J Antimicrob Chemother. 2004;53:4–9. doi: 10.1093/jac/dkh029. [DOI] [PubMed] [Google Scholar]
- 4.Gangl E, Utkin I, Gerber N, Vouros P. Structural elucidation of metabolites of ritonavir and indinavir by liquid chromatography-mass spectrometry. J Chromatogr A. 2002;974:91–101. doi: 10.1016/s0021-9673(02)01243-8. [DOI] [PubMed] [Google Scholar]
- 5.Hsu A, Granneman GR, Witt G, Locke C, Denissen J, Molla A, et al. Multiple-dose pharmacokinetics of ritonavir in human immunodeficiency virus-infected subjects. Antimicrob Agents Chemother. 1997;41:898–905. doi: 10.1128/aac.41.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, et al. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos. 1998;26:552–61. [PubMed] [Google Scholar]
- 7.Kumar GN, Dykstra J, Roberts EM, Jayanti VK, Hickman D, Uchic J, et al. Potent inhibition of the cytochrome P-450 3A-mediated human liver microsomal metabolism of a novel HIV protease inhibitor by ritonavir: A positive drug-drug interaction. Drug Metab Dispos. 1999;27:902–8. [PubMed] [Google Scholar]
- 8.Soars MG, Grime K, Riley RJ. Comparative analysis of substrate and inhibitor interactions with CYP3A4 and CYP3A5. Xenobiotica. 2006;36:287–99. doi: 10.1080/00498250500446208. [DOI] [PubMed] [Google Scholar]
- 9.Kirby BJ, Collier AC, Kharasch ED, Whittington D, Thummel KE, Unadkat JD. Complex drug interactions of HIV protease inhibitors 1: inactivation, induction, and inhibition of cytochrome P450 3A by ritonavir or nelfinavir. Drug Metab Dispos. 2011;39:1070–8. doi: 10.1124/dmd.110.037523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirby BJ, Collier AC, Kharasch ED, Dixit V, Desai P, Whittington D, et al. Complex drug interactions of HIV protease inhibitors 2: in vivo induction and in vitro to in vivo correlation of induction of cytochrome P450 1A2, 2B6, and 2C9 by ritonavir or nelfinavir. Drug Metab Dispos. 2011;39:2329–37. doi: 10.1124/dmd.111.038646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kirby BJ, Collier AC, Kharasch ED, Whittington D, Thummel KE, Unadkat JD. Complex drug interactions of the HIV protease inhibitors 3: effect of simultaneous or staggered dosing of digoxin and ritonavir, nelfinavir, rifampin, or bupropion. Drug Metab Dispos. 2012;40:610–6. doi: 10.1124/dmd.111.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. 2005;5:6–13. doi: 10.1038/sj.tpj.6500285. [DOI] [PubMed] [Google Scholar]
- 13.Fleming I. Vascular cytochrome p450 enzymes: physiology and pathophysiology. Trends Cardiovasc Med. 2008;18:20–5. doi: 10.1016/j.tcm.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 14.Hashizume T, Imaoka S, Mise M, Terauchi Y, Fujii T, Miyazaki H, et al. Involvement of CYP2J2 and CYP4F12 in the metabolism of ebastine in human intestinal microsomes. J Pharmacol Exp Ther. 2002;300:298–304. doi: 10.1124/jpet.300.1.298. [DOI] [PubMed] [Google Scholar]
- 15.Lafite P, Dijols S, Zeldin DC, Dansette PM, Mansuy D. Selective, competitive and mechanism-based inhibitors of human cytochrome P450 2J2. Arch Biochem Biophys. 2007;464:155–68. doi: 10.1016/j.abb.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu KH, Kim MG, Lee DJ, Yoon YJ, Kim MJ, Shon JH, et al. Characterization of ebastine, hydroxyebastine, and carebastine metabolism by human liver microsomes and expressed cytochrome P450 enzymes: major roles for CYP2J2 and CYP3A. Drug Metab Dispos. 2006;34:1793–7. doi: 10.1124/dmd.106.010488. [DOI] [PubMed] [Google Scholar]
- 17.Matsumoto S, Hirama T, Matsubara T, Nagata K, Yamazoe Y. Involvement of CYP2J2 on the intestinal first-pass metabolism of antihistamine drug, astemizole. Drug Metab Dispos. 2002;30:1240–5. doi: 10.1124/dmd.30.11.1240. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto S, Yamazoe Y. Involvement of multiple human cytochromes P450 in the liver microsomal metabolism of astemizole and a comparison with terfenadine. Br J Clin Pharmacol. 2001;51:133–42. doi: 10.1111/j.1365-2125.2001.01292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CA, Neul D, Clouser-Roche A, Dalvie D, Wester MR, Jiang Y, et al. Identification of novel substrates for human cytochrome P450 2J2. Drug Metab Dispos. 2010;38:347–56. doi: 10.1124/dmd.109.030270. [DOI] [PubMed] [Google Scholar]
- 20.Xu M, Ju W, Hao H, Wang G, Li P. Cytochrome P450 2J2: distribution, function, regulation, genetic polymorphisms and clinical significance. Drug Metab Rev. 2013;45:311–52. doi: 10.3109/03602532.2013.806537. [DOI] [PubMed] [Google Scholar]
- 21.Paine MF, Khalighi M, Fisher JM, Shen DD, Kunze KL, Marsh CL, et al. Characterization of interintestinal and intraintestinal variations in human CYP3A-dependent metabolism. J Pharmacol Exp Ther. 1997;283:1552–62. [PubMed] [Google Scholar]
- 22.Lin YS, Dowling AL, Quigley SD, Farin FM, Zhang J, Lamba J, et al. Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol Pharmacol. 2002;62:162–72. doi: 10.1124/mol.62.1.162. [DOI] [PubMed] [Google Scholar]
- 23.Lee CA, Jones JP, 3rd, Katayama J, Kaspera R, Jiang Y, Freiwald S, et al. Identifying a selective substrate and inhibitor pair for the evaluation of CYP2J2 activity. Drug Metab Dispos. 2012;40:943–51. doi: 10.1124/dmd.111.043505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obach RS, Reed-Hagen AE. Measurement of Michaelis constants for cytochrome P450-mediated biotransformation reactions using a substrate depletion approach. Drug Metab Dispos. 2002;30:831–7. doi: 10.1124/dmd.30.7.831. [DOI] [PubMed] [Google Scholar]
- 25.Barre J, Chamouard JM, Houin G, Tillement JP. Equilibrium dialysis, ultrafiltration, and ultracentrifugation compared for determining the plasma-protein-binding characteristics of valproic acid. Clin Chem. 1985;31:60–4. [PubMed] [Google Scholar]
- 26.Yang Y, Grubb MF, Luk CE, Humphreys WG, Josephs JL. Quantitative estimation of circulating metabolites without synthetic standards by ultra-high-performance liquid chromatography/high resolution accurate mass spectrometry in combination with UV correction. Rapid Commun Mass Spectrom. 2011;25:3245–51. doi: 10.1002/rcm.5220. [DOI] [PubMed] [Google Scholar]
- 27.Brunton L, Chabner B, Knollman B. Goodman and Gilman's The Pharmacological Basis of Therapeutics. McGraw-Hill Professional; 2010. [Google Scholar]
- 28.Ernest CS, 2nd, Hall SD, Jones DR. Mechanism-based inactivation of CYP3A by HIV protease inhibitors. J Pharmacol Exp Ther. 2005;312:583–91. doi: 10.1124/jpet.104.075416. [DOI] [PubMed] [Google Scholar]
- 29.Denissen JF, Grabowski BA, Johnson MK, Buko AM, Kempf DJ, Thomas SB, et al. Metabolism and disposition of the HIV-1 protease inhibitor ritonavir (ABT-538) in rats, dogs, and humans. Drug Metab Dispos. 1997;25:489–501. [PubMed] [Google Scholar]
- 30.Hrycay EG, Bandiera SM. Cytochrome P454 Enzymes. In: Gad SC, editor. Preclinical Development Handbook: ADME and Biopharmaceutical Properties. Wiley&Sons; 2008. [Google Scholar]
- 31.Ingelman-Sundberg M, Sim SC, Gomez A, Rodriguez-Antona C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. 2007;116:496–526. doi: 10.1016/j.pharmthera.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Bierman WF, Scheffer GL, Schoonderwoerd A, Jansen G, van Agtmael MA, Danner SA, et al. Protease inhibitors atazanavir, lopinavir and ritonavir are potent blockers, but poor substrates, of ABC transporters in a broad panel of ABC transporter-overexpressing cell lines. J Antimicrob Chemother. 2010;65:1672–80. doi: 10.1093/jac/dkq209. [DOI] [PubMed] [Google Scholar]
- 33.Kharasch ED, Bedynek PS, Park S, Whittington D, Walker A, Hoffer C. Mechanism of ritonavir changes in methadone pharmacokinetics and pharmacodynamics: I. Evidence against CYP3A mediation of methadone clearance. Clin Pharmacol Ther. 2008;84:497–505. doi: 10.1038/clpt.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DiCenzo R, Peterson DR, Cruttenden K, Mariuz P, Rezk NL, Hochreiter J, et al. Effects of minocycline and valproic acid coadministration on atazanavir plasma concentrations in human immunodeficiency virus-infected adults receiving atazanavir-ritonavir. Antimicrob Agents Chemother. 2008;52:3035–9. doi: 10.1128/AAC.00194-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ouellet D, Hsu A, Qian J, Lamm JE, Cavanaugh JH, Leonard JM, et al. Effect of fluoxetine on pharmacokinetics of ritonavir. Antimicrob Agents Chemother. 1998;42:3107–12. doi: 10.1128/aac.42.12.3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Filppula AM, Neuvonen M, Laitila J, Neuvonen PJ, Backman JT. Autoinhibition of CYP3A4 Leads to Major Role of CYP2C8 in Imatinib Metabolism: Variability in CYP2C8 Activity May Alter Plasma Concentrations and Response. Drug Metab Dispos. 2012 doi: 10.1124/dmd.112.048017. [DOI] [PubMed] [Google Scholar]