Abstract
Purpose
Simultaneous dual labeling to visualize specific RNA and protein content within the same formalin-fixed paraffin embedded (FFPE) section can be technically challenging and usually impossible, because of variables such as tissue fixation time and pretreatment methods to access the target RNA or protein. Within a specific experiment, ocular tissue sections can be a precious commodity. Thus, the ability to easily and consistently detect and localize cell-specific expression of RNA and protein within a single slide would be advantageous. In this study, we describe a simplified and reliable method for combined in situ hybridization (ISH) and immunohistochemistry (IHC) for detection of mRNA and protein, respectively, within the same FFPE ocular tissue.
Methods
Whole mouse eyes were prepared for 5 micron FFPE sections after fixation for 3, 24, 48 or 72 h. Customized probes from Advanced Cell Diagnostics to detect mRNA for vascular endothelial growth factor (VEGF), hypoxia-inducible factor 1-alpha (HIF-1α), and hypoxia-inducible factor 2-alpha (HIF-2α) were used for ISH. Various parameters were tested using the novel RNAscope method for ISH and optimized for compatibility with subsequent IHC for glial fibrillary acidic protein (GFAP) or GS-lectin within the same tissue section. Dual fluorescent visualization of Fast Red ISH and Alexa Fluor 488 IHC signal was observed with confocal microscopy.
Results
A fixation time of 72 h was found to be optimal for ISH and subsequent IHC. The RNAscope probes for VEGF, HIF-1α, and HIF-2α mRNA all gave a strong Fast Red signal with both 48 h and 72 h fixed tissue, but the optimal IHC signal for either GFAP or GS-lectin within a retinal tissue section after ISH processing was observed with 72 h fixation. A pretreatment boiling time of 15 min and a dilution factor of 1:15 for the pretreatment protease solution were found to be optimal and necessary for successful ISH visualization with 72 h FFPE ocular tissue.
Conclusions
The protocol presented here provides a simple and reliable method to simultaneously detect mRNA and protein within the same paraffin-embedded ocular tissue section. The procedure, after preparation of FFPE sections, can be performed over a 2-day or 4-day period. We provide an optimization strategy that may be adapted for any RNAscope probe set and antibody for determining retinal or ocular cell-specific patterns of expression.
Introduction
The research community currently lacks a robust and convenient method to simultaneously visualize the presence and distribution of mRNA transcripts and protein products in cells and tissues. Although distinct detection techniques exist for mRNA (in situ hybridization [ISH]) and for proteins (immunohistochemistry [IHC]), dual processing of a histological sample to preserve tissue and cellular morphology is technically difficult and usually limited to frozen sections or insect tissue [1-3]. Loss of structural integrity of formalin-fixed paraffin embedded (FFPE) tissue slices is usually the result of the harsh reagents and conditions initially used for either IHC or ISH, resulting in material of insufficient quality for subsequent downstream assays on the same tissue section. Thus, a simplified and reliable method for the simultaneous detection of mRNA and protein in FFPE tissue is warranted.
In the area of ocular research, the precise cellular localization and colocalization of transcripts and protein products within the cytologically complex, multilaminar neurosensory retina is desirable. Development of a method to use the same ocular tissue section for both IHC and ISH may also reduce the burden on animal welfare and expense by decreasing the number of animals required for each study. The RNAscope probes and signal amplification method used in ISH are based on the branched DNA (bDNA) method described by Collins et al. and Player et al. [4,5]. The bDNA method utilizes single sequence-specific target probes that hybridize to the target DNA or RNA of interest. The RNAscope target probes (Z probes) have been modified to minimize nonspecific hybridization and maximize signal amplification [6]. A Z probe consists of a 25 bp stretch of oligonucleotides that is complementary to a region of the mRNA of interest (visualized as the lower horizontal of the Z), a spacer sequence (visualized as the diagonal of the Z) and a 14 bp linker tail (visualized as the upper horizontal of the Z). Two adjacent 14 bp linker tails from separate Z probes are required for the binding of a preamplifier. Thus, a Z probe pair (ZZ) must align and hybridize to a contiguous 50 bp region of the target mRNA as a prerequisite to form a 28 bp sequence-specific landing site for the preamplifier. The preamplifier can bind up to 20 amplifiers, which in turn can each bind 20 alkaline phosphatase (AP)–conjugated label probes that are subsequently visualized with Fast Red substrate. A mRNA target of 1 kb can bind 20 ZZ probes, yielding an amplification potential of 8,000 label probes, thereby increasing the sensitivity of ISH.
Successful and reproducible hybridization of ZZ probes on FFPE ocular tissue can be challenging. In addition, manipulation methods that allow access for antigens to bind specific antibodies on FFPE slides for optimal IHC are generally incompatible with ISH procedures. The reverse is also true; pretreatment methods that favor probe hybridization for ISH are usually not suitable for downstream IHC studies. Various pretreatment methods have been suggested for optimizing RNAscope ZZ probe hybridization [6] and have been used successfully for ISH on rodent (Mongolian gerbil) gastric tissue [7], but to our knowledge, a standardized method for ocular tissue has not been defined. In this study, we adapt and optimize the RNAscope method of ISH for FFPE mouse retinal tissue sections and modify an IHC protocol for subsequent protein analysis within the same tissue section. We also provide a simple and specific optimization strategy for ISH using the RNAscope ZZ probes for rodent FFPE eye sections.
Methods
Animals
Breeding pairs of C57BL/6 mice were purchased from The Charles River Laboratory (Hollister, CA). Experiments were designed and performed in accordance with the guidelines provided in the ARVO Statement for the use of Animals in Ophthalmic and Vision Research and with the approval of the Institutional Animal Care and Use Committee of Oregon Health & Science University. At postnatal day 13, animals were sacrificed by CO2 euthanasia and subsequent cervical dislocation. Both eyes were enucleated and fixed in 10% neutral buffered formalin for 72 h at room temperature. Fixed eyes were paraffin embedded, sectioned at 5 µm and mounted to slides (Superfrost® Plus, Fisher Scientific, Waltham, MA).
Reagents
1) RNAscope® 2.0 Red FFPE Kit (Cat # 310036, Advanced Cell Diagnostics, Hayward, CA)
2) Target RNA oligo probes specific for the mRNA of mouse vascular endothelial growth factor (VEGF120), hypoxia-inducible factor 1-alpha (HIF-1α), and hypoxia-inducible factor 2-alpha (HIF-2α) were synthesized by Advanced Cell Diagnostics. Probe details are as follows:
VEGF: Catalog # 312931 (Advanced Cell Diagnostics), Accession # NM_001025257.3. Probe region covers 3,106 bp spanning nucleotides 124–3,230
HIF-1α: Catalog # 313821 (Advanced Cell Diagnostics), Accession # NM_010431.2. Probe region covers 973 bp spanning nucleotides 1,182–2,154
HIF-2α: Catalog # 314371 (Advanced Cell Diagnostics), Accession # NM_010137.3. Probe region covers 400 bp spanning nucleotides 3,112–3,511
3) 700 ml 1X Pretreatment Solution 2 (proprietary formulation, Advanced Cell Diagnostics)
4) Preatreatment Solution 3 (proprietary formulation, Advanced Cell Diagnostics). Recommended dilution is 1:15 in phosphate buffered saline (PBS)
5) 3 l of 1X Wash Buffer (proprietary formulation, Advanced Cell Diagnostics)
6) 20X SSC buffer (3M NaCl, 300 mM sodium citrate)
7) 2.0 Amp 1 solution (proprietary formulation, Advanced Cell Diagnostics)
8) 2.0 Amp 2 solution (proprietary formulation, Advanced Cell Diagnostics)
9) 2.0 Amp 3 solution (proprietary formulation, Advanced Cell Diagnostics)
10) 2.0 Amp 4 solution (proprietary formulation, Advanced Cell Diagnostics)
11) 2.0 Amp 5-Red solution (proprietary formulation, Advanced Cell Diagnostics)
12) 2.0 Amp 6-Red solution (proprietary formulation, Advanced Cell Diagnostics)
13) Tris-buffered saline (TBS; 50 mM Tris-Cl, 150 mM NaCl, pH 7.5)
14) Blocking solution (40 µl goat serum, 0.02 g bovine serum albumin and 75 µl TritonX-100 in 20 ml TBS)
15) Slowfade antifade reagent (Invitrogen, Grand Island, NY)
In situ hybridization
[This is a modification of the protocol provided by Advanced Cell Diagnostics, made suitable for subsequent IHC in retinal tissue.]
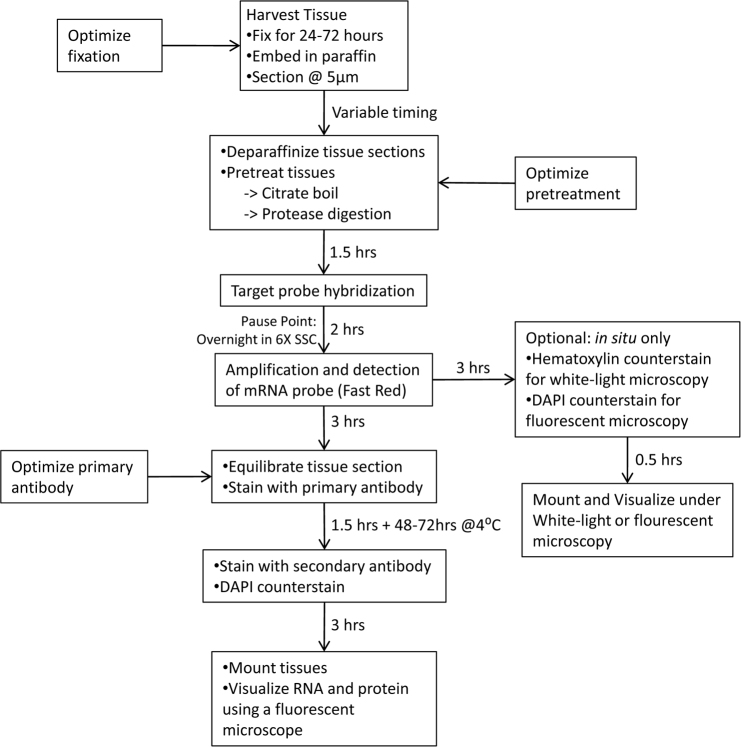
All steps are performed at room temperature unless otherwise indicated. An outline of the dual ISH and IHC protocol is shown in Figure 1.
Figure 1.
Overview of the protocol for combined in situ hybridization and immunohistochemistry in formalin-fixed paraffin embedded ocular tissue. The flowchart demonstrates the various stages of the protocol, including estimations of the time needed for each step.
1. Bake the slides face up for 1 h at 60 °C.
2. Deparaffinize the tissue sections.
a. Incubate the slides twice in fresh xylene for 5 min each.
b. Incubate the slides twice in fresh 100% ethanol for 5 min each.
In preparation for Step 4, heat 700 ml 1X Pretreatment Solution 2 to a boil.
3. Create a hydrophobic barrier around the tissue using an ImmEdge pen (Cat # H-4000, Vector Labs, Burlingame, CA).
4. Ensure the temperature of the 1X Pretreatment Solution 2 is between 100 and 104 °C. Add the slides in a slide rack to the boiling solution for 15 min.
[Note: We have eliminated the Pretreatment 1 step, as this is required only for horseradish peroxidase (HRP) detection methods.]
5. Wash the slides twice in deionized water (diH2O) for 5 min each.
6. Cover the tissue sections with diluted Pretreatment Solution 3 (1:15 dilution in PBS) and incubate in a hybridization oven at 40 °C for 30 min.
[Note: Pretreatment 3 is a protease solution; if it is used undiluted, the incubation time can be shortened to 10−20 min, but there is a risk of over-permeabilization of the tissue. Thus, we recommend using a diluted solution and increasing the incubation time up to 40 min to optimize results.]
7. Wash the slides twice in diH2O for 5 min each. Warm the mRNA target probe solution to 40 °C in preparation for Step 8.
8. Decant the H2O and overlay the tissue with the prewarmed mRNA target probe solution. Incubate in a hybridization oven for 2 h at 40 °C.
[Note: Although we used the Slide Chamber Unit with an MJ Research Thermocycler (MJ Research, St. Bruno, Quebec, Canada) for our hybridization steps, we found that any humidified oven is sufficient. A key point is to keep the oven or chamber humidified and to ensure that the tissue section contained within the hydrophobic barrier is completely covered with probe solution.]
9. Wash the slides twice in 1X Wash Buffer for 2 min each.
OPTIONAL STOPPING POINT: Incubate the slides in 6X SSC (diluted from 20X) for up to 24 h at 4 °C.
10. Wash the slides once for 1 min in 1X Wash Buffer.
[Note: This step can be eliminated if the optional stopping point is not used.]
[Note: The 2.0 Amp 1–6 solutions are supplied in eye drop vials. To cover the tissue in Steps 11–21, we squeezed one or two drops over each tissue section contained within the hydrophobic barrier. Ensure that the whole tissue section is completely covered, and make note of any slides that look dry after an incubation step, as this may influence final signal intensity. If making a comparison with treated/disease versus control slides, we would recommend repeating the probe set for any slides that become dry.]
11. Cover the tissue with 2.0 Amp 1 solution and incubate in the oven for 30 min at 40 °C.
12. Wash the slides twice for 2 min each in Wash Buffer.
13. Cover the tissue with 2.0 Amp 2 solution and incubate for 15 min at 40 °C.
14. Wash the slides twice for 2 min each in Wash Buffer.
15. Cover the tissue with 2.0 Amp 3 solution and incubate for 30 min at 40 °C.
16. Wash the slides twice for 2 min each in Wash Buffer.
17. Cover the tissue with 2.0 Amp 4 solution and incubate for 15 min at 40 °C.
18. Wash the slides twice for 2 min each with Wash Buffer.
19. Cover the tissue with 2.0 Amp 5-Red solution and incubate for 30 min at room temperature in a humidified chamber.
20. Wash the slides twice for 2 min each with Wash Buffer.
21. Cover the tissue with 2.0 Amp 6-Red solution and incubate for 15 min at room temperature in a humidified chamber.
22. Wash the slides twice for 2 min each with Wash Buffer.
23. Prepare the Fast Red solution by mixing Red-B and Red-A at a ratio of 1(Red B):59(Red A). These solutions are provided with the RNAscope® 2.0 Red FFPE Kit.
24. Decant the Wash Buffer from the slides and cover the tissue with the Fast Red solution. Incubate at room temperature and monitor the color change through a microscope (a 10× objective is sufficient). Optimal staining time will depend on the amount of “positive” red staining observed without a strong background. Depending on the abundance of your target transcript within the tissue section, this could be between 1 and 20 min. After 20 min, nonspecific red staining will appear over most of the tissue.
25. When slides are fully developed, immediately decant the Fast Red solution and wash twice for 2 min each in diH2O with gentle agitation. Proceed to the dual-immunohistochemistry protocol.
Dual immunohistochemistry
All steps are performed at room temperature unless otherwise indicated.
1. After the ISH procedure, wash the slides three times for 5 min each in 1X TBS.
2. Block the slides in blocking solution for 1–2 h.
3. Dilute the primary antibody of interest in blocking solution as appropriate. Decant the blocking solution, add the diluted primary antibody, and incubate for 72 h at 4 °C. To reduce evaporation, place the slides in a humidified chamber with ample primary antibody solution.
[Note: For the images presented in Figure 2 and Figure 3, we used: rabbit anti-cow glial fibrillary acidic protein (GFAP; DAKO, Carpinteria, CA) [1:200] and GS Lectin Alexa Fluor 488 (Invitrogen, Grand Island, NY) [1:200].]
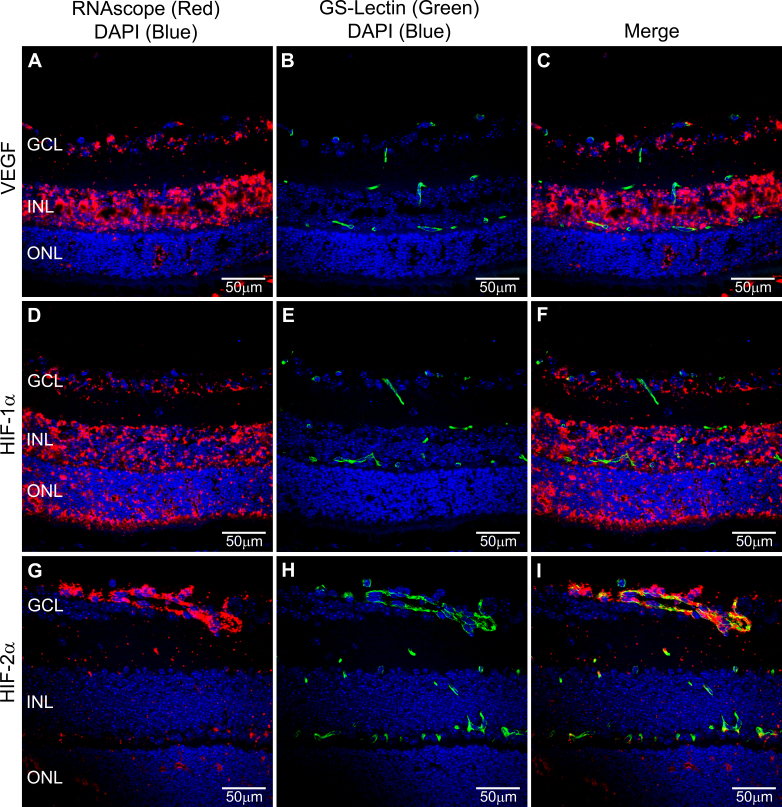
Figure 2.
Dual RNAscope in situ hybridization (ISH) and GS-lectin immunohistochemistry (IHC). Representative images of dual RNAscope ISH (red) and GS-lectin IHC (B, C, E, F, H, I, endothelial cells, green) using mRNA probes for VEGF (A, C), HIF-1α (D, F) and HIF-2α (G, I). Nuclei were labelled with 4’,6-diamidino-2-phenylindole (DAPI) to distinguish the nuclear layers of the retina. All images were obtained using a 40× objective. Labels: ganglion cell layer (GCL), inner nuclear layer (INL), outer nuclear layer (ONL).
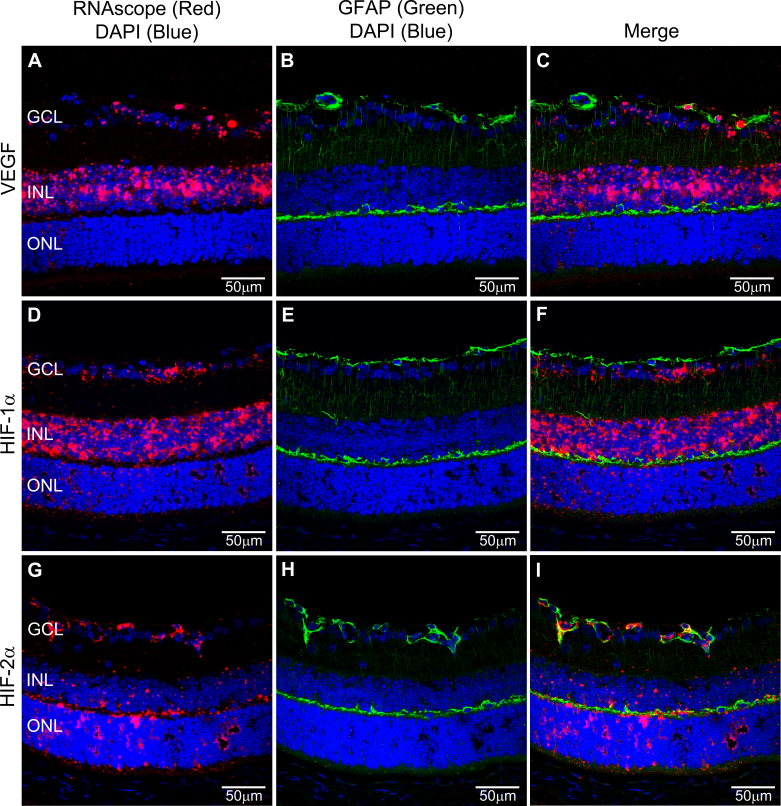
Figure 3.
Dual RNAscope in situ hybridization (ISH) and GFAP immunohistochemistry (IHC). Representative images of dual RNAscope ISH (red) and GFAP IHC (B, C, E, F, H, I, Müller cells, green) using mRNA probes for VEGF (A, C), HIF-1α (D, F) and HIF-2α (G, I). Nuclei were labelled with 4’,6-diamidino-2-phenylindole (DAPI) to distinguish the nuclear layers of the retina. All images were obtained using a 40× objective. Labels: ganglion cell layer (GCL), inner nuclear layer (INL), outer nuclear layer (ONL).
4. Wash the slides three times for 5 min each in 1X TBS.
5. Dilute the secondary antibody in blocking solution at a predetermined concentration. Decant the TBS, add the diluted secondary antibody, and incubate for 2–3 h at room temperature in a humidified chamber.
[Note: For Figure 3, we used: goat anti-rabbit Alexa Fluor 488 (Invitrogen) [1:200]. No secondary antibody is required for the directly conjugated GS-lectin (Figure 2); thus, this step was eliminated.]
6. Wash the slides three times for 5 min each in 1X TBS.
7. Optional: Apply 4’6-diamidino-2-phenylindole (DAPI), diluted 1:50,000 in H2O, for 3 min to visualize nuclei (wavelength = 405 nm).
8. Wash the slides three times for 5 min each in 1X TBS.
9. Mount the slides using Slowfade antifade reagent (Invitrogen). Decant the TBS wash solution and cover the tissue with Slowfade. Gently drop a coverslip onto the slide and seal with nail polish.
10. Image the slides immediately or store them at 4 °C.
Imaging
Images were obtained using fluorescent confocal microscopy (Olympus Fluoview 1000, Olympus Corporation, Center Valley, PA). The Fast Red signal was visualized using the 594 nm laser. [Note: Certain AP substrates that allow colorimetric detection, such Fast Red and Fast Blue, can also be visualized fluorescently [8]. This allows detection of dual or multiple target transcripts.]
Optimization of the protocol and troubleshooting
The protocol we have presented is a modification of the protocol recommended by Advanced Cell Diagnostics and was specifically adapted to preserve retinal detail for ISH and subsequent IHC. Although all our probe sets worked best after 72 h fixation, optimization may be required for each specific mRNA target and downstream antibody detection. Our experience suggested that the fixation times (3, 24, 48, or 72 h) and Pretreatment 2 and Pretreatment 3 incubation times should be the first steps altered for optimization. We also recommend testing various concentrations of Pretreatment 3, from undiluted to a 1:15 dilution in PBS.
Results
The detailed protocol presented in the Methods and outlined in the Figure 1 flow chart provided cell-specific positive staining for all probes analyzed. In addition, the protocol allowed for subsequent IHC for GS-lectin (Figure 2) and GFAP (Figure 3). Note in Figure 2 the overlap of the HIF-2α ISH signal and lectin labeling of the developing retinal vasculature within the ganglion cell layer (GCL). This procedure can be adapted to detect other retina-specific mRNAs and proteins following the presented optimization protocol.
In our experience, the most important factors to obtain successful staining were the fixation time and the concentration of the Pretreatment 3 solution. We found the most successful fixation time for this protocol to be 72 h. This length of time enabled the tissue to withstand the harsh conditions of the ISH protocol while still enabling detection of proteins by standard IHC. Certain antibodies may require further optimization and favor a shorter fixation. We obtained successful ISH staining using tissues fixed for as little as 3 h; however, the shorter fixation led to increased tissue degradation after incubation with Pretreatment 2 and 3. Thus, if a shorter fixation time is necessary for a particular probe set and corresponding antibody, shorter incubation times with Pretreatments 2 and 3 are recommended. The dilution of the Pretreatment 3 solution is another important variable, as this heat-mediated enzymatic digestion step can destroy tissue morphology at high concentrations. We found that dilution of Pretreatment 3 to 1:15 in PBS and incubation for 30 min at 40 °C gave the best ISH probe signal while maintaining tissue integrity for subsequent morphological and IHC analysis.
It is also possible to vary the times associated with each of the pretreatment steps. As stated in our Methods, Pretreatment 1 was unnecessary, as we used a Fast Red detection system, eliminating the need to quench endogenous peroxidases associated with a horseradish peroxidase (HRP) detection system. We found the boil step using Pretreatment 2 solution to be critical for successful ISH, and therefore the full 15 min boil in undiluted Pretreatment 2 solution is recommended. If undiluted Pretreatment 3 solution is used, the incubation time should be shortened from 30 min to 10–20 min.
Discussion
The retina is sensitive to the harsh conditions of standard ISH, resulting in compromised morphology and diffuse staining in paraffin-embedded tissue. Using the RNAscope method of ISH, we describe a dual ISH and IHC protocol that is suitable for simultaneously detecting mRNA and proteins in cytologically preserved murine retina. The RNAscope method of ISH differs from most standard techniques and is based on the bDNA method that is described in detail in the Introduction [6]. The main difference in this new technology lies in the probe design, which enhances sensitivity while maintaining specificity of hybridization to target sequences.
The success of sensitive and reproducible ISH using the RNAscope methodology opens the possibility of designing probes to shorter regions and the localization of alternatively spliced variants of the same gene. Typically, probes are designed to a target sequence of about 1,000 bp (20 ZZ probe pairs) for an optimum hybridization signal. A target sequence of less than 300 bp (six ZZ probe pairs) is not recommended, as the chance of off-target hybridization may be increased, along with further susceptibility to failure if RNA degradation has occurred. Successful hybridization and signal visualization have been reported, however, with as few as three ZZ probe pairs, corresponding to a target region of 150 bp [6]. If the target sequence is abundantly expressed, we postulate that high-quality ISH can be achieved on very short targets (between 150 and 300 bp) by designing shorter ZZ probe pairs, where each probe is 18 bp and not 25 bp, to span the splice junctions that are unique to specific mRNA isoforms.
We modified the RNAscope ISH protocol to allow subsequent protein analysis within the same tissue section. Specifically, we employed the unique property of Fast Red to fluoresce [8] under 594 nm laser stimulation, thereby eliminating the need to purchase fluorescent-labeled probes for ISH, and adapted the ISH protocol to accommodate subsequent IHC reactions without compromising protein integrity or tissue morphology. This approach yielded clean signals with minimal background. We demonstrated the applicability of these methods by localizing mRNA transcribed from genes pertinent to hypoxic disease (VEGF, HIF-1α, and HIF-2α) to cells within ocular tissue, with subsequent IHC to localize the transcripts to specific retinal cells within the same tissue section.
Concurrent localization of mRNA and the encoded protein, and comparison of the relative abundance of both transcripts and proteins under different conditions, is valuable when trying to correlate transcriptional regulation with post-translational control for a gene of interest. Depending on cell type, only about 40% of cellular protein levels can be directly attributed to mRNA abundance [9]. Thus, this method may be useful to study how manipulations in mRNA levels (by siRNAs or micro-RNAs, for example) impact protein abundance.
Although our method is optimized for mouse ocular tissue, this protocol could be easily adapted to study the temporal and spatial patterns of expression of a plethora of transcripts associated with complex degenerative disease (such as age-related macular degeneration) or neoplastic disease (ocular melanoma) in archived FFPE human ocular tissue. We have provided detailed steps for optimization of fixation and pretreatment conditions to expand this method to other tissues and potentially other organisms, which will accommodate much wider research and clinical applications.
Acknowledgments
This work was supported by the National Eye Institute (grants R01EY019042 to BA and JTS, and R01EY018625 to CWM), the Clayton Foundation for Research, and Research to Prevent Blindness.
References
- 1.Nehmé B, Henry H, Mouginot D. Combined fluorescent in situ hybridization and immunofluorescence: Limiting factors and a substitution strategy for slide-mounted tissue sections. J Neurosci Methods. 2011;196:281–8. doi: 10.1016/j.jneumeth.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Newton SS, Dow A, Terwilliger R, Duman R. A simplified method for combined immunochemistry and in-situ hybridization in fresh-frozen, cryocut mouse brain sections. Brain Res Brain Res Protoc. 2002;9:214–9. doi: 10.1016/s1385-299x(02)00148-4. [DOI] [PubMed] [Google Scholar]
- 3.Toledano H, D’Alterio C, Lonza-Coll M, Jones DL. Dual fluorescence detection of protein and RNA in drosophila tissues. Nat Protoc. 2012;7:1808–17. doi: 10.1038/nprot.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins ML, Irvine B, Tyner D, Fine E, Zayati C, Chang C, Horn T, Ahle D, Detmer J, Shen LP, Kolberg J, Bushnell S, Urdea MS, Ho DD. A branched DNA signal amplification assay for quantification of nucleic acid targets below 100 molecules/ml. Nucleic Acids Res. 1997;25:2979–84. doi: 10.1093/nar/25.15.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Player AN, Shen LP, Kenny D, Antao VP, Kolberg JA. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J Histochem Cytochem. 2001;49:603–12. doi: 10.1177/002215540104900507. [DOI] [PubMed] [Google Scholar]
- 6.Wang F, Flanagan J, Su N, Wang L, Bui S, Nielson A, Wu X, Vo H, Ma X, Luo Y. RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14:22–29. doi: 10.1016/j.jmoldx.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sørdal Ø, Qvigstad G, Nordum IS, Gustafsson B, Waldum HL. In situ hybridization in human and rodent tissue by the use of a new and simplified method. Appl Immunohistochem Mol Morphol. 2013;21:185–9. doi: 10.1097/PAI.0b013e31825a0048. [DOI] [PubMed] [Google Scholar]
- 8.Lauter G, Söll I, Hauptmann G. Two-color fluorescent in situ hybridization in the embryonic zebrafish brain using differential detection systems. BMC Dev Biol. 2011;11:1–11. doi: 10.1186/1471-213X-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–32. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]