Significance
IL-10 is an immune-regulatory cytokine with pro- and anti-inflammatory functions. Through its B cell-stimulating capacities, IL-10 contributes to the differentiation, activation and survival of B cells. Thus, it has been linked with autoimmune disorders, including systemic lupus erythematosus (SLE). Here, we demonstrate T cells as a source of increased IL-10 expression in SLE. Reduced DNA methylation of the IL10 gene allows for transcription-factor recruitment. Increased phosphorylation of the transcription factor Stat3 in SLE T cells results in epigenetic remodeling and trans-activation of IL10, allowing for IL-10 expression. Thus, our observations offer molecular targets in the search for pathophysiologic mechanisms and target-directed treatment options in SLE.
Abstract
The immune-regulatory cytokine IL-10 plays a central role during innate and adaptive immune responses. IL-10 is elevated in the serum and tissues of patients with systemic lupus erythematosus (SLE), an autoimmune disorder characterized by autoantibody production, immune-complex formation, and altered cytokine expression. Because of its B cell-promoting effects, IL-10 may contribute to autoantibody production and tissue damage in SLE. We aimed to determine molecular events governing T cell-derived IL-10 expression in health and disease. We link reduced DNA methylation of the IL10 gene with increased recruitment of Stat family transcription factors. Stat3 and Stat5 recruitment to the IL10 promoter and an intronic enhancer regulate gene expression. Both Stat3 and Stat5 mediate trans-activation and epigenetic remodeling of IL10 through their interaction with the histone acetyltransferase p300. In T cells from SLE patients, activation of Stat3 is increased, resulting in enhanced recruitment to regulatory regions and competitive replacement of Stat5, subsequently promoting IL-10 expression. A complete understanding of the molecular events governing cytokine expression will provide new treatment options in autoimmune disorders, including SLE. The observation that altered activation of Stat3 influences IL-10 expression in T cells from SLE patients offers molecular targets in the search for novel target-directed treatment options.
IL-10 is an immune-regulatory cytokine that plays a central role in innate and adaptive immune responses (1, 2). Immune cells ubiquitously express IL-10 with T and B cells, natural killer (NK) cells, mast cells, eosinophils, dendritic cells, and monocytes/macrophages as major sources. IL-10 modulates T-cell responses through the inhibition of major histocompatibility complex class II expression, limited costimulation, and reduced proinflammatory cytokine expression from antigen-presenting cells. Conversely, IL-10 promotes B-cell differentiation, proliferation, survival, and antibody production. Thus, IL-10 has been implicated in the pathophysiology of autoimmune disorders (1–3).
Systemic lupus erythematosus (SLE) is an autoimmune disease of unknown etiology. In SLE, immune responses are directed against cells, tissues, and organs. Autoantibody production by B cells and plasma cells, the accumulation of immune complexes in tissues, and excessive cytokine production contribute to autoimmune pathology (4). A growing body of literature suggests increased IL-10/IL-10 receptor interactions contributing to SLE (1–3). Studies document increased IL-10 serum levels in SLE patients and lupus-prone mice correlating with disease activity, antibody production, and organ damage (2, 3, 5–9). The molecular mechanisms governing IL10 are incompletely understood. IL-10 expression is controlled on the transcriptional and posttranscriptional levels. IL10 is trans-regulated by a number of factors, including signal transducer and activator of transcription molecules Stat1, Stat3, Stat4, and Stat5 (1, 2). We and others demonstrated that, in addition to the IL10 promoter, an enhancer in the fourth intron, referred to as intronic Stat-responsive element (I-SRE), regulates IL-10 in murine NK cells (through Stat4) and human T cells (through Stat5) (10, 11). Recruitment of Stat proteins to the I-SRE induces epigenetic remodeling. The molecular mechanisms, however, remained unknown (10, 11).
Here, we demonstrate that DNA methylation controls Stat recruitment to IL10 regulatory elements. In SLE T cells, reduced DNA methylation allows for transcription-factor recruitment. Both Stat3 and Stat5 trans-activate the IL10 promoter and the I-SRE. Furthermore, Stat3 and Stat5 interact with the histone acetyltransferase p300, instructing epigenetic remodeling. Increased Stat3 phosphorylation in SLE T cells enhances its recruitment to regulatory elements and the replacement of Stat5 at the I-SRE, resulting in trans-activation and epigenetic remodeling. Correction of increased Stat3 phosphorylation and the resulting imbalance between Stat3 and Stat5 could provide novel treatment strategies in SLE.
Results
IL-10 Expression Is Increased in SLE T Cells.
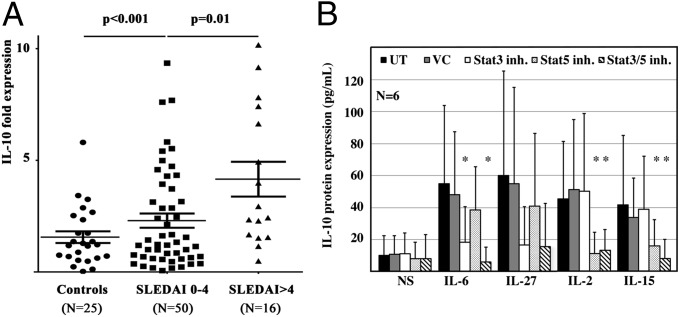
IL-10 may contribute to SLE. The cellular sources, however, are incompletely understood. We asked whether IL-10 expression is increased in T cells from SLE patients. Indeed, we detected higher IL-10 mRNA expression in ex vivo isolated T cells from inactive [SLE disease activity index (SLEDAI) 0–4] and active SLE patients (SLEDAI >4) (Fig. 1A) compared with controls, reflecting disease activity in individual patients (Fig. S1A). Also, serum IL-10 protein levels were increased in active SLE patients (Fig. S1B) and correlated with disease activity (Fig. S1C). Our observations indicate that T cells are a source of IL-10 in SLE patients with T cell-derived IL-10 expression following disease activity.
Fig. 1.
Cytokine-induced IL-10 expression in T cells is Stat3- or Stat5-dependent. (A) IL-10 mRNA expression is increased in ex vivo isolated T cells from SLE patients (n = 66) compared with controls. T cells from active SLE patients, defined as individuals with an SLE disease activity index (SLEDAI) >4, express more IL-10 compared with inactive SLE patients (SLEDAI <4). (B) Primary human T cells were cultured over 24 h in the absence (NS) or presence of cytokines (IL-6, IL-27, IL-2, IL-15). IL-10 protein expression was monitored in cell-culture supernatants from all groups. Absolute values are displayed in A, and the relative increase is given in B. To monitor the effects of Stat transcription factors, some cells were treated with Stat3 or Stat5 inhibitors or their combination. Vehicle controls (VC) were included. IL-6 induces IL-10 protein expression primarily through Stat3. Stimulation with IL-27 induces IL-10 expression through mainly Stat3 but also Stat5. IL-2 and IL-15 both strongly induce IL-10 protein expression through Stat5. UT, untreated.
Cytokine-Induced IL-10 Expression Is Regulated by Stat3 and Stat5.
Stat transcription factors regulate IL10. The molecular mechanisms governing IL-10 expression remain unknown. To investigate the effects of Stat3 and Stat5, we stimulated primary human T lymphocytes with cytokines that activate Stat3 (IL-6 and IL-27) or Stat5 (IL-2 and IL-15). To induce surface expression of high-affinity cytokine receptors on T cells, they had been prestimulated for 3 d with anti-CD3 and anti-CD28 antibodies, followed by a 24-h “starvation” period. To investigate the involvement of Stat3 or Stat5 in IL-10 expression, chemical inhibitors were used (Fig. 1 B and C). IL-6 induced IL-10 primarily through Stat3 because concomitant chemical blockade of Stat3 but not Stat5 abrogated IL-10 expression. Stimulation with IL-27 induced IL-10 through both Stat3 and Stat5. Stat3 blockade had more pronounced effects on IL-10 expression compared with Stat5 blockade, suggesting a more central role of Stat3 in response to IL-27. IL-2 and IL-15 both induced IL-10 protein expression, which was primarily mediated by Stat5.
Bioinformatic Analysis of IL10.
To map in silico predicted Stat-responsive elements (SREs) to their exact genomic location and previously reported DNase hypersensitivity sites, we performed a web-based alignment of mammalian IL10 genes using the University of California, Santa Cruz Genome Browser (http://genome.ucsc.edu/cgi-bin/hgTracks?db=hg19&position=chr1%3A206940947-206945839). The IL10 gene has a well-defined 5′ proximal promoter spanning ∼1,000 base pairs. IL10 spans five exons and four introns, followed by a 3′ untranslated region (UTR) (Fig. S2). Sequence conservation is particularly high within the proximal promoter, the six mRNA-coding regions (five exons and the 3′ UTR) and two noncoding regions in the third and the fourth intron, all mapping DNase hypersensitivity regions.
Stat3 and Stat5 Govern IL-10 Expression.
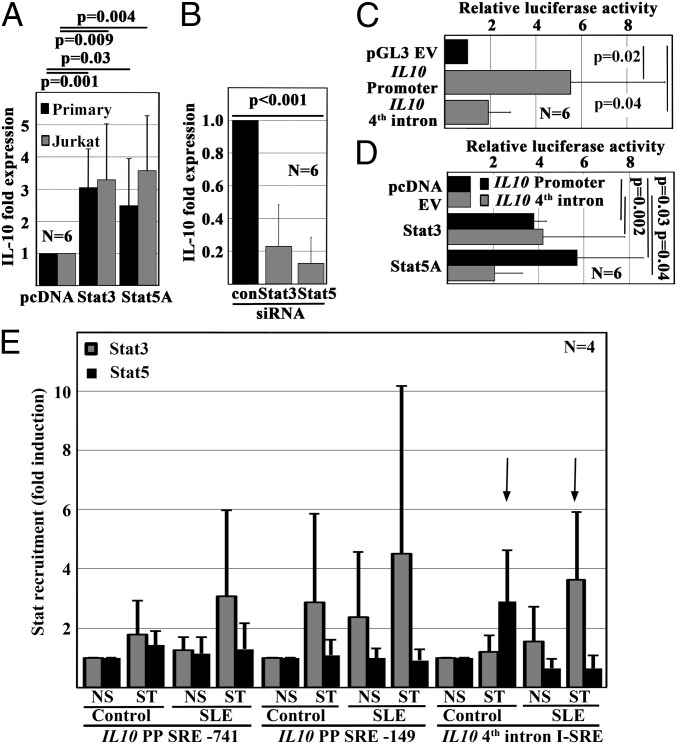
To investigate effects of Stat3 and Stat5 on IL10, we forced their expression in primary human and in Jurkat T cells (Fig. 2A). Both forced expression of Stat3 or Stat5 induced IL-10 mRNA expression in human T cells. Thus, we performed Stat3 or Stat5 knockdown studies in Jurkat T cells, which resulted in reduced IL-10 mRNA expression (Fig. 2B). Therefore, we aimed to determine the involvement of promoter SREs and the I-SRE in the regulation of IL10. Using an online tool (TF search engine: www.cbrc.jp/research/db/TFSEARCH.html), we predicted two SREs within the IL10 promoter −741 and −149 base pairs upstream of the transcriptional start site (SRE-741 and SRE-149) (Fig. S3A) in addition to the previously reported I-SRE in the fourth intron (Fig. S3A). Next, we generated luciferase reporter constructs as follows: (i) a 943 base pair-spanning construct harboring the two promoter SREs and (ii) a 630 base pair-spanning construct harboring the I-SRE (Fig. S3B). Both constructs exhibited increased luciferase activity compared with an empty pGL3 vector (Fig. 2C). Forced expression of either Stat3 or Stat5 resulted in increased luciferase activity in both constructs. Of note, Stat3 effects on the I-SRE were higher compared with Stat5 (Fig. 2D). The difference, however, did not meet statistical significance. Deletion of the SRE-149 within the IL10 promoter or the I-SRE both reduced luciferase activity, which then also failed to be increased in response to Stat3 or Stat5 (Fig. S3C).
Fig. 2.
T cell-derived IL-10 expression is controlled by Stat3 and Stat5. (A) Stat3 or Stat5 expression was forced in ex vivo isolated primary human or unstimulated Jurkat T cells. Both Stat3 and Stat5 significantly induced IL-10 mRNA expression compared with cells transfected with pcDNA empty vectors. (B) In agreement with this observation, Stat3 or Stat5 knockdown with siRNAs resulted in abrogated IL-10 mRNA expression in Jurkat T cells. (C) Transfected into Jurkat T cells, luciferase reporter constructs containing the 5′ proximal promoter sequence or an intronic enhancer element (fourth intron) exhibit increased luciferase activity when compared with an empty pGL4 vector (pGL3 EV). (D) Both Stat3 and Stat5 increased luciferase activity of either of the two reporter constructs in Jurkat T cells. (E) As assessed by ChIP, Stat3 and Stat5 differentially recruit to specific SREs in response to T-cell receptor (TCR) stimulation in primary human T cells from controls and SLE patients. Of note, T cells from SLE patients exhibit two distinct differences to controls: (i) increased recruitment of Stat3 under resting conditions and (ii) the replacement of Stat5 with Stat3 in an intronic enhancer element (I-SRE).
Stat3 and Stat5 Recruit Differentially to SREs.
Both Stat3 and Stat5 induce IL-10. Stat3, however, exhibited higher efficacy in the trans-activation of the I-SRE compared with Stat5. Thus, we aimed to determine whether Stat3 and Stat5 differentially recruit to the IL10 promoter and the I-SRE in human T cells. Performing ChIP, we mapped Stat3 and Stat5 recruitment to the IL10 proximal promoter and I-SRE (Fig. 2F). In T cells from controls, Stat3 recruited to the proximal promoter (SRE-149) in response to stimulation with anti-CD3 and anti-CD28 antibodies whereas Stat5 recruitment mostly localized to the I-SRE. In SLE T cells, Stat3 recruited to both promoter SREs (SRE-741 and SRE-149). Interestingly, Stat3 replaced Stat5 at the I-SRE in SLE T cells.
T Cells from SLE patients Exhibit Increased Stat3 Phosphorylation.
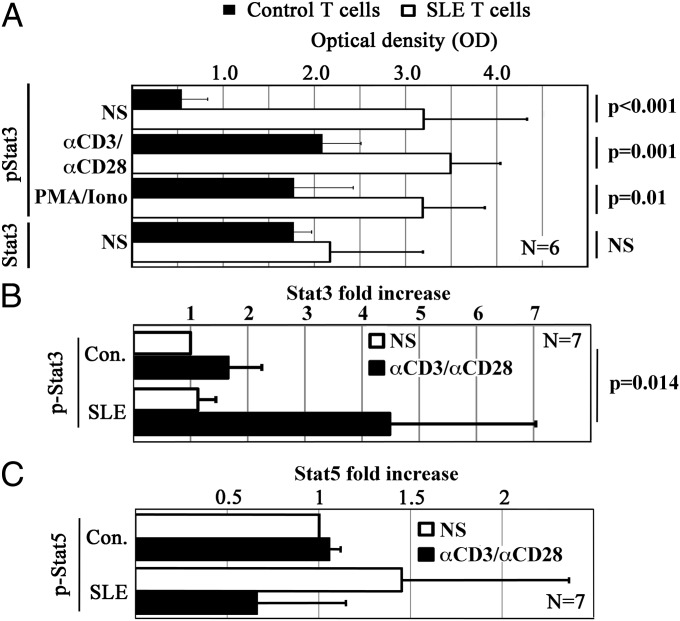
Stat transcription factors undergo activation through phosphorylation. To determine whether increased Stat3 phosphorylation (pStat3) results in the replacement of Stat5 at the I-SRE of SLE T cells, we performed intracellular Stat3/pStat3 ELISAs (Pierce). SLE T cells exhibited comparable amounts of total Stat3 compared with controls. In contrast, pStat3 was significantly increased in resting SLE T cells, and after stimulation (Fig. 3 A and B). Of interest, especially unstimulated SLE T cells had increased amounts of pStat3. Next, we aimed to determine whether potentially reduced Stat5 phosphorylation (pStat5) might partially be responsible for its replacement by pStat3 at the I-SRE. We performed intracellular transcription factor staining, followed by flow cytometry. No significant differences were detected between ex vivo isolated T cells from SLE patients and controls (Fig. 3C). Taken together, our findings indicate an imbalance between pStat3 and pStat5 in SLE T cells that may contribute to increased IL-10 expression.
Fig. 3.
T cells from SLE patients exhibit increased Stat3 phosphorylation. (A) Primary human T cells were cultured over 6 h in the absence (NS) or presence of anti-CD3 and anti-CD28 antibodies (αCD3/αCD28) or phorbol 12-myristate 13-acetate and ionomycine as indicated. (A) Phospho-Stat3 (Tyr705) (pStat3) and total Stat3 levels were measured using intracellular ELISA kits. Although total Stat3 levels were comparable in T cells from SLE patients and controls, pStat3 was significantly increased in T cells from SLE patients under resting conditions and in response to T-cell stimulation. (B) Phospho-Stat3 (Tyr705) in response to stimulation with anti-CD3 and anti-CD28 antibodies was additionally assessed by intracellular staining, confirming the results from A. (C) Phospho-Stat5 (Tyr694) was monitored in resting and stimulated T cells from SLE patients and controls by intracellular staining. No significant differences were determined between the groups.
Reduced DNA Methylation Promotes IL-10 Expression.
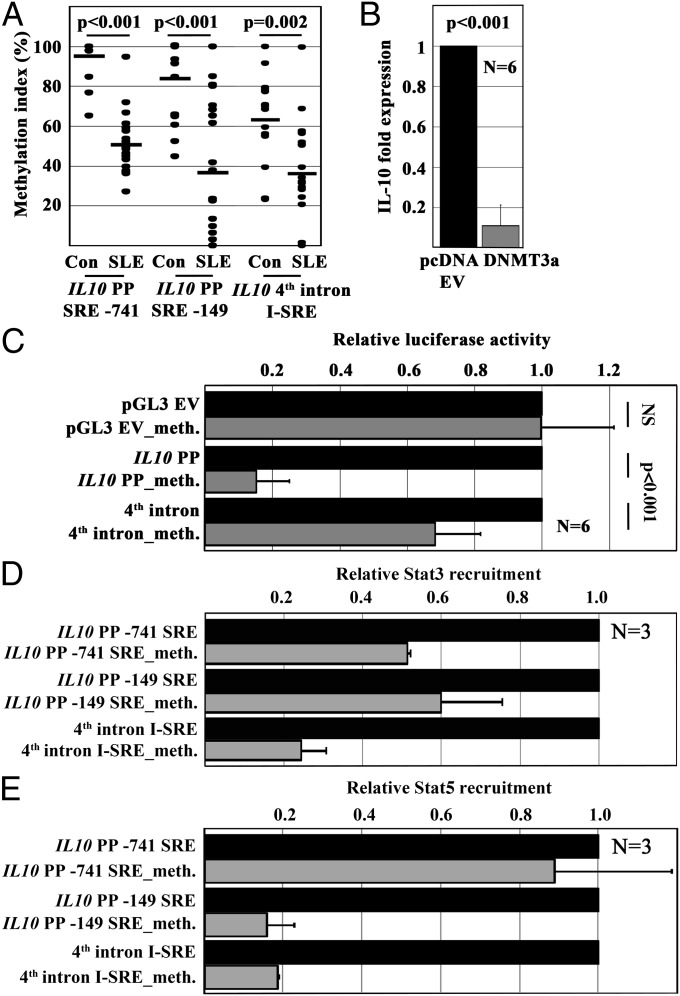
Reduced DNA methylation of the IL10 promoter and the fourth intron correlate with gene expression. Thus, we asked whether the IL10 promoter and the I-SRE exhibit reduced DNA methylation, allowing for transcription factor/DNA interactions. Indeed, SLE T cells exhibited reduced DNA methylation within the proximal promoter and the I-SRE (Fig. 4A). To determine the potential influence of DNA methylation on IL-10 expression, we forced the expression of DNA methyltransferase (DNMT)3A, inducing de novo DNA methylation. Indeed, methylation of IL10 reduced gene expression (Fig. 4B). To target effects of DNA methylation on IL10 regulation, we methylated our luciferase constructs with M.Sssl (Zymo Research). DNA methylation resulted in reduced luciferase activity of both the promoter and the enhancer constructs (Fig. 4C), which was mediated by impaired Stat3 or Stat5 recruitment, as assessed by Reporter-ChIP (Fig. 4 D and E). In agreement with the aforementioned ChIP experiments in human T cells, Stat5 was not recruited to the SRE-741.
Fig. 4.
Reduced DNA methylation allows for Stat recruitment in SLE T cells. (A) DNA methylation of regulatory regions within the IL10 5′ proximal promoter (SRE-741 and SRE-149) and an intronic enhancer element (I-SRE) is significantly reduced in T cells from SLE patients compared with controls. (B) Increased DNA methylation through forced expression of DNMT3a results in significantly reduced IL-10 mRNA expression. (C) Enzymatic methylation of reporter constructs of IL10 regulatory elements results in reduced luciferase activity. (D and E) Reduced luciferase activity is caused by impaired Stat3 (D) or Stat5 (E) recruitment to methylated SREs. Of note, in agreement with our findings from Fig. 2F, Stat5 is not recruited to the SRE-741 in the IL10 promoter.
Stat3 and Stat5 Mediate Histone Methylation Through Their Interaction with p300.
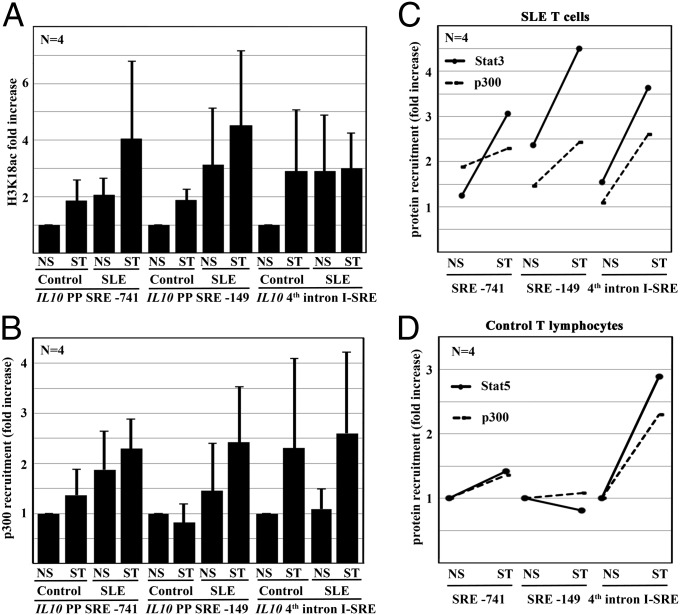
T-cell activation results in histone acetylation around the IL10 I-SRE. The exact molecular mechanisms, however, remained unclear. We aimed to investigate whether histone acetylation, as assessed by histone H3 acetylation at lysine K18 (H3K18ac), follows Stat recruitment. Indeed, in response to stimulation with anti-CD3 and anti-CD28 antibodies, T cells from controls and SLE patients exhibited increased H3K18ac of the IL10 promoter. Of note, ex vivo isolated SLE T cells already exhibited H3K18ac levels that were achieved in control T cells only after stimulation (Fig. 5A). Interestingly, H3K18ac of the I-SRE was increased in control T cells only in response to stimulation. Ex vivo isolated T cells from SLE patients already exhibited H3K18ac, which was not further increased in response to stimulation.
Fig. 5.
IL10 undergoes histone Hk18 acetylation through p300 in response to TCR stimulation. (A) IL10 undergoes epigenetic modifications in response to TCR stimulation with anti-CD3 and anti-CD28 antibodies as assessed by ChIP. Both the proximal promoter and an intronic enhancer element exhibit increased H3K18ac. Of note, H3K18 in T cells from SLE patients is strikingly higher compared with controls. (B) The transcriptional coactivator p300 that has histone acetyltransferase activity recruits to the same regions. P300 recruitment is enhanced in T cells from SLE patients. (C and D) Stat3 (C, in SLE T cells) or Stat5 (D, in controls) recruitment maps p300 recruitment in response to TCR stimulation. NS, not stimulated; ST, simulated.
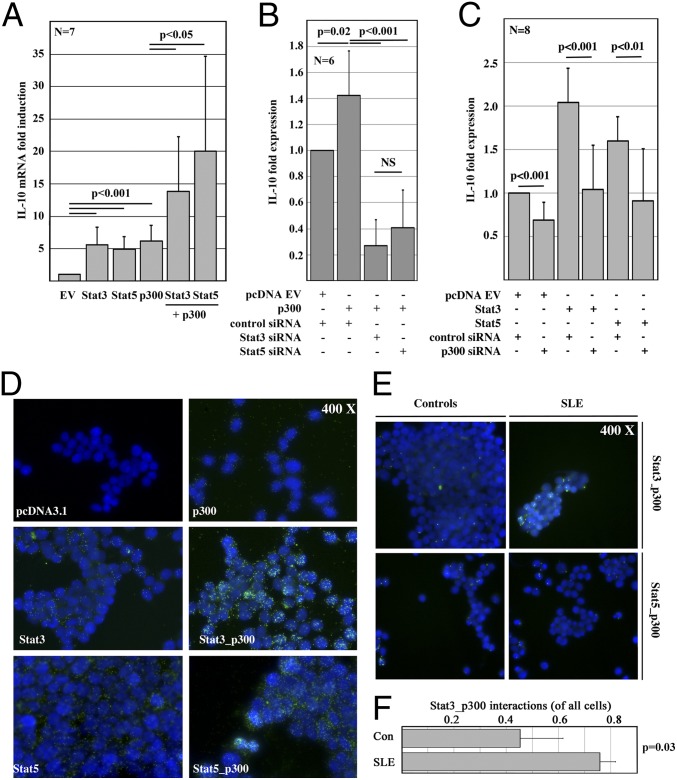
Studies suggested Stat transcription factors interacting with the histone acetyltransferase p300. Thus, we mapped Stat3 and Stat5 recruitment with p300 (Fig. 5 B–D). Indeed, Stat3 in SLE T cells (Fig. 5C) and Stat5 in control T cells (Fig. 5D) colocalized with p300. To explore potentially synergistic effects between Stat3 or Stat5 and p300, we forced the expression of either Stat transcription factor in the absence or presence of p300 (Fig. 6A). Indeed, forced p300 expression significantly increased the transcription of IL-10. The combination of Stat3 or Stat5 with p300 had synergistic effects on IL-10 expression. Thus, we aimed to establish functional interactions between Stat transcription factors and p300. We forced the expression of p300, at the same time knocking down Stat3 or Stat5 with siRNAs (Fig. 6B). As before, p300 increased IL-10 expression. Interestingly, either Stat3 or Stat5 knockdown reduced the effects of p300. This finding was in agreement with our observations in response to forced Stat3 or Stat5 expression with or without p300 knockdown (Fig. 6C). Stat3 or Stat5 induced IL-10 expression that was reduced as a result of p300 knockdown, suggesting an interaction and functional synergy between Stat3 or Stat5 and p300. To investigate whether Stat3 or Stat5 mediates p300 recruitment, we forced the expression of Stat3 or Stat5 monitoring p300 recruitment to the IL10 gene. Indeed, Stat3 or Stat5 expression in human T cells resulted in increased recruitment of p300 to SRE-149 and the I-SRE (Fig. S4A) that was reflected by H3K18ac (Fig. S4B). Interestingly, Stat3 did not recruit p300 to SRE-741. In agreement with these observations, inhibition of p300 with C646 resulted in impaired IL-10 expression in response to stimulation with Stat-inducing cytokines (Fig. S4C). Experiments in T cells from healthy controls, which were stimulated with anti-CD3 and anti-CD28 antibodies in the absence or presence of Stat3 or Stat5 inhibitors confirmed differential Stat-dependent corecruitment of p300 (Fig. S5). Although p300 corecruits with Stat3 to the IL10 promoter (SRE-149), it corecruits to the I-SRE with Stat5. In both cases, p300 mediates H3K18ac. Of note, inhibition of Stat5 did not affect p300 corecruitment to the IL10 promoter (SRE-149), which was occupied by Stat3. Comparably minor effects of Stat5 inhibition on p300 corecruitment to the I-SRE suggest a partial replacement with Stat3 secondary to an imbalance between Stat5 and Stat3 in analogy to SLE T cells (Fig. S5).
Fig. 6.
Stat3 and Stat5 mediate chromatin remodeling through their interaction with p300. (A) Forced expression of Stat3, Stat5, or p300 induces IL-10 mRNA expression in Jurkat T cells. Stat3 or Stat5, in combination with p300, shows synergistic effects on IL-10 mRNA expression. (B) Effects of forced p300 expression are reversed by knockdown of Stat3 or Stat5 with siRNAs. (C) Effects of forced Stat3 or Stat5 expression are reversed by p300 knockdown with siRNAs. (D) Proximity ligation assays (PLAs) in HEK 293T cells indicate a physical interaction between Stat3 or Stat5 and p300. Although forced expression of p300 or Stat3/Stat5 alone results in a weak color reaction, indicating low levels of endogenous p300 and Stat3/Stat5 expression, the combination of forced p300 and Stat3/Stat5 expression results in a strong signal. Of note, the signal in response to the combination of forced p300 and Stat5 expression was weaker compared with Stat3 and p300. (E) PLAs in TCR-stimulated T cells from controls and SLE patients confirmed an interaction between Stat3 or Stat5 and p300. Furthermore, the interactions between Stat3 but not Stat5 and p300 were increased in SLE T cells. (F) The fraction of PLA signal-positive cells indicating Stat3/p300 interactions is given in T cells from five SLE patients and six controls. EV, empty vector.
To test our hypothesis that T-cell receptor (TCR) stimulation mediates interactions between Stat3 or Stat5 with p300, we performed proximity ligation assays (PLAs; Olink) that indicate interactions between proteins. We transfected HEK293T cells with either an empty pcDNA3.1 vector, p300, Stat3, or Stat5 expression plasmids or their combination, monitoring for protein interactions (Fig. 6 D–F). Indeed, forced expression of p300, Stat3, or Stat5 resulted in a weak signal, reflecting endogenous Stat3/5 and p300 expression of HEK293T cells (Fig. 6D). The combination of forced Stat3 or Stat5 expression together with p300 resulted in a strong signal, indicating interactions between Stat transcription factors and p300. To investigate whether Stat3 or Stat5 also interact with p300 in human T cells, we performed PLA assays in T cells from controls or SLE patients. Indeed, TCR-stimulated cells exhibited interactions between Stat3 or Stat5 and p300 (Fig. 6 E and F). In agreement with our colocalization (Figs. 2F and 5) and Stat activation (Fig. 2 B and C) experiments, interactions between Stat3 and p300 were enhanced in SLE T cells. These observations indicate a replacement of Stat5 with Stat3 at the I-SRE, mediating trans-activation and chromatin remodeling through the histone acetyltransferase p300.
Discussion
The immune-regulatory cytokine IL-10 controls the growth, differentiation, and activity of immune cells. In various cells, including monocytes/macrophages and T lymphocytes, IL-10 reduces the expression of proinflammatory cytokines, thus inhibiting effector phenotypes (1, 2). Some effects of IL-10 on B cells, however, appear contradictory. IL-10 promotes B-cell survival, proliferation, differentiation, and antibody production (1, 2). Increased IL-10 expression in SLE patients correlates with disease activity and antibody production, and IL-10 blockade corrected dysregulated cytokine responses (12) and mediated clinical improvement in a subset of SLE patients (13). Given the immune-regulatory effects of IL-10 on T cells, enhanced IL-10 expression in SLE is seemingly conflicting with the well-established overexpression of IL-6 and IL-17A in SLE (14–16). Recently, however, this contradiction has been at least partially resolved. Reduced expression of the IL-10 receptor on SLE T cells alters the effects of IL-10 on activated or memory T cells that are major sources of proinflammatory cytokines. As a result, IL-10 fails to inhibit proinflammatory cytokine expression in SLE T cells (17).
Sources of IL-10 are manifold, including B and T cells, monocytes/macrophages, NK cells, and others. In SLE, monocytes and B cells have been reported as major sources of IL-10 (1, 2). Here, we demonstrate enhanced IL-10 expression in ex vivo isolated T cells from SLE patients. In individual patients, T cell-derived IL-10 expression followed disease activity, suggesting a role of T cell-derived IL-10 in disease pathology. This observation is of special interest, given the effects of IL-10 on differentiation, activity, and survival of B cells, promoting antibody production and immune complex generation (1, 18). Regardless of its role regulating the immune system and contributing to a number of autoimmune and infectious disorders, the molecular mechanism governing IL10 remains poorly understood. Tsuji-Takayama et al. reported IL-2-dependent IL-10 expression in human regulatory T cells, which was mediated by Stat5 recruitment to a Stat-binding motif in the fourth intron of IL10 (I-SRE) (10). At the same time, we demonstrated that IL-2 and IL-12 synergistically induce IL-10 in murine NK cells (11). IL-10 expression was dependent on Stat4 recruitment to the same region. In both reports, Stat recruitment to the I-SRE resulted in histone acetylation (10, 11). The molecular mechanisms mediating chromatin remodeling, however, remained elusive.
The involvement of Stat transcription factors in the regulation of IL10 is of special interest because Stat3 has been demonstrated to be increased in SLE T cells (19, 20). Conflicting data exist regarding Stat5 in SLE (21, 22). Although attenuated IL-2 expression in SLE T cells suggests reduced Stat5 activation (23–25), one study demonstrated increased Stat5 phosphorylation in SLE (22). In T cells from SLE patients and lupus-prone MRL/lpr mice, pStat3 is increased (19, 26). Stat3 plays a central role in the regulation of proinflammatory effector cytokines, including IL-17A, promoting effector T-cell phenotypes (27, 28). Furthermore, Stat3 is involved in IL-10 induction, which resulted in the proposal of Stat3-blocking strategies in autoimmune disorders and cancer (1, 2, 12, 13). None of those studies, however, targeted the molecular events induced by Stat3 at the IL10 gene. We aimed to determine the role of Stat3 and Stat5 in the regulation of IL10 in T cells and their potential involvement in the molecular immune-pathology of SLE. Differentially inhibiting Stat3 or Stat5, we investigated their contribution to signal-specific IL-10 expression. Applying reporter constructs, we documented trans-regulatory effects of Stat3 and Stat5 on IL10. One intriguing finding in this context is that Stat3 and Stat5 transcription factors differentially recruit to select SREs: although Stat3 mainly recruits to the IL10 promoter in control T cells (at SRE-149), Stat5 mainly recruits to the I-SRE. In SLE T cells, Stat3 recruitment to the promoter is increased, also involving SRE-741. Furthermore, Stat3 replaces Stat5 at the I-SRE. Considering increased pStat3 in both resting and activated SLE T cells, the replacement of Stat5 is most likely caused by competition for the I-SRE. Because forced Stat3 or Stat5 expression in T cells had comparable effects on IL10, increased IL-10 expression in SLE T cells is most likely caused by increased pSat3 rather than increased activity of Stat3 over Stat5.
Although the majority of studies focused on transcriptional events, additional mechanisms govern gene expression. Epigenetic mechanisms govern gene expression without altering the DNA sequence (29, 30). DNA methylation regulates the recruitment of transcription factors to regulatory elements. Histone modifications control chromatin structure through nucleosome rearrangement. Usually, the histone code reflects DNA methylation with activating histone modifications in regions with low DNA methylation. The net result is a dynamic accessibility of DNA to transcriptional regulators, determining cell-, tissue-, and disease-specific expression of genes (29, 30). SLE T cells exhibit distinct epigenetic anomalies, including reduced DNA methylation and increased histone acetylation (8, 30–32). Thus, we wondered whether DNA methylation of IL10 is reduced in SLE T cells, affecting transcription factor recruitment. Indeed, in agreement with recent findings in a Chinese cohort, DNA methylation of the IL10 promoter was reduced in our study population (8). Furthermore, we for the first time, to our knowledge, demonstrate reduced DNA methylation of an intronic enhancer in SLE T cells, allowing for increased Stat transcription factor recruitment.
We and others previously documented that IL10 undergoes epigenetic remodeling in response to stimulation of immune cells (10, 11, 33). The molecular events resulting in histone modifications, however, remained unknown. Reports indicated that Stat transcription factors might interact with the histone acetyltransferase p300 (34, 35). Indeed, we were able to detect H3K18ac in response to T-cell stimulation. Applying state-of-the-art technologies, we dissected functional interactions between Stat transcription factors and p300, mediating p300 recruitment to IL10 regulatory regions in the IL10 promoter (Stat3) and an intronic enhancer element (Stat3 and Stat5) instructing chromatin remodeling (H3K18ac). Thus, we for the first time, to our knowledge, have deciphered Stat3- and Stat5-mediated molecular events instructing epigenetic remodeling of IL10 (Fig. S6). In SLE T cells, trans-activation and chromatin remodeling are enhanced as a result of increased Stat3 phosphorylation, resulting in a replacement of Stat5 at the I-SRE. Enhanced recruitment of Stat3 to the IL10 promoter and the I-SRE mediates the recruitment of histone acetyltransferase p300 to those regions, instructing epigenetic remodeling (H3K18ac). Our findings provide previously unidentified molecular events, contributing to increased IL-10 expression in SLE T cells. Targeting the imbalance between phosphorylated Stat3 and Stat5 in SLE T cells may be a promising candidate in the search for novel, target-directed therapeutic approaches correcting dysregulated cytokine expression. The translational importance of our observations is underscored by promising results from the application of Stat3 inhibitors in select autoimmune disorders and cancers.
Materials and Methods
More detailed information on the applied methodology can be accessed in SI Materials and Methods.
Human Subjects.
SLE patients included in our studies were diagnosed according to the American College of Rheumatology classification criteria (36) and gave written informed consent. Healthy age-, sex-, and ethnicity-matched individuals were chosen as controls (SI Materials and Methods, Demographic Information). Inactive patients were defined by an SLE disease activity index (SLEDAI) between 0 and 4.
Cytokine Stimulation and Stat Inhibition.
Primary human T cells were cultured in RPMI 1640 with 10% (vol/vol) FCS in the absence or presence of Stat3 or Stat5 inhibitors (Stat3 inhibitor VI, CAS 501919–59-1, Stat5 CAS 285986–31-4; EMD Millipore) or vehicle control. As indicated, cytokines were added to some wells. Effects on IL-10 secretion were assessed after 24 h.
Semiquantitative Real-Time PCR.
Semiquantitative real-time PCR (qRT-PCR) was performed using SybrGreen site-specific primers on an ABI StepOnePlus Real time PCR system. Results were normalized to 18S. Primer sequences for qRT-PCR are summarized in SI Materials and Methods, Primers.
Expression Plasmids and T-Cell Transfection.
Expression plasmids for human DNMT3a have been described previously (24). Reporter constructs spanning the proximal 946 bp of the human IL10 promoter or a 630 bp-spanning fragment of the fourth intron with enhancer activity were PCR-amplified and cloned into luciferase vector pGL3-Basic (Promega). Site-directed mutagenesis of SREs was performed (Stratagene). Human T cells were transfected with expression plasmids using the Amaxa transfection system (Lonza) or Lipofectamine (Life Technologies) as indicated.
Luciferase Assays.
Ex vivo isolated primary human or unstimulated Jurkat T cells were transfected with the indicated plasmids, using the Amaxa transfection system (Lonza). Five hours after transfection, cells were collected and lysed, and luciferase activity was quantified using the Promega Dual Luciferase Assay System (Promega).
Methyl-CpG-DNA Immunoprecipitation.
Methyl-CpG-DNA immunoprecipitation (MeDIP) assays were carried out following the manufacturer’s instructions (Zymo Research). Equal amounts of methylated (100%) human CpG-DNA and demethylated human CpG-DNA (Zymo Research) were included as “input” and negative control.
Methylation of Reporter Plasmids.
To investigate the effects of DNA methylation on IL10 promoter activity, we methylated reporter constructs and the empty pGL3 plasmid using CpG-DNA methylase (Zymo Research).
Cotransfection of T cells with Stat3 or Stat5 Expression Plasmids and p300 siRNA.
Jurkat T cells were transfected with Stat3 or Stat5 expression plasmids, control siRNA, or p300-specific siRNA (OriGene). Later, cells were transfected with p300 expression plasmids and Stat3 or Stat5 siRNAs to reconfirm our findings following the same protocols.
ChIP.
Anti-H3K18ac (Abcam), anti-p300 (Santa Cruz), anti-Stat3 (EMD Millipore), and anti-Stat5 (Abcam) antibodies and normal rabbit IgG were obtained from Upstate (EMD Millipore). ChIP was carried out according to the manufacturer’s instructions (Invitrogen, Life Technologies). ChIP-DNA was subject to real-time qPCR. Where indicated, cells were pretreated with C646, a selective small-molecule inhibitor of the p300 histone acetyltransferase (Origene) according to the manufacturer’s protocols.
Intracellular Stat Phosphorylation Assays.
To determine intracellular Stat3 levels and Stat3 phosphorylation (Tyr705), Thermo Scientific Pierce STAT3 In-Cell ELISA Kits were applied following the manufacturer’s protocols. Intracellular staining for phosphorylated Stat3 (Tyr705) (BD Biosciences) or phosphorylated Stat5 (Tyr694) (BD Biosciences) was carried out in resting or stimulated primary human T cells from controls and SLE patients with phycoerythrin (PE)-labeled antibodies (clones 4/P-Stat3, and 47/Stat5) as indicated.
Proximity Ligation Assay.
HEK 293T cells were transfected with expression plasmids for Stat3, Stat5, p300, or the combination of Stat3 or Stat5 and p300. Cells were harvested and subjected to the Duolink proximity ligation assay following the manufacturer’s instructions (Olink). Cells were mounted with DAPI-containing medium (Olink) and read on a fluorescence microscope (Zeiss). The number of PLA signals per cell was quantified using ImageJ software (http://rsb.info.nih.gov/ij/disclaimer.html).
Statistical Analysis.
A paired two-tailed Student t test was used for statistical analysis. The Pearson product moment correlation coefficient (r) was used to determine the correlation between IL-10 mRNA levels and individual SLE disease activity indices (SLEDAIs).
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grant R01 AI 42269 (to G.C.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1408023111/-/DCSupplemental.
References
- 1.Hedrich CM, Bream JH. Cell type-specific regulation of IL-10 expression in inflammation and disease. Immunol Res. 2010;47(1-3):185–206. doi: 10.1007/s12026-009-8150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hofmann SR, Rösen-Wolff A, Tsokos GC, Hedrich CM. Biological properties and regulation of IL-10 related cytokines and their contribution to autoimmune disease and tissue injury. Clin Immunol. 2012;143(2):116–127. doi: 10.1016/j.clim.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Peng H, et al. Role of interleukin-10 and interleukin-10 receptor in systemic lupus erythematosus. Clin Rheumatol. 2013;32(9):1255–1266. doi: 10.1007/s10067-013-2294-3. [DOI] [PubMed] [Google Scholar]
- 4.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 5.Houssiau FA, et al. Serum interleukin 10 titers in systemic lupus erythematosus reflect disease activity. Lupus. 1995;4(5):393–395. doi: 10.1177/096120339500400510. [DOI] [PubMed] [Google Scholar]
- 6.Enghard P, Langnickel D, Riemekasten G. T cell cytokine imbalance towards production of IFN-gamma and IL-10 in NZB/W F1 lupus-prone mice is associated with autoantibody levels and nephritis. Scand J Rheumatol. 2006;35(3):209–216. doi: 10.1080/03009740500417791. [DOI] [PubMed] [Google Scholar]
- 7.Mellor-Pita S, et al. Monocytes and T lymphocytes contribute to a predominance of interleukin 6 and interleukin 10 in systemic lupus erythematosus. Cytometry B Clin Cytom. 2009;76(4):261–270. doi: 10.1002/cyto.b.20468. [DOI] [PubMed] [Google Scholar]
- 8.Zhao M, et al. Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J Biomed Biotechnol. 2010;2010:931018. doi: 10.1155/2010/931018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koenig KF, et al. Serum cytokine profile in patients with active lupus nephritis. Cytokine. 2012;60(2):410–416. doi: 10.1016/j.cyto.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Tsuji-Takayama K, et al. The production of IL-10 by human regulatory T cells is enhanced by IL-2 through a STAT5-responsive intronic enhancer in the IL-10 locus. J Immunol. 2008;181(6):3897–3905. doi: 10.4049/jimmunol.181.6.3897. [DOI] [PubMed] [Google Scholar]
- 11.Grant LR, et al. Stat4-dependent, T-bet-independent regulation of IL-10 in NK cells. Genes Immun. 2008;9(4):316–327. doi: 10.1038/gene.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lauwerys BR, Garot N, Renauld JC, Houssiau FA. Interleukin-10 blockade corrects impaired in vitro cellular immune responses of systemic lupus erythematosus patients. Arthritis Rheum. 2000;43(9):1976–1981. doi: 10.1002/1529-0131(200009)43:9<1976::AID-ANR8>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Llorente L, et al. Clinical and biologic effects of anti-interleukin-10 monoclonal antibody administration in systemic lupus erythematosus. Arthritis Rheum. 2000;43(8):1790–1800. doi: 10.1002/1529-0131(200008)43:8<1790::AID-ANR15>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 14.Apostolidis SA, Crispín JC, Tsokos GC. IL-17-producing T cells in lupus nephritis. Lupus. 2011;20(2):120–124. doi: 10.1177/0961203310389100. [DOI] [PubMed] [Google Scholar]
- 15.Apostolidis SA, et al. The dysregulation of cytokine networks in systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31(10):769–79. doi: 10.1089/jir.2011.0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauen T, Hedrich CM, Tenbrock K, Tsokos GC. cAMP responsive element modulator: A critical regulator of cytokine production. Trends Mol Med. 2013;19(4):262–269. doi: 10.1016/j.molmed.2013.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui HD, et al. Interleukin-10 receptor expression and signalling were down-regulated in CD4⁺ T cells of lupus nephritis patients. Clin Exp Immunol. 2011;165(2):163–171. doi: 10.1111/j.1365-2249.2011.04424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Llorente L, et al. Role of interleukin 10 in the B lymphocyte hyperactivity and autoantibody production of human systemic lupus erythematosus. J Exp Med. 1995;181(3):839–844. doi: 10.1084/jem.181.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harada T, et al. Increased expression of STAT3 in SLE T cells contributes to enhanced chemokine-mediated cell migration. Autoimmunity. 2007;40(1):1–8. doi: 10.1080/08916930601095148. [DOI] [PubMed] [Google Scholar]
- 20.Pflegerl P, et al. Epidermal loss of JunB leads to a SLE phenotype due to hyper IL-6 signaling. Proc Natl Acad Sci USA. 2009;106(48):20423–20428. doi: 10.1073/pnas.0910371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale MB, Krutzik PO, Samra SS, Crane JM, Nolan GP. Stage dependent aberrant regulation of cytokine-STAT signaling in murine systemic lupus erythematosus. PLoS ONE. 2009;4(8):e6756. doi: 10.1371/journal.pone.0006756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Guo Y, Bao C, Shen N. Multidimensional single cell based STAT phosphorylation profiling identifies a novel biosignature for evaluation of systemic lupus erythematosus activity. PLoS ONE. 2011;6(7):e21671. doi: 10.1371/journal.pone.0021671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crispín JC, Tsokos GC. Transcriptional regulation of IL-2 in health and autoimmunity. Autoimmun Rev. 2009;8(3):190–195. doi: 10.1016/j.autrev.2008.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hedrich CM, et al. cAMP response element modulator alpha controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc Natl Acad Sci USA. 2012;109(41):16606–166–11. doi: 10.1073/pnas.1210129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedrich CM, Rauen T, Tsokos GC. cAMP-responsive element modulator (CREM)α protein signaling mediates epigenetic remodeling of the human interleukin-2 gene: implications in systemic lupus erythematosus. J Biol Chem. 2011;286(50):43429–43436. doi: 10.1074/jbc.M111.299339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bethunaickan R, Berthier CC, Zhang W, Kretzler M, Davidson A. Comparative transcriptional profiling of 3 murine models of SLE nephritis reveals both unique and shared regulatory networks. PLoS ONE. 2013;8(10):e77489. doi: 10.1371/journal.pone.0077489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Z, O’Shea JJ. Th17 cells: A new fate for differentiating helper T cells. Immunol Res. 2008;41(2):87–102. doi: 10.1007/s12026-007-8014-9. [DOI] [PubMed] [Google Scholar]
- 28.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 29.Ballestar E. An introduction to epigenetics. Adv Exp Med Biol. 2011;711:1–11. doi: 10.1007/978-1-4419-8216-2_1. [DOI] [PubMed] [Google Scholar]
- 30.Hedrich CM, Tsokos GC. Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol Med. 2011;17(12):714–724. doi: 10.1016/j.molmed.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Costa-Reis P, Sullivan KE. Genetics and epigenetics of systemic lupus erythematosus. Curr Rheumatol Rep. 2013;15(9):369. doi: 10.1007/s11926-013-0369-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Hedrich CM, et al. Dynamic DNA methylation patterns across the mouse and human IL10 genes during CD4+ T cell activation: Influence of IL-27. Mol Immunol. 2010;48(1-3):73–81. doi: 10.1016/j.molimm.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paulson M, et al. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J Biol Chem. 1999;274(36):25343–25349. doi: 10.1074/jbc.274.36.25343. [DOI] [PubMed] [Google Scholar]
- 35.Pfitzner E, Jähne R, Wissler M, Stoecklin E, Groner B. p300/CREB-binding protein enhances the prolactin-mediated transcriptional induction through direct interaction with the transactivation domain of Stat5, but does not participate in the Stat5-mediated suppression of the glucocorticoid response. Mol Endocrinol. 1998;12(10):1582–1593. doi: 10.1210/mend.12.10.0180. [DOI] [PubMed] [Google Scholar]
- 36.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.