Significance
Aneuploidy refers to the gain or loss of individual chromosomes within a cell. Typically, aneuploidy is associated with detrimental consequences at both the cellular and organismal levels. However, reports of high levels of aneuploidy in the brain and liver suggested that aneuploidy might play a positive role in these organs. Here we use single cell sequencing to determine the prevalence of aneuploidy in somatic tissues. We find that aneuploidy is a rare occurrence in the liver and brain and is no more prevalent in these tissues than in skin. Our results demonstrate high karyotypic stability in somatic tissues, arguing against a role for aneuploidy in organ function and reinforcing its adverse effects at the cellular and organismal levels.
Abstract
Whole-chromosome copy number alterations, also known as aneuploidy, are associated with adverse consequences in most cells and organisms. However, high frequencies of aneuploidy have been reported to occur naturally in the mammalian liver and brain, fueling speculation that aneuploidy provides a selective advantage in these organs. To explore this paradox, we used single cell sequencing to obtain a genome-wide, high-resolution assessment of chromosome copy number alterations in mouse and human tissues. We find that aneuploidy occurs much less frequently in the liver and brain than previously reported and is no more prevalent in these tissues than in skin. Our results highlight the rarity of chromosome copy number alterations across mammalian tissues and argue against a positive role for aneuploidy in organ function. Cancer is therefore the only known example, in mammals, of altering karyotype for functional adaptation.
DNA copy number alterations, deviations from the diploid DNA content, can vary in length from a few base pairs to entire genomes. When the entire genome is duplicated, cells are referred to as polyploid. Aneuploidy refers to a change in the copy number of individual chromosomes. The changes in gene copy number caused by these karyotype alterations typically change expression of the affected genes (1–3). Therefore, copy number alterations can have profound effects on cellular and organismal physiology. This is especially true for aneuploidy, in which the relative dosage of many genes is altered.
In multicellular organisms, aneuploidy can be present either in all cells, termed “constitutional aneuploidy,” or only in select cells, called “somatic aneuploidy.” Constitutional aneuploidy typically has adverse consequences for the organism. In humans a third copy of chromosome 21 causes Down syndrome, and a third copy of any other autosome almost always results in embryonic or childhood lethality (4). Supporting the adverse effects of aneuploidy at the organismal level, chromosome copy number changes are also associated with impaired fitness at the cellular level. Aneuploid yeast and mammalian cells exhibit slower proliferation and proteotoxic stress (2, 3, 5, 6).
Somatic aneuploidy also has adverse consequences for the organism. In humans, mutations in BUB1B cause the disease mosaic variegated aneuploidy (MVA) (7). BUB1B encodes BubR1, a component of the spindle assembly checkpoint required for accurate chromosome segregation (8). Cells of MVA patients are frequently aneuploid for one or more chromosomes, and MVA patients exhibit growth retardation, developmental defects, and childhood cancers. Mice homozygous for a hypomorphic mutation of BUB1B (BUB1BH/H) show similar phenotypes (9). Cancer is another example of a disease characterized by aneuploidy. In this case, however, changes in gene copy number are thought to increase the fitness of tumor cells relative to untransformed cells because of selection for oncogene gains and tumor suppressor gene losses (10).
In light of the many reports describing significant adverse effects of aneuploidy on cellular and organismal fitness, it is surprising that two organs, the liver and brain, are reported to harbor high levels of aneuploidy. Hepatocytes, the primary cell type of the liver, are diploid in neonates. However, hepatocytes become polyploid during early postnatal development by initiating mitosis but failing cytokinesis (11, 12). Importantly, a recent reexamination of hepatocyte ploidy by FISH reported aneuploidy in more than 50% of hepatocytes (13, 14). Duncan et al. (13) suggested that polyploid hepatocytes underwent aberrant mitoses with high levels of chromosome mis-segregation, thereby generating aneuploidy among polyploid cells. The authors further speculated that aneuploidy endows hepatocytes with phenotypic variability and adaptability upon exposure to various noxious agents and metabolic stresses (15).
Rehen et al. (16) were first to report elevated aneuploidy in the mammalian brain. Using spectral karyotyping (SKY), they reported that 33% of embryonic mouse neural progenitor cells harbored loss or gain of one or more chromosomes. Subsequent studies using FISH indicated that approximately 20% of adult mouse and human brain cells were aneuploid (16–20). These aneuploid cells were believed to arise from chromosome mis-segregation events in neural progenitor cells and were shown to integrate into brain circuitry (21, 22). These high levels of aneuploidy were proposed to provide the brain with its notable phenotypic diversity while simultaneously predisposing the organ to neurodegeneration (23–26).
The reports of aneuploidy in the liver and brain suggest that, in these tissues, aneuploidy may not compromise organ function but instead provide beneficial phenotypic plasticity. Indeed a positive role for aneuploidy is well documented in experimental evolution studies in microorganisms (27, 28). The reports of high levels of aneuploidy in the brain and liver thus raised the exciting possibility that these organs somehow avoid the adverse consequences of aneuploidy and instead use these changes to their benefit, all while avoiding oncogenic transformation. Here, we use single cell sequencing to provide a genome-wide, high-resolution assessment of aneuploidy in mammalian tissues. Unlike previous reports, we find that the prevalence of aneuploidy in liver and brain is low and comparable to the frequency in skin. Our findings argue against a positive role for somatic chromosome copy number alterations in tissue function and instead reinforce their adverse consequences for cellular and organismal fitness.
Results
Detecting Somatic Aneuploidy by Single Cell Sequencing.
The studies reporting aneuploidy in the brain and liver used FISH and SKY to quantify aneuploidy. Both methods are prone to artifacts. In FISH, probe stretching, probe clustering, failed hybridization, or off-target hybridization can cause signals to be lost or gained inappropriately. Even a low frequency of artifacts for a single chromosome can lead to a gross overestimation of aneuploidy when extrapolated across all chromosomes. SKY can also overestimate aneuploidy because the spreading procedure can move chromosomes between nuclei. We therefore used single cell sequencing as an alternative method to assess somatic aneuploidy in brain and liver. We dissociated tissues from mice and humans, amplified the genomic DNA of single cells, sequenced the amplified DNA, and inferred copy number from sequencing read depth (Fig. 1A). When possible, we isolated tissues from males because monosomy of the X chromosome provides an internal control for identifying copy number alterations. We avoided permeabilization and fixation of target cells to minimize interference with whole-genome amplification. In the liver, we isolated nuclei instead of whole cells to facilitate detection of copy number changes in a polyploid setting.
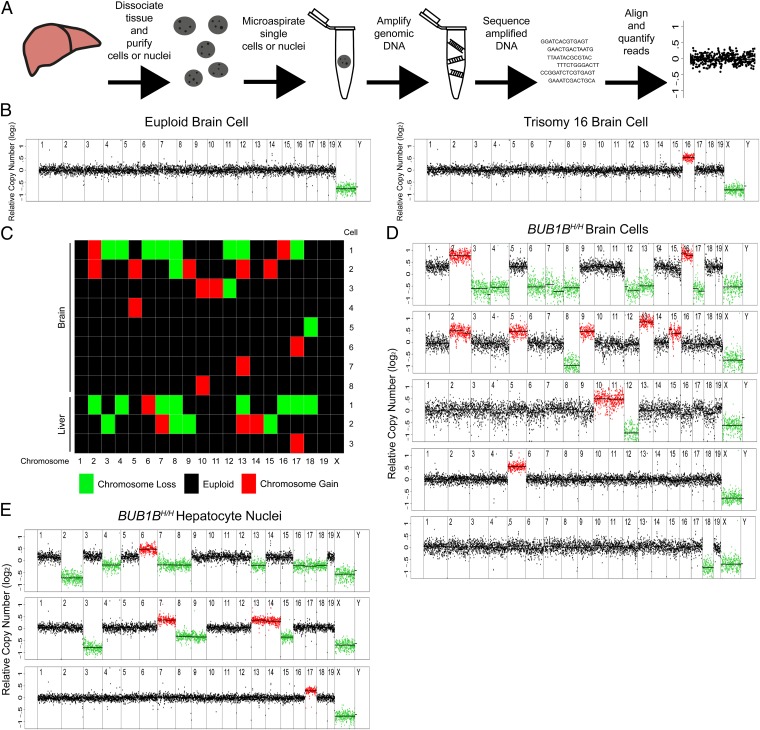
Fig. 1.
Validating detection of aneuploidy by single cell sequencing. (A) Overview of the method used to detect copy number alterations in single cells by whole-genome sequencing. (B) Segmentation plots of a euploid brain cell (Left) and a trisomy 16 brain cell (Right), isolated from male mouse embryos euploid and trisomic for chromosome 16, respectively. Segmentation plots show copy number of single cells relative to a euploid reference on a log2 scale. Segments above threshold for gain are colored in red, segments below threshold for loss are colored in green. (C) Heat map of chromosome losses and gains in aneuploid brain cells and liver nuclei from BUB1BH/H mice. (D) Segmentation plots of five aneuploid cells from a BUB1BH/H mouse brain. (E) Segmentation plots of three aneuploid nuclei from a BUB1BH/H mouse liver.
For each cell, on average, 8.5 × 106 reads aligned to the reference genome, corresponding to 0.1× coverage. We quantified reads in 500-kb windows and used a hidden Markov model to generate segments of defined copy number relative to a euploid reference (29). We did not analyze the Y chromosome for copy number alterations because of its low amount of unique sequence.
Approximately 10% of sequenced cells exhibited aberrantly high intrachromosomal sequence depth variability (Fig. S1A, Top and Upper Middle), as might arise from incomplete fragmentation of genomic DNA or biased amplification of fragments during whole-genome amplification. To identify these cells and exclude them from the analysis, we measured the SDs of corrected read copies (log2 based) within sliding windows (30 adjacent 500-kb bins) across a single chromosome, averaged the SDs for each chromosome, computed the mean average SD across the three most variable autosomes, and excluded cells for which this variability score (VS) exceeded 0.34 (SI Materials and Methods and Fig. S1 B and C). In total, we excluded 10.1% of sequenced cells (Fig. S1D). This metric does not bias against aneuploidy, because variability is measured within each chromosome. Consistent with this, the average VS of all aneuploid cells we identified was equivalent to the average VS of all euploid cells (Fig. S1A, Lower Middle and Bottom, and S1C).
We validated several aspects of our approach to ensure that this method reliably detects somatic aneuploidy. First we confirmed that loss or gain of a chromosome caused a change in relative read depth that exceeded background fluctuations in read depth. We sequenced single brain cells from trisomy 16 mouse embryos (3) and were indeed able to detect an additional copy of chromosome 16 in all cells from the trisomy 16 embryo (Fig. 1B, Right). We next determined whether the method was able to detect aneuploidies of unknown composition. We sequenced single brain cells from an adult male BUB1BH/H mouse, because the BUB1B mutation is reported to cause aneuploidy in one-third of cells (9). Indeed, 8 of the 21 brain cells we analyzed [38.1%, 95% confidence interval (CI) 18.1%–61.6%] were aneuploid for one or more chromosomes (Fig. 1 C and D).
To determine whether we were able to detect aneuploidy in polyploid cells, we sequenced hepatocyte nuclei from an adult male BUB1BH/H mouse. Here, 3 of the 16 hepatocyte nuclei we analyzed (18.8%, 95% CI 4%–45.6%) harbored one or more chromosomes with read depths indicating loss or gain in a tetraploid nucleus (Fig. 1 C and E). We conclude that our method can detect single as well as multiple chromosome gains or losses in diploid and polyploid cells.
Last, we tested whether our approach selected against aneuploid cells because of differences in viability during tissue dissociation or operator bias during microaspiration. We mixed equal volumes of trisomy 16 and euploid brain, dissociated the tissue together, and sequenced single cells from this mixture. We found that five of seven cells (71.4%, 95% CI 29%–96.3%) were trisomic for chromosome 16, indicating that our procedure does not discriminate against aneuploid cells.
Together, these experiments show that single cell sequencing can detect simple and complex aneuploidies in an unbiased manner. Furthermore, by compiling the relative copy numbers (log2 ratios) for all aneuploid chromosomes and X chromosome monosomies, we were able to establish cutoffs for detecting copy number variants in all subsequently sequenced diploid and polyploid cells (SI Materials and Methods).
Prevalence of Aneuploidy in Skin.
There are no reports of elevated aneuploidy in skin. Therefore, we sequenced single cells from skin to establish a baseline level of somatic aneuploidy. We isolated keratinocytes by dissociating epidermis and selecting uncornified cells. In the mouse we found one aneuploid cell (trisomy 12) among 37 cells analyzed, indicating that 2.7% of mouse keratinocytes are aneuploid (95% CI 0.1%–14.2%; Figs. S2 and S3 and Table S1). To examine aneuploidy in human skin, we isolated keratinocytes from epidermis obtained upon autopsy of a 48-y-old woman and a 68-y-old man. Of the 53 keratinocytes analyzed, none were aneuploid (Fig. S2 and Table S1). We conclude that aneuploidy in human skin is low (95% CI 0–6.7%; Table S1).
Prevalence of Aneuploidy in Brain.
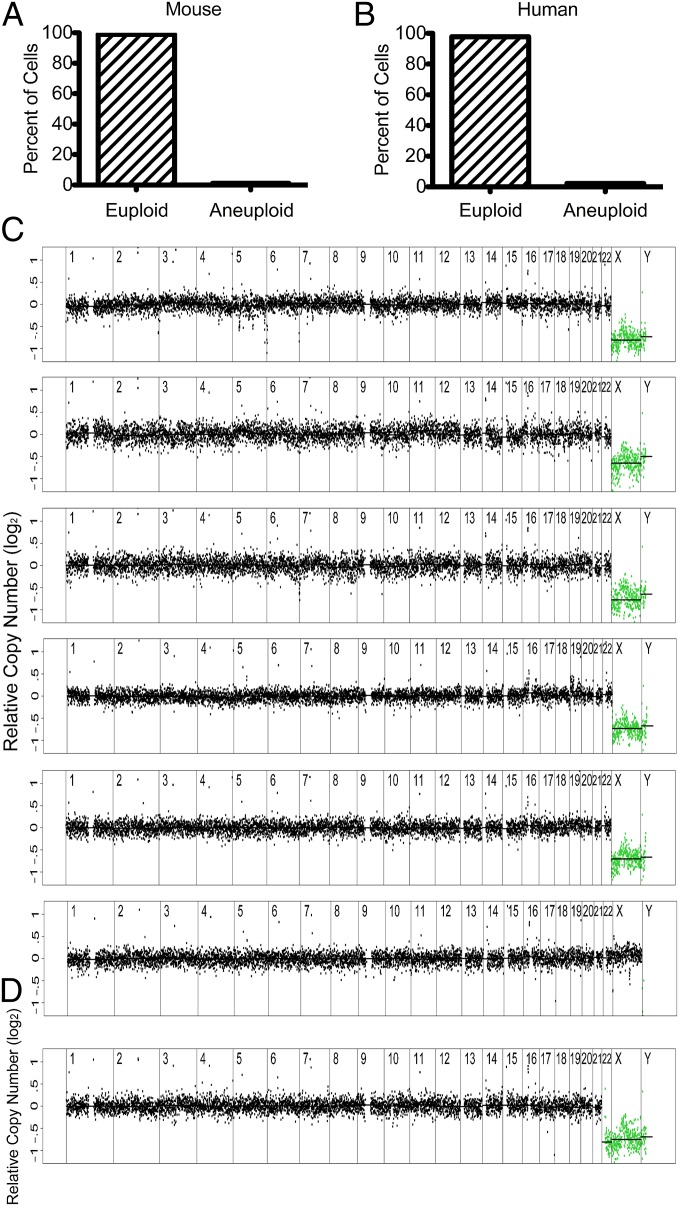
In the mammalian brain, aneuploidy is reported to be highest in neural progenitor cells, with a prevalence of 33% (16). To investigate aneuploidy in embryonic neural progenitor cells, we isolated brains from mouse embryos expressing the neural progenitor marker nestin-GFP. Of the 36 cells analyzed, none were aneuploid (Fig. S2 and Table S1). To assess the degree of aneuploidy in the adult mouse brain, we dissociated gray matter from cerebral cortex and enriched for neurons and glia by density gradient sedimentation. Single cell gene expression analysis revealed that the cell suspension obtained by this method contained 75% neurons, 12.5% glia, and 12.5% unknown cell types (Fig. S4A). Of the 43 cells analyzed, we identified one aneuploid cell (trisomy 15; Figs. S2 and S4B). Thus, only 2.3% of mouse brain cells are aneuploid (95% CI 0.1%–12.3%; Table S1).
To specifically examine neurons, we immunostained adult mouse brain cells with an antibody against the neuronal nuclear marker NeuN and enriched for NeuN-positive cells by FACS. Of 18 cells sequenced, we had to exclude 9 cells because of high variability in read depth between adjacent genomic windows (Fig. S1 B–D). Of the 9 cells analyzed, none were aneuploid (Fig. S2). We presume that the high VS in many of the neurons isolated by this method was due to permeabilization, fixation, and/or immunostaining interfering with whole-genome amplification. As an alternate approach, we purified DRD2-expressing medium spiny neurons of the basal ganglion from an adult mouse carrying a DRD2-GFP transgene. We sequenced 10 cells, all of which could be analyzed and none of which were aneuploid (Fig. S2). We conclude that the sequence variability observed in the neurons purified by NeuN immunostaining was due to the immunostaining procedure interfering with sample preparation. Importantly, our analysis of 19 neurons did not reveal a single aneuploid cell (95% CI 0–17.6%; Fig. S2 and Table S1). This result is not significantly different from that for the mixed brain cell population. In total, the prevalence of aneuploidy in the mouse brain is 1% (95% CI 0–5.6%; Fig. 2A).
Fig. 2.
Prevalence of aneuploidy in brain. (A and B) Prevalence of euploid and aneuploid cells in mouse (A) and human (B) brain. (C) Segmentation plots of euploid human brain cells. (D) Segmentation plot of an aneuploid human brain cell harboring a monosomy for chromosome 22.
To extend these studies to humans, we dissociated gray matter of frontal lobe obtained during autopsy of four individuals: a 52-y-old man, 68-y-old man, 70-y-old man, and a 48-y-old woman. None of these individuals had a history of neurologic disease or evidence of brain pathology upon autopsy. Of the 89 cells analyzed, all but two cells were euploid (Fig. 2C and Fig. S2). Of the aneuploid cells, one harbored a monosomy for chromosome 22 (Fig. 2D). The other cell had a read depth on chromosome 18 consistent with trisomy in a tetraploid cell (Fig. S4C). Because cell suspensions were extremely dilute, we believe that this indeed represents a tetraploid cell with chromosome loss rather than two cells isolated together, one of which was aneuploid. We conclude that the prevalence of aneuploidy in the adult human brain is 2.2% (95% CI 0.3%–7.9%; Fig. 2B and Table S1), significantly less than the more than 20% aneuploidy reported by prior studies (16–20).
Prevalence of Aneuploidy in Liver.
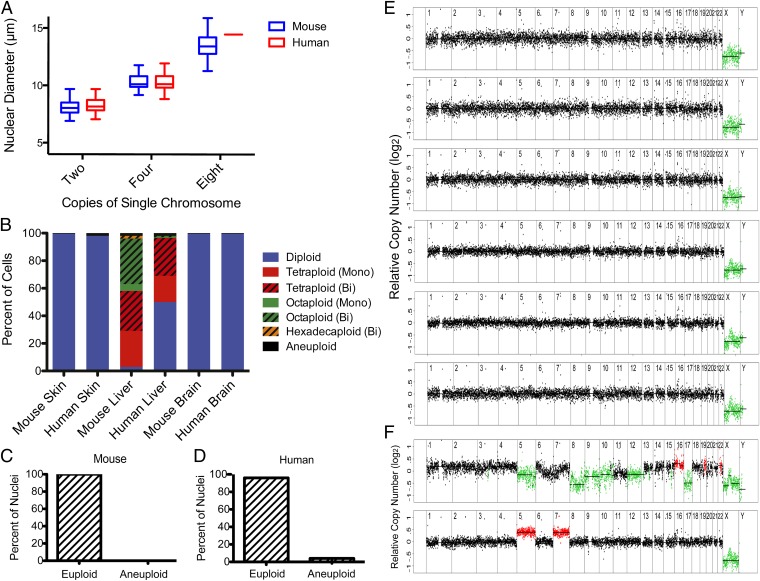
To determine the degree of aneuploidy in the liver, we first assessed the ploidy distribution in this organ using nuclear size measurements coupled with dual-color, single-chromosome FISH. In liver, but not other tissues, we observed many binucleate cells and many cells with more than two copies of a single chromosome. The elevated copy number states were associated with increases in nuclear diameter (Fig. 3A). Because polyploidy increases nuclear diameter, the increases in chromosome copy number in hepatocytes likely reflect polyploidy. We thus classified cells having 2, 4, 8, or 16 copies of a single chromosome as being diploid, tetraploid, octaploid, or hexadecaploid, respectively. In mouse liver, the majority of hepatocytes were polyploid, with tetraploid cells occurring most frequently (Fig. 3B). Approximately half of tetraploid cells were binucleate, whereas the majority of octaploid and hexadecaploid cells were binucleate. In human liver, ploidy ranged from diploid to octaploid, with diploid and tetraploid cells occurring most often and with similar frequency (Fig. 3B).
Fig. 3.
Prevalence of aneuploidy in liver. (A) Increase in chromosome copy number correlates with an increase in nuclear diameter in mouse and human hepatocytes. Note that only a single octaploid human hepatocyte nucleus was identified. (B) Prevalence of cells of different ploidy for a single chromosome in mouse and human skin (keratinocytes), liver (hepatocytes), and brain (neurons and glia). Polyploid hepatocytes are indicated as being mononucleate (mono) or binucleate (bi). For example, tetraploid (bi) describes a cell with two nuclei, each of which contains two copies of a single chromosome. n = 200 cells per tissue. (C and D) Prevalence of euploid and aneuploid nuclei in mouse (C) and human (D) liver. (E) Segmentation plots of euploid human hepatocyte nuclei. (F) Segmentation plots of a human hepatocyte nucleus for which a karyotype could not be determined (Upper) and a tetraploid human hepatocyte nucleus harboring pentasomy for chromosomes 5 and 7 (Lower).
Aneuploidy is more difficult to detect in polyploid cells because the adjusted read depth on the aneuploid chromosome is smaller. As described above, we were able to detect aneuploidy in tetraploid hepatocyte nuclei from BUB1BH/H mice (Fig. 1E). Importantly, in all presumably diploid cells we analyzed from other tissues, only one brain cell harbored a chromosome with an adjusted read depth consistent with a single chromosome gain or loss in a tetraploid cell. Thus, the chromosome changes we observed in BUB1BH/H hepatocyte nuclei likely represent actual aneuploidy rather than background fluctuations in read depth. Detection of aneuploidy in octaploid nuclei is more challenging because the adjusted read depth from a single chromosome loss or gain is further reduced. Indeed, 14% of all diploid cells analyzed harbored one or more chromosomes with an average read depth that would be consistent with a chromosome gain or loss in an octaploid cell. Thus, chromosome gains or losses in an octaploid cell cannot be distinguished from background fluctuations in read depth. We therefore did not consider fluctuations of this magnitude as aneuploidy. This did not significantly affect the outcome of our analysis because octaploid hepatocyte nuclei are rare (Fig. 3B and Fig. S5A).
Having established cut-offs for detecting aneuploidy in a polyploidy setting, we isolated nuclei from mouse liver. To enrich for hepatocytes, we preferentially picked nuclei with diameters consistent with tetraploidy (Fig. S5A). Of the 66 cells analyzed, we did not identify a single aneuploid nucleus (Fig. 3C, Fig. S2, and Table S1). We next sequenced commercially available human hepatocytes prepared from a 46-y-old man and a 51-y-old man. Neither of these individuals had liver disease; however, the second individual had an extensive drug and alcohol history. The purity of these hepatocyte preparations exceeds 70%. Of the 39 hepatocytes analyzed from the first donor, all cells except one were euploid (Fig. 3E and Fig. S2). Although we could not assign a karyotype to the other cell owing to extensive fluctuation of segments, it met our quality control criteria and was considered aneuploid (Fig. 3F, Upper). Of the 61 nuclei analyzed from the second donor, all but three cells were euploid (Fig. 3E and Fig. S2). All three aneuploid nuclei were tetraploid with gains of one or two chromosomes (Fig. 3F, Lower, and Fig. S5B). In total, the prevalence of aneuploidy in human hepatocytes is 4% (95% CI 1.1%–9.9%; Fig. 3D). Thus, single cell sequencing indicates that less than 5% of hepatocytes are aneuploid (Table S1).
Discussion
Here we provide the first assessment (to our knowledge) of chromosome copy number alterations in multiple tissues at genome-wide, single cell resolution. Our sequencing results suggest that the prevalence of aneuploid cells in the mammalian brain and liver is less than 5% (Table S1). This observation is in agreement with a recent single-neuron sequencing study that, despite identifying a high frequency of subchromosome copy number variants, did not observe high levels of whole-chromosome aneuploidy in neurons (30). Prior reports used SKY and FISH to assess the prevalence of aneuploidy and found it to exceed 50% and 20% in the liver and brain, respectively (13, 14, 16–20). We attribute this difference to drawbacks associated with using FISH to detect somatic aneuploidy. Indeed, using dual-color, single-chromosome FISH we too observed low levels of aneuploidy for a single chromosome that would, when extrapolated across all chromosomes, indicate that over 40% of hepatocytes and 10% of brain cells are aneuploid (Fig. 3B). However, we also observed similar rates of aneuploidy in skin. That we had to exclude up to one-third of cells from analysis because of discrepancies in signal number between two probes targeting the same chromosome illustrates the limitations of FISH. The rare cases of signal gain or loss for both probes could just as likely represent two simultaneous hybridization artifacts. FISH artifacts that are interpreted as aneuploidy even when occurring at low frequency can lead to gross overestimation of aneuploidy when used to infer the prevalence of aneuploidy across the genome. We conclude that single cell sequencing is a superior method for characterizing somatic karyotypic changes.
Whether the rare cells harboring copy number alterations that we did identify in normal tissues represent incipient transformation remains to be determined. Furthermore, it is important to note that we only analyzed skin, brain, and liver tissue that had no evidence of disease. It is possible that aneuploidy is elevated in disease states other than cancer. Indeed, it is interesting to note that proteotoxic stress is a hallmark of both aneuploidy and neurodegenerative disease (6, 31).
The Implications of Low Aneuploidy for Hepatocyte Proliferation.
The low prevalence of aneuploidy in normal tissues means that either the events producing such aberrations occur rarely or the cells harboring such changes are eliminated. Both possibilities are likely to occur. The wide array of detrimental phenotypes associated with constitutional aneuploidy in vitro and in vivo suggests that aneuploid somatic cells may indeed be selected against when arising amid a euploid population. On the other hand, our sequencing of BUB1BH/H mice indicates that cells can survive despite multiple copy number alterations, although their survival in these mice may be due to the presence of fewer euploid cells and thus reduced selective pressure (9).
The low level of aneuploidy has particular implications for our understanding of liver biology. Hepatocytes are not the only example of polyploidy in mammals. Trophoblast giant cells, which constitute the placenta, and megakaryocytes, the precursors of platelets, are also polyploid. However, in these two cases, and indeed in cases in other organisms, polyploidy is associated with a terminally differentiated state and cell cycle arrest (32). Hepatocytes are an exception to this rule. Upon liver damage, hepatocytes reenter the cell cycle and proliferate extensively (33). This is a nontrivial task because polyploid cells, in addition to having multiple copies of their genome, have multiple centrosomes. When a cell with multiple centrosomes enters mitosis, a multipolar spindle forms that, if uncorrected, randomly segregates chromosomes to produce highly aneuploid cells. Studies of cancer cells have shown that multiple centrosomes can cluster to form bipolar spindles (34). However, the process of centrosome clustering produces many merotelic kinetochore–microtubule attachments, where a kinetochore attaches to microtubules emanating from both spindle poles. Chromosomes with merotelic attachments are frequently mis-segregated (35). It is difficult to imagine that the liver culls the appreciable fraction of aneuploid hepatocytes expected to arise from these faulty mitoses. This implies that hepatocytes have either enhanced mechanisms for detecting and correcting merotelic attachments or use a unique mechanism for segregating chromosomes in the presence of multiple centrosomes. Determining how hepatocytes prevent or correct merotelic attachments will be an important future question.
The Implications of Low Levels of Aneuploidy for Neural Diversity.
The brain is notable for its remarkable diversity, both at the cellular and functional level. There are more than 10,000 different types of neurons, and even neurons of the same type can differ in their expression of synapse components and in their thresholds for excitation and firing (23). Diversity at the cellular level presumably translates into diversity at the functional level. There is remarkable interindividual heterogeneity in cognition, intellect, and behavior. For example, even isogenic mice exhibit remarkable diversity in stress responses (36). The report of high levels of aneuploidy in the brain led to extensive speculation that random aneuploidies endowed the brain with cellular and thus functional diversity (23). However, our study shows that somatic aneuploidy is much less common than previously reported. Given the low prevalence of aneuploidy in normal tissues, it is difficult to imagine that such changes play a positive role in organ function. We infer that normal mammalian tissues do not use aneuploidy as a means of phenotypic diversification or adaptation.
Materials and Methods
Tissue Sources.
Frozen human liver was obtained from the Brigham and Women’s Hospital Tissue and Blood Repository. Human hepatocytes were purchased from Life Technologies. Fresh human frontal lobe and epidermis were obtained upon autopsy at Massachusetts General Hospital.
Single Cell Sequencing and Copy Number Analysis.
Tissues were dissociated into single cells. Single cells and nuclei were isolated by microaspiration, and genomic DNA was amplified using the GenomePlex Single Cell Whole Genome Amplification Kit (Sigma). Amplified DNA was purified, barcoded, pooled, and sequenced on an Illumina HiSeq2000. Sequencing reads were aligned using BWA (0.6.1). HMMcopy (0.1.1) was used to estimate gene copy number in 500-kb bins (29). Cells with high variability in copy number across bins were excluded from analysis.
Supplementary Material
Acknowledgments
We thank the Massachusetts Institute of Technology BioMicro Center for sequencing; the Koch Institute Swanson Biotechnology Center for flow cytometry and histology; the Whitehead Institute Keck Microscopy Facility for imaging; Jan van Deursen, David Scadden, and David Housman for providing mouse strains; Matthew Frosch, Daniel Mordes, and Inma Cobos Sillero for providing autopsy samples; Zachary Crook for technical assistance; Mark Daly for helpful discussions; and David Housman, Frank Solomon, Mark Daly, and members of the A.A. laboratory for their critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) Physical Sciences Oncology Center Grant 5-U54-CA143874, a senior scholar award from the Ellison Medical Foundation, Koch Institute Support Grant P30-CA14051 from the National Cancer Institute, and by grants from the Howard Hughes Medical Institute and the Kathy and Curt Marble Cancer Research Fund. The Neuropathology Core of the Massachusetts Alzheimer Disease Research Center is supported by NIH Grant P50 AG005134. K.A.K. is supported by National Institute of General Medical Sciences Training Grant T32GM007753.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP041670).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1415287111/-/DCSupplemental.
References
- 1.Kahlem P, et al. Transcript level alterations reflect gene dosage effects across multiple tissues in a mouse model of down syndrome. Genome Res. 2004;14(7):1258–1267. doi: 10.1101/gr.1951304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 3.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322(5902):703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassold TJ, Jacobs PA. Trisomy in man. Annu Rev Genet. 1984;18:69–97. doi: 10.1146/annurev.ge.18.120184.000441. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y-C, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144(4):499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev. 2012;26(24):2696–2708. doi: 10.1101/gad.207407.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanks S, et al. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet. 2004;36(11):1159–1161. doi: 10.1038/ng1449. [DOI] [PubMed] [Google Scholar]
- 8.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36(7):744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 10.Davoli T, et al. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell. 2013;155(4):948–962. doi: 10.1016/j.cell.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guidotti J-E, et al. Liver cell polyploidization: A pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278(21):19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 12.Margall-Ducos G, Celton-Morizur S, Couton D, Brégerie O, Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120(Pt 20):3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 13.Duncan AW, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467(7316):707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncan AW, et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142(1):25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duncan AW, et al. Aneuploidy as a mechanism for stress-induced liver adaptation. J Clin Invest. 2012;122(9):3307–3315. doi: 10.1172/JCI64026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rehen SK, et al. Chromosomal variation in neurons of the developing and adult mammalian nervous system. Proc Natl Acad Sci USA. 2001;98(23):13361–13366. doi: 10.1073/pnas.231487398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rehen SK, et al. Constitutional aneuploidy in the normal human brain. J Neurosci. 2005;25(9):2176–2180. doi: 10.1523/JNEUROSCI.4560-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pack SD, et al. Individual adult human neurons display aneuploidy: Detection by fluorescence in situ hybridization and single neuron PCR. Cell Cycle. 2005;4(12):1758–1760. doi: 10.4161/cc.4.12.2153. [DOI] [PubMed] [Google Scholar]
- 19.Yurov YB, et al. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PLoS One. 2007;2(6):e558. doi: 10.1371/journal.pone.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faggioli F, Wang T, Vijg J, Montagna C. Chromosome-specific accumulation of aneuploidy in the aging mouse brain. Hum Mol Genet. 2012;21(24):5246–5253. doi: 10.1093/hmg/dds375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang AH, et al. Chromosome segregation defects contribute to aneuploidy in normal neural progenitor cells. J Neurosci. 2003;23(32):10454–10462. doi: 10.1523/JNEUROSCI.23-32-10454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsbury MA, et al. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci USA. 2005;102(17):6143–6147. doi: 10.1073/pnas.0408171102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muotri AR, Gage FH. Generation of neuronal variability and complexity. Nature. 2006;441(7097):1087–1093. doi: 10.1038/nature04959. [DOI] [PubMed] [Google Scholar]
- 24.Iourov IY, Vorsanova SG, Liehr T, Yurov YB. Aneuploidy in the normal, Alzheimer’s disease and ataxia-telangiectasia brain: Differential expression and pathological meaning. Neurobiol Dis. 2009;34(2):212–220. doi: 10.1016/j.nbd.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Arendt T, Brückner MK, Mosch B, Lösche A. Selective cell death of hyperploid neurons in Alzheimer’s disease. Am J Pathol. 2010;177(1):15–20. doi: 10.2353/ajpath.2010.090955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kennedy SR, Loeb LA, Herr AJ. Somatic mutations in aging, cancer and neurodegeneration. Mech Ageing Dev. 2012;133(4):118–126. doi: 10.1016/j.mad.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313(5785):367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yona AH, et al. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci USA. 2012;109(51):21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ha G, et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012;22(10):1995–2007. doi: 10.1101/gr.137570.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McConnell MJ, et al. Mosaic copy number variation in human neurons. Science. 2013;342(6158):632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 32.Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol. 2011;27:585–610. doi: 10.1146/annurev-cellbio-092910-154234. [DOI] [PubMed] [Google Scholar]
- 33.Zou Y, et al. Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS One. 2012;7(2):e30675. doi: 10.1371/journal.pone.0030675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460(7252):278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gregan J, Polakova S, Zhang L, Tolić-Nørrelykke IM, Cimini D. Merotelic kinetochore attachment: causes and effects. Trends Cell Biol. 2011;21(6):374–381. doi: 10.1016/j.tcb.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakovcevski M, Schachner M, Morellini F. Individual variability in the stress response of C57BL/6J male mice correlates with trait anxiety. Genes Brain Behav. 2008;7(2):235–243. doi: 10.1111/j.1601-183X.2007.00345.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.