Significance
Normal cells rarely missegregate chromosomes, but the majority of cancer cells have a chromosomal instability (CIN) phenotype that makes errors more common and results in abnormal chromosomal content (aneuploidy). Although aneuploidy promotes transformation via gain of oncogenes and loss of tumor suppressors, it also slows cell proliferation and disrupts metabolic homeostasis. Aneuploidy therefore represents a liability as well as a source of selective advantage for cancer cells. We provoked CIN in murine T cells by weakening the spindle-assembly checkpoint and then studied the consequences. We found that CIN dramatically accelerates cancer in a genetically predisposed background and that the resulting aneuploid cancers are metabolically deranged, a vulnerability that may open new avenues to treating aneuploid cancers.
Keywords: chromosomal instability, mouse models, CIN, tumor metabolism, T-cell acute lymphoblastic lymphoma
Abstract
Aneuploidy is a hallmark of human solid cancers that arises from errors in mitosis and results in gain and loss of oncogenes and tumor suppressors. Aneuploidy poses a growth disadvantage for cells grown in vitro, suggesting that cancer cells adapt to this burden. To understand better the consequences of aneuploidy in a rapidly proliferating adult tissue, we engineered a mouse in which chromosome instability was selectively induced in T cells. A flanked by Lox mutation was introduced into the monopolar spindle 1 (Mps1) spindle-assembly checkpoint gene so that Cre-mediated recombination would create a truncated protein (Mps1DK) that retained the kinase domain but lacked the kinetochore-binding domain and thereby weakened the checkpoint. In a sensitized p53+/− background we observed that Mps1DK/DK mice suffered from rapid-onset acute lymphoblastic lymphoma. The tumors were highly aneuploid and exhibited a metabolic burden similar to that previously characterized in aneuploid yeast and cultured cells. The tumors nonetheless grew rapidly and were lethal within 3–4 mo after birth.
Aneuploidy is a hallmark of oncogenesis, affecting two out of three cancers (1). Aneuploidy arises during mitosis as a result of chromosomal instability (CIN) (2–4). The frequent occurrence of CIN in solid human tumors suggests a fundamental link between aneuploidy and cancer (5). However, primary mouse embryonic fibroblasts (MEFs) carrying a supernumerary chromosome have decreased proliferative potential, as do cells isolated from Down syndrome patients (6, 7) implying that chromosome imbalance imposes a physiological burden that lowers fitness, at least in untransformed cells (6, 8–10). In some mouse models, CIN appears to have a significant impact on lifespan at the organismal level, with increased aneuploidy decreasing life expectancy and vice versa (11–13). However, how the fitness cost imposed by CIN is balanced by its potential to promote oncogenic transformation remains poorly understood.
Mouse models of CIN involving conditional or hypomorphic mutations in spindle-assembly checkpoint (SAC) genes provide a means to study aneuploidy and assess its impact on cell fitness and oncogenesis. The SAC detects the presence of maloriented or detached kinetochores during mitosis and arrests cells in metaphase until all pairs of sister chromatids achieve the bioriented geometry that is uniquely compatible with normal disjunction (14–16). The SAC constitutes a signaling cascade [comprising monopolar spindle 1 (Mps1), Bub, Mad, CenpE, and RZZ proteins] that blocks activation of the anaphase-promoting complex, and thus mitotic progression, until all chromosomes are properly aligned (17). In the mouse, germ-line deletion of SAC genes results in early embryonic lethality, whereas heterozygous knockout of Mad2 and other SAC genes generates relatively weak tumor phenotypes late in life (2–4). Paradoxically, some SAC mutations (e.g., CenpE heterozygosity) appear to be both tumor predisposing and tumor suppressing, depending on the context (18). Hypomorphic BubR1 mutations also have the unexpected property of promoting progeria (11).
Conditional mutations typically yield tumor phenotypes more representative of human disease than germ-line mutations (19), but conditional alleles have been little studied in the case of the spindle checkpoint. We therefore engineered a conditional flanked by Lox (FLOX) mutation into Mps1, a gene thought to function upstream in the SAC pathway (20) and then selectively truncated the protein by expressing Cre recombinase in T cells. The Mps1 truncation (deletion in the kinetochore domain; Mps1DK) removes the kinase-targeting domain but leaves the rest of the protein intact. We show that expression of this truncated protein causes chromosomal instability in MEFs and aneuploidy in two different Cre-expressing mouse lines. Mps1 truncation in combination with heterozygous p53 deletion leads to early-onset lymphoblastic lymphoma and consequent death. In lymphoma cells, changes in the expression of metabolic, splicing, and DNA-synthesis genes are very similar to changes previously identified in aneuploid yeast and cultured murine cells (6, 8) and appear to constitute a hallmark of chromosomal imbalance.
Results
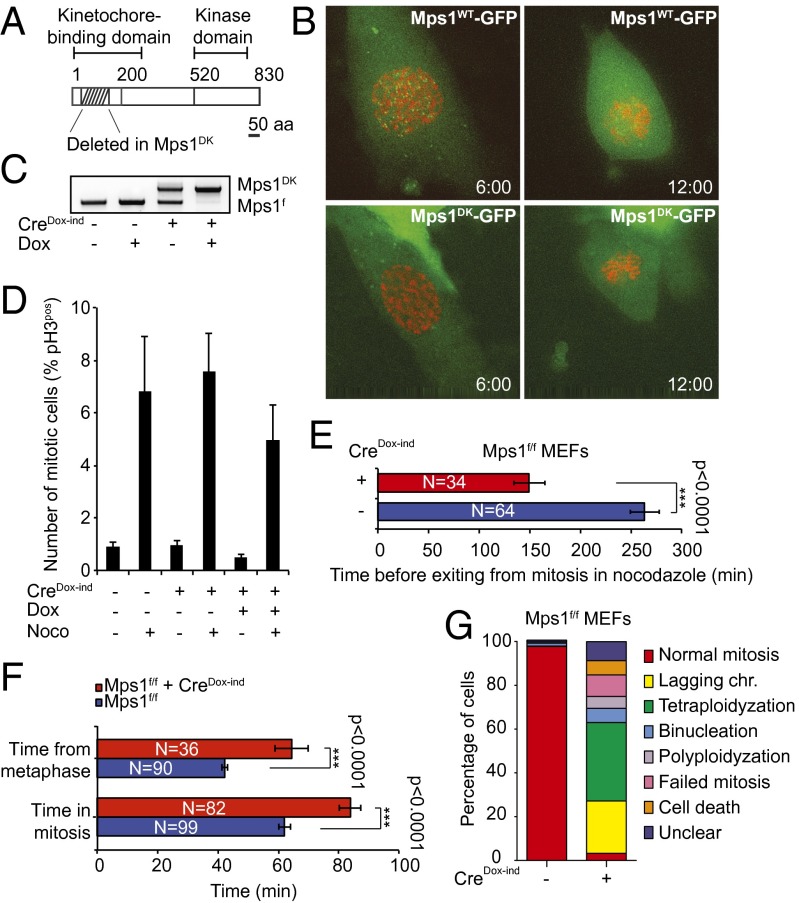
To provoke CIN in a tissue-restricted fashion, we engineered a conditional Mps1f truncation allele by flanking exons 3 and 4 of the Mps1 locus with lox-sites; correct targeting of Mps1 in mouse ES cells was confirmed by Southern blotting and RT-PCR (Fig. S1 A–C and SI Materials and Methods). Upon expression of Cre recombinase (21), the Mps1f allele generates a truncated Mps1 kinase lacking residues 47–154, a domain involved in kinetochore binding (Fig. 1A) (22). When expressed in MEFs as a GFP fusion, the Mps1DK protein had the anticipated molecular weight but, unlike wild-type Mps1-GFP, did not accumulate to the same levels on prometaphase kinetochores [Fig. 1B (compare upper and lower panels), Fig. S1 D and E, and Movies S1 and S2]. We conclude that the Mps1DK mutation impairs but does not prevent kinetochore binding, a conclusion supported by overexpression studies in human MCF 10A cells (Fig. S1 F and G and Movies S3 and S4).
Fig. 1.
Mps1 truncation leads to mitotic delay, severe abnormalities, and a weakened SAC. (A) Schematic representation of the Mps1 truncation allele. (B) Time-lapse image stills showing clear kinetochore localization of retrovirally transduced wild-type GFP-Mps1 in prometaphase (Upper) and reduced binding of GFP-Mps1DK to kinetochores (Lower) in MEFs. DNA was labeled with retroviral H2B-Cherry. (C) PCR detecting the truncation/deletion alleles for Mps1 in genomic DNA isolated from control or Cre-infected Mps1f/f MEFs. (D) Average mitotic index of Dox-inducible, Cre-transduced MEFs after 6 h of nocodazole treatment. The mitotic index is the percentage of mitotic cells as measured by phospho-histone H3 (pH3) staining. Error bars show the SEM of at least four biological replicates. (E) Average time of mitotic exit for nocodazole-arrested control Mps1f/f (blue) or Cre-infected Mps1f/f (red) MEFs. (F) Average duration of mitosis (Upper) and time from metaphase to cytokinesis (Lower) of Mps1f/f control (blue) and Cre-infected (red) MEFs as determined by time-lapse microcopy. Error bars show the SEM of the number of cells indicated within the bar. (G) Distribution of mitotic phenotypes for control and Cre-infected Mps1f/f MEFs as observed by time-lapse microscopy. Tetraploidization: a seemingly normal cell failed cytokinesis, resulting in one large tetraploid cell; binucleation: a seemingly normal cell failed cytokinesis, resulting in one cell with two nuclei; polyploidization: a seemingly tetraploid or polyploid cell failed cytokinesis, resulting in a polyploid cell; failed mitosis: a combination of mitotic errors.
The Mps1DK Truncation Weakens the SAC and Causes CIN.
To determine the consequences of Mps1 mutation for chromosome segregation at a cellular level, we isolated Mps1f/f embryos, generated MEFs, and transduced them with retroviruses expressing doxycycline (Dox)-inducible Cre (GFP-T2A-Cre). We found that exposure of these cells to Dox resulted in highly efficient switching, yielding Mps1DK MEFs within 24–48 h (Fig. 1C, compare lanes 1 and 2 with lane 4). Some recombination also was observed in the absence of Dox (Fig. 1C, lane 3). When treated with the spindle poison nocodazole, both control Mps1f/f and Mps1DK MEFs arrested in mitosis, showing that both cell types can respond to spindle disassembly (Fig. 1D). However, when time-lapse imaging was used to assay the duration of arrest, Mps1DK/DK cells were observed to exit mitosis 150 ± 16 min after DNA condensation, in contrast to 264 ± 15 min in control cells (congenic Mps1f/f cells not exposed to Cre), a significant difference (P < 0.0001) (Fig. 1E). The observation that cells expressing Mps1DK are unable to sustain mitotic arrest in the presence of spindle damage suggests that the SAC is impaired but not inactivated by the Mps1DK mutation (23) and confirms our goal in creating the allele.
Time-lapse imaging in the absence of nocodazole showed that H2B-Cherry–transduced Mps1DK MEFs spent ∼40% longer in mitosis than control cells (Fig. 1F and Fig. S1H). In addition, they frequently contained lagging chromosomes (Fig. 1 F and G and Movies S5, S6, and S7; control cells are shown in Movies S8 and S9), and half of all the cells failed to form a proper metaphase plate (Movie 10), resulting in polyploidy (Fig. 1G and Movie S11). Although this extended mitosis might appear to be a paradoxical phenotype for a SAC hypomorph, it has been shown that other SAC proteins both promote and sense chromosome–microtubule attachment and that partial inactivation of these proteins actually lengthens mitosis because the residual SAC function is able to sense incomplete attachment (22). We conclude that the Mps1DK mutation causes a partial loss of checkpoint function and also impairs kinetochore–microtubule attachment, preventing Mps1DK cells from stably arresting in the presence of spindle poisons and missegregating chromosomes under normal growth conditions (22).
Mps1DK Provokes Aggressive T-Cell Acute Lymphoblastic Lymphoma in a p53 Heterozygous Background.
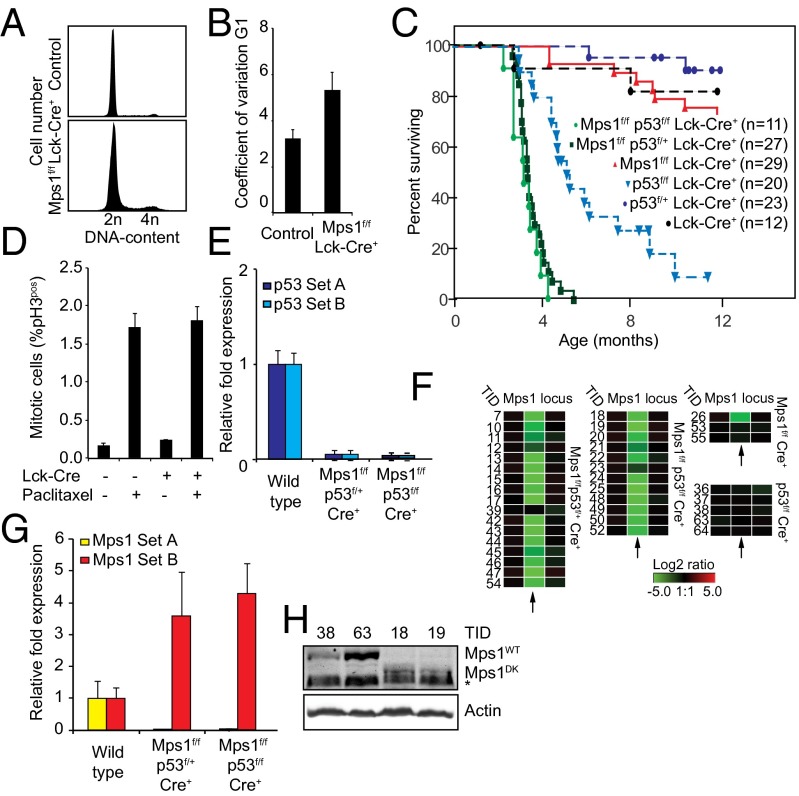
The Mps1DK truncation was introduced into a highly mitotic, nonessential adult tissue by crossing Mps1f/f mice with animals bearing a T-cell–specific Lck-Cre transgene (24). PCR revealed efficient switching of Mps1f to Mps1DK in T cells from 8- to 10-wk-old animals (Fig. S2A) concomitant with changes in the DNA content of G1 thymocytes (compare peak width and coefficient of variation for G1 peaks in Mps1DK and wild-type animals in Fig. 2 A and B), but life span was unaffected (Fig. 2C, red line). When Mps1f/f Lck-Cre+ and wild-type control mice were injected with paclitaxel, a microtubule-stabilizing drug that interferes with spindle assembly, elevated levels of mitotic cells were observed 5 h later in both genotypes (Fig. 2D). This result is consistent with data from MEFs showing that Mps1DK-expressing cells arrest in the presence of spindle damage.
Fig. 2.
Mps1 truncation provokes aneuploidy in vivo and decreases T-ALL latency in a p53-compromised background. (A) Distribution of DNA content in control and Mps1DK T cells. At least 10,000 T cells were counted. (B) Coefficient of variation values for the DNA content within G1 peaks in cell-cycle profiles of Mps1DK/DK and Cre− T cells. Error bars show the SEM of more than five biological replicates for experimental animals or more than two replicates for control animals. (C) Kaplan–Meier curves showing overall survival of indicated genotypes. (D) Average mitotic index (% pH3) of thymocytes isolated from paclitaxel- or control-injected mice 4–6 h posttreatment. Error bars show SEM of more than five biological replicates in experimental animals or more than two replicates in control animals. (E) qPCRs showing complete loss of expression of p53 (p53 probes A and B) in lymphomas of indicated genotypes. Error bars show SEM for at least three tumors per genotype. (F) aCGH data showing the loss of the kinetochore-binding sequence in Mps1 in tumors with the indicated genotypes. Each rectangle represents a single aCGH probe value. Three probes values are shown: one probe recognizing the kinetochore-binding domain (Center) and two probes flanking the 5′ (Left) and 3′ (Right) sides of the deleted region. Log2 ratios less than −5 indicate complete loss of the indicated probe. Numbers on the left refer to tumor identification numbers (TID) (Dataset S1). (G) qPCRs showing full conversion of Mps1WT to Mps1DK in tumors with the indicated genotypes. Primer set A recognizes the sequence deleted in Mps1DK, and set B detects a fragment in the kinase domain. (H) Mps1 protein levels in p53f/f (TIDs 38 and 63 and full-length Mps1) and Mps1DK p53f/f tumors (TIDs 18 and 19) showing the conversion of full-length to truncated Mps1 in the latter genotype. The asterisk indicates a background band recognized in all lysates and runs just below Mps1DK that is detected only in TIDs 18 and 19.
To assay tumor formation in a sensitized environment, we generated Mps1f/f Lck-Cre+ animals heterozygous for a FLOX-p53 allele (25). Loss of p53 suppresses aneuploidy-associated apoptosis in multiple cell types and is oncogenic in thymocytes (26–28). Mps1f/fp53f/+ Lck-Cre+ mice rapidly developed T-cell acute lymphoblastic lymphomas (T-ALL); ∼50% of animals had died of the disease by age 3.5 mo, and 100% had died by age 5 mo (Fig. 2C, dark green line). In contrast, heterozygosity at Mps1 had little effect on the survival of p53-null mice: Mps1f/+p53f/f Lck-Cre+ animals had survival curves indistinguishable from those of p53f/f Lck-Cre+ mice (Fig. S2B). In addition, Mps1 wild-type p53f/+ Lck-Cre+ mice rarely developed disease, and tumor-free survival was indistinguishable from that of Lck-Cre+ control animals (Fig. 2C, compare dark blue and black lines) (29). Thus, the Mps1DK mutation is strongly oncogenic in T cells on a p53f/+ background.
Mps1DK promoted loss of heterozygosity (LOH) at the p53 locus: the PCR product corresponding to p53Δ was substantially more abundant than the product corresponding to wild-type p53 in DNA from Mps1f/fp53f/+ Lck-Cre+ tumors (Fig. S2C, compare tumors 9–17 with 18–21). Quantitative PCR (qPCR) data were consistent with this finding: p53 mRNA was virtually undetectable in tumors recovered from Mps1f/f p53f/+ animals (Fig. 2E). To characterize the LOH event, we extracted probe values from array-based comparative genomic hybridization (aCGH) for 17 tumors arising in Mps1f/f p53f/+ animals (Fig. S2D). In all but two animals (tumors 12 and 39), hybridization to p53 probes was low, similar to that of p53-null tumors from Mps1f/f p53f/f animals (compare Fig. S2 D and E). Hybridization to neighboring probes was unaffected, suggesting that the wild-type copy of p53 had been replaced by p53Δ through either CIN or mitotic recombination. We conclude that Mps1 truncation facilitates p53 LOH, a highly oncogenic event in thymocytes (29–31).
The protumorigenic effects of Mps1 truncation do not appear to involve p53 LOH alone. Tumor induction was significantly faster in Mps1f/f p53f/+ Lck-Cre+ (and Mps1f/f p53f/f Lck-Cre+) mice than in p53f/f Lck-Cre+ mice, with death of 50% of the former animals by age 3.5 mo as opposed to 5 mo for the latter (P < 0.0001) (Fig. 2C). Analysis of mRNA and genomic DNA confirmed efficient Cre-mediated deletion of p53 in tumors having either genotype (Fig. 2E and Fig. S2 C–E). Moreover, accelerated tumorigenesis in Mps1f/f p53f/f double-mutant animals relative to p53f/f animals was confirmed with a second Cre driver, mouse mammary tumor virus (MMTV)-Cre, which is transcribed in both T cells and the mammary gland (Fig. S2F) (32). However, no mammary tumors were observed in these animals, presumably because T-ALL developed before breast cancers could emerge.
To show that tumors comprised cells in which the Mps1f loci had been excised and thus that lymphomagenesis was not driven by p53 loss alone (a concern because p53 is such a strong tumor suppressor in T cells), we measured the efficiency of Cre-mediated recombination at the Mps1 genomic locus. We assayed Mps1 mRNA levels using PCR and aCGH, and we performed Western blotting. In tumors isolated from Mps1f/f p53f/f and Mps1f/f p53f/+ mice, bands corresponding to Mps1DK were the predominant amplified products (Fig. S2G and Dataset S1) in both Lck-Cre+ and MMTV-Cre+ backgrounds. In the aCGH data, we observed nearly complete loss of hybridization to sequences excised by Cre-mediated recombination of the Mps1f/f locus (Fig. 2F). qPCR of tumor RNA also confirmed the loss of Mps1 expression: Probes selective for the nonmutated 3′ domain (Mps1 probe set B; Fig. 2G and Fig. S2H) yielded a strong qPCR product, whereas probes corresponding the 5′ region of Mps1 deleted in Mps1DK (Mps1 probe set A) were ∼20-fold less abundant in tumors than in wild-type thymus DNA. Moreover, RT-PCR followed by Sanger sequencing confirmed the presence of correctly recombined Mps1DK transcript (in tumors 13 and 23) and full-length Mps1 in p53f/f tumors (tumors 36 and 63) (Fig. S2I and Dataset S2). qPCR also showed that Mps1DK is overexpressed in tumors by approximately fourfold relative to wild-type Mps1 in parental cells, presumably because the elevated expression of the hypomorphic allele confers a selective advantage on cells. Finally, by Western blotting we could detect a protein band corresponding to the expected length of the Mps1DK protein in Mps1f/f p53f/f Lck-Cre+ tumor samples (tumors 18 and 19; Fig. 2H). We conclude that the Mps1DK allele was maintained in the great majority of T cells throughout the development of a tumor, and thus the acceleration in tumorigenesis observed in double-mutant animals reflects ongoing synergy between Mps1 and p53 mutations.
Mps1DK-Driven Tumors Exhibit Recurring Chromosomal Abnormalities.
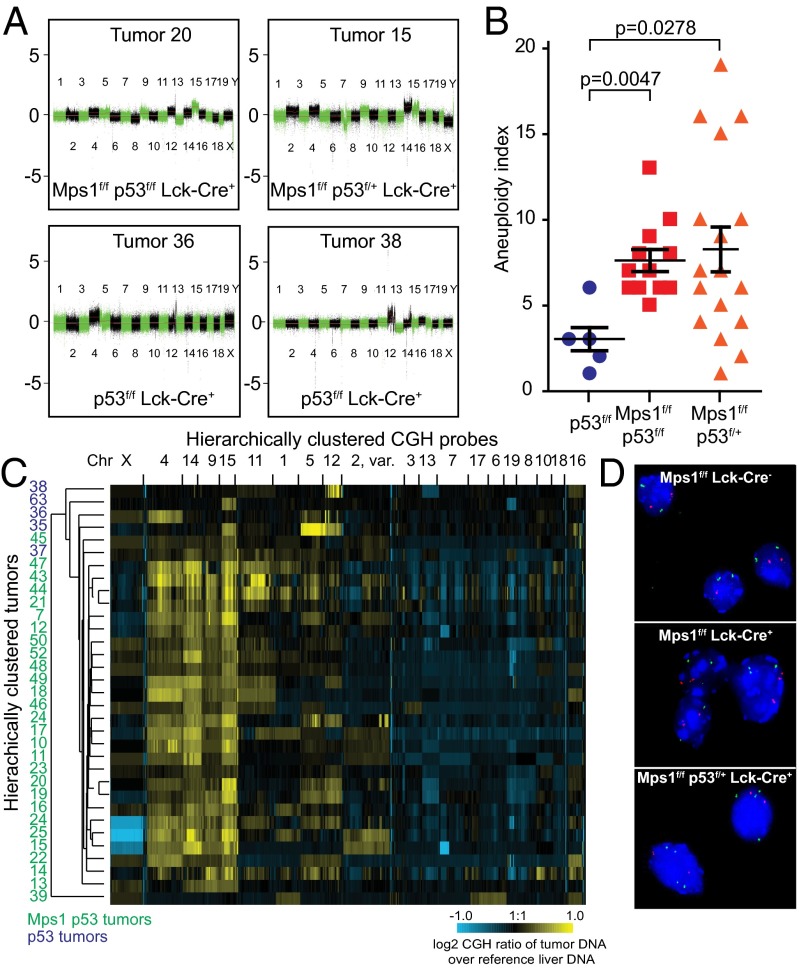
To determine the extent of aneuploidy in Mps1DK T-ALL, we used aCGH to quantify chromosome copy number across the genome and interphase FISH to quantify chromosome number in single cells. aCGH revealed frequent loss and gain events for multiple chromosomes (four representative plots are shown in Fig. 3A, and normalized aCGH data are summarized in Dataset S3). As a simple measure of CIN, we summed the total number of chromosome gain and loss events in each tumor to create an “aneuploidy index.” The aneuploidy index ranged from 3 to 19 in 30 tumor samples examined and was significantly higher in tumors arising in Mps1f/f p53f/f or Mps1f/f p53f/+ animals (average indices of 8.2 and 7.6, respectively) than in p53f/f animals (average index 2.4; P = 0.047 and P = 0.028, respectively) (Fig. 3B). Similarly, unsupervised single-linkage hierarchical clustering of cumulative aCGH data showed that tumors from Mps1f/f mice heterozygous (Mps1f/f p53f/f) or homozygous (Mps1f/f p53f/+) for p53 deletion clustered together and had more chromosomal abnormalities (Fig. 3C; green numbers) than tumors from p53f/f animals that had less severe aneuploidy (Fig. 3C; blue numbers). Probes lying on the same chromosomes coclustered across all tumor samples, demonstrating that a significant fraction of the aneuploidy in these tumors involved gain and loss of whole chromosomes.
Fig. 3.
Mps1 truncation leads to CIN and clonally stable karyotypes in tumors. (A) Representative aCGH profiles for four tumors showing whole-chromosome instability. (B) Aneuploidy index (total number of whole chromosomes gained and lost as assessed by aCGH) for tumors with the indicated genotypes. (C) Single-linkage hierarchical cluster analysis for individual tumors (top to bottom) and CGH probes (left to right). Clustering was separated visually into 20 clear groups. (D) Representative interphase FISH images of control (Top), Mps1DK T cells (Middle), and Mps1DK T-ALL cells (Bottom) showing copy numbers for Chr15 (green) and Chr17 (red).
Amplification of Chr15 was particularly frequent in aCGH data, regardless of genotype, and is known to be common feature of mouse T-ALLs (7, 33, 34). In addition, amplification of Chr4 and Chr14 and to a lesser extent Chr9 was observed in many tumors, and Chr13 and Chr19 were commonly deleted, suggesting that those chromosomes carry genes important for transformation or aneuploid tumor progression. Interphase FISH confirmed aneuploidy in nontransformed Mps1-mutant thymocytes and heterogeneity in chromosome number within a single tumor. For example, Chr15 and Chr17 were aneuploid in a greater number of Mps1f/f Lck-Cre+ thymocytes than in wild-type thymocytes (an average of 5% vs. 11% of cells for Chr15 and 6% vs. 12% of cells for Chr17) (Fig. S2J). In Mps1f/f p53f/+ Lck-Cre+ T-ALL tumors we observed Chr15 trisomy in up to 80% cells, but the fraction of cells involved differed among animals (Fig. 3D and Fig. S2J).
To identify common focal loss and gain events, we performed genome-wide cumulative segmental gain or loss analysis (SGOL) comparing Mps1DK-driven and p53f/f Lck-Cre+ tumors. For both tumor classes, SGOL revealed strong deletion peaks on Chr6 and Chr14, consistent with a unique pattern of recombined T-cell receptor α/β loci. These results strongly suggest that tumors arose from a single parental T cell (Fig. S3 A and B). We can reconcile the SGOL and FISH data by hypothesizing that T-ALLs are clonal early in their development [at the time of T-cell receptor (TCR) rearrangement] but that ongoing CIN results in subsequent chromosome loss and gain. Selection is expected to maintain some aneuploidies, e.g., Chr15 amplification, whereas other chromosomes (e.g., Chr17) might be subjected to ongoing loss and gain.
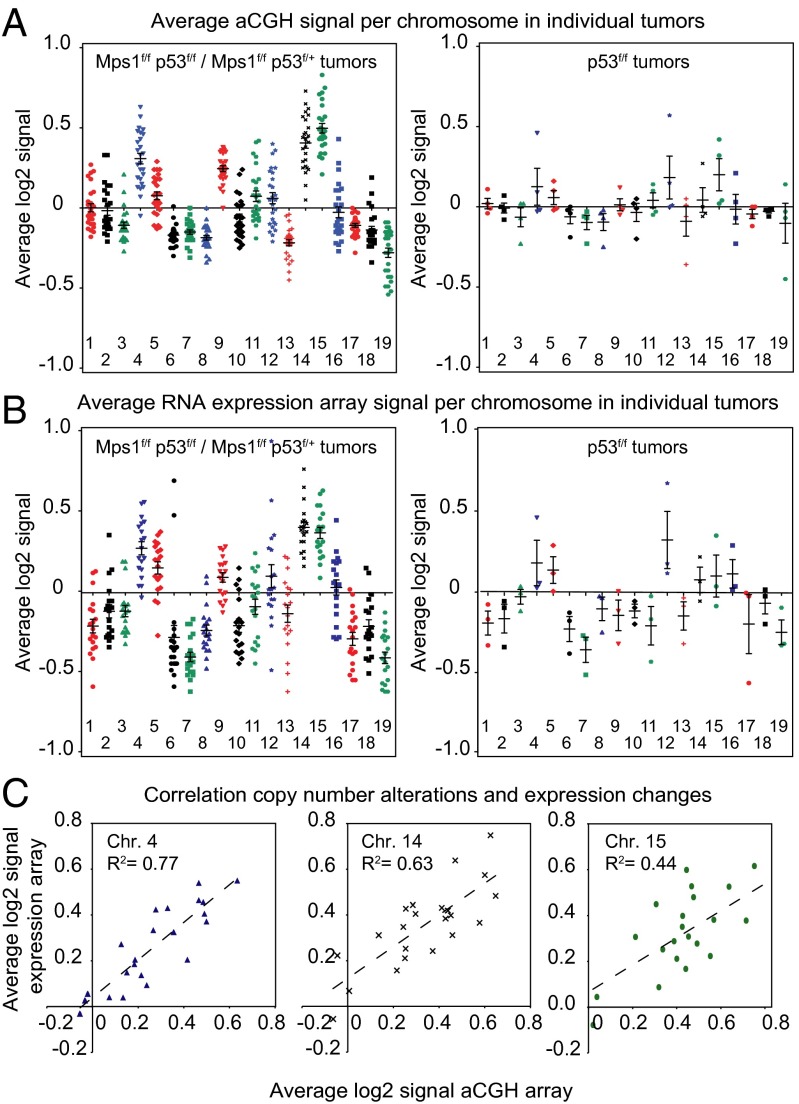
Recent studies on budding yeast and MEFs carrying supernumerary chromosomes have shown proportional increases in gene copy number and transcription (6, 8). To determine if these increases are present in tumors driven by Mps1 truncation, we used Illumina expression arrays to analyze the transcriptomes of 22 tumor samples that previously had been studied by aCGH as well as thymus DNA isolated from 6-wk-old wild-type mice. A strong correlation between mRNA and gene copy number was observed when aCGH values and expression levels were sorted by chromosomal position (Fig. S3 C and D). When we calculated the average expression changes per individual chromosome for each tumor (using healthy thymic samples as a control) and then compared the value with the aCGH intensity (Fig. 4 A and B and Dataset S4), the correlation between expression and copy number was R2 = 0.44–0.76 for the most commonly aneuploid chromosomes (Chr4, Chr14, and Chr15). (Fig. 4C and Fig. S4 show correlation for all chromosomes.) We conclude that in tumors, as in cultured cells, chromosomal imbalances on average are translated into increases and decreases in transcription, and thus there is little or no dosage compensation.
Fig. 4.
Gene copy number results in proportional transcription changes in aneuploid tumors. (A and B) Average aCGH (A) or RNA expression values (B) were calculated per chromosome for each tumor and plotted for each tumor group. Each individual symbol represents the average value of that chromosome in one tumor; black crosses show the mean and SEM across all tumors per chromosome. (C) Linear regression plots showing the correlation strength (coefficient of correlation, R2) between copy number changes (aCGH) and expression changes (expression arrays) for frequently gained Chr4, Chr14, and Chr15.
Mps1DK Tumors Show Evidence of Aneuploidy-Induced Stress.
To identify genes significantly over- and underexpressed in T-ALLs, we sorted genes by their cumulative expression changes across all samples (annotated in Fig. S5A and Dataset S4). For Chr4, Chr14, and Chr15, the majority of genes had a positive cumulative score; the reverse was true for Chr19, reflecting the correlation between changes in transcription and gene copy number. Among the outliers we found several that exhibited an inverse correlation between copy number and expression including the keratins Krt, 5, 7, 8, and 18, Epsi1, and Chdr1. These genes are expressed in the thymic cortex and not in tumor cells, and their lower expression in mutant animals likely reflects T-ALL–mediated depletion of cortical tissue. A second set of outlier genes was significantly overexpressed relative to other genes on the same chromosome. This set includes genes involved in cell metabolism (Srm, Gln3, Cox6a, Drospha, and Adk), cellular stress (Serp2 and Hsf1), cell cycle (Recql4, Cdkn2a, Skp2, and Tnc), and epigenetic regulation (Prmt5 and Cbx5). Surprisingly, Myc, a known oncogene in T-ALL (7), was not among the strongest positive outliers on Chr15. In future work it should be possible to use Mps1DK-driven aneuploidy to identify new oncogenes or tumor suppressors involved in T-ALL as well as genes involved in cell survival in the presence of CIN.
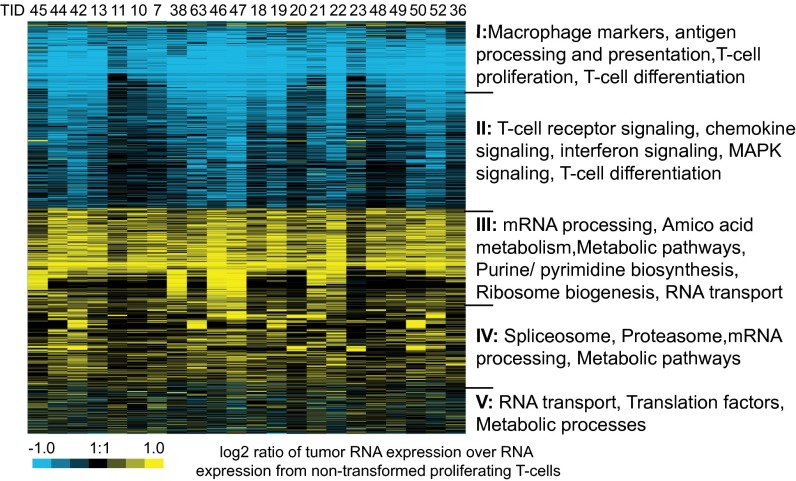
To begin to identify biological pathways altered by aneuploidy, we compared probes that were up- or down-regulated at least 1.5-fold in >15% of tumors analyzed (4 of 22); we identified ∼3,300 genes by this analysis. We then used WebGestalt (35) to find Gene Ontogeny (GO) categories that were significantly enriched (using a Bonferroni corrected P value < 0.05). The most commonly deregulated pathways were TCR signaling, mRNA processing, cell cycle, and pathways involved in cellular metabolism (Fig. S3E). When we performed hierarchical clustering of deregulated genes (Fig. 5), dividing clusters into five groups based on whether genes were strongly or weakly up- or down-regulated, T-cell differentiation and signaling factors were down-regulated; this result is consistent with histological data showing that tumors are poorly differentiated. In contrast, pathways involved in cellular metabolisms (GO terms for “metabolic pathways,” “RNA metabolic pathways,” “spliceosome,” “translation factors,” “nucleotide synthesis,” and others) were up-regulated. Misregulation of these processes previously has been associated with aneuploid stress in cultured mammalian cells and budding yeast (6, 8, 36, 37). We conclude that dysregulation of metabolic pathways is a common feature of CIN in multiple organisms and cell types, including rapidly growing tumors.
Fig. 5.
Mps1DK-driven CIN activates genes regulating various aspects of cellular metabolism. Single-linkage hierarchical clustering of transcriptome data shows the most significant deregulated pathways per group. Groups I–V were grouped by visual inspection. Full gene lists and enrichment analyses are in Dataset S4.
Discussion
In this article we report the development and analysis of mice in which CIN and consequent aneuploidy are induced in a rapidly proliferating but nonessential adult tissue by conditionally truncating the SAC kinase Mps1. We show that the Mps1 truncation, which deletes a kinetochore-targeting domain but leaves kinase activity and other functions intact, can provoke but not sustain a checkpoint arrest in the presence of spindle poison; it also causes chromosome misalignment and generates lagging chromosomes, consistent with the dual role of SAC proteins in promoting and sensing kinetochore attachment. When mutated in murine T cells, Mps1 truncation causes aneuploidy, but this aneuploidy is insufficient for efficient oncogenic transformation. However, in a predisposed p53 heterozygous-deletion background, Mps1 truncation results in rapidly growing T-ALL. We observe frequent p53 LOH in Mps1f/f p53f/+ Lck-Cre+ animals, consistent with the known role of p53 as a potent tumor suppressor in T cells. However, p53 LOH cannot be the only oncogenic event in T-ALLs, because tumor latency is significantly shorter in Mps1f/f p53f/+ Lck-Cre+ animals than in p53f/f Lck-Cre+ animals. aCGH also reveals higher levels of aneuploidy in compound-mutant animals than in single mutants. Taken together, these data suggest that Mps1 mutation causes p53 LOH and that the combined Mps1-p53 mutant genotype results in more efficient gain and loss of cancer genes than p53 mutation alone.
We interpret aCGH and interphase FISH data to show that T-ALLs are initially clonal (based on TCR rearrangement) but that ongoing CIN results in tumors with recurrent aneuploidies in Chr4, Chr14, Chr15, and Chr19 as well as sporadic changes in other chromosomes. The net result is changes in the expression of oncogenes and tumor suppressors driving T-ALL as well as changes in as yet unidentified pathways involved in tolerizing cells to aneuploidy. A significant body of literature has emerged over the past few years examining the consequences of aneuploidy for the physiology of yeast and cultured murine fibroblasts. These studies have revealed recurrent up-regulation of pathways involved in mRNA processing and other metabolic processes (6, 8, 37). Our data show that T-ALLs generated by Mps1 truncation and p53 loss also exhibit transcriptional signatures similar to those of aneuploid MEFs (6) and untransformed tissues (36). Thus, T-ALLs cells can grow rapidly in the face of frequent chromosome loss and gain, but CIN nonetheless imposes a burden on tumor cells similar to that observed in MEFs and budding yeast cells. This finding is potentially significant, because proteotoxic stress and metabolic dysregulation have become important targets for cancer therapy and may represent a means to kill highly aneuploid cancers selectively.
Materials and Methods
Analysis of Mice.
Mice used in this study had a mixed C57BL/6 and 129/Sv D3 genetic background. Mps1 conditional mice harboring a deletion of residues 47–154 were generated as described in SI Materials and Methods. p53 conditional-knockout mice were obtained from Anton Berns (Department of Genetics, The Netherlands Cancer Institute, Amsterdam, The Netherlands) (25), Lck-Cre transgenic mice from Taconic (38), and MMTV-Cre mice (39) from the MIT mouse repository. Mice were intercrossed to obtain the described strains and genotyped as described previously (25, 39). (Genotyping PCR primers are given in Dataset S5.) For survival studies, mice were monitored for tumor development weekly starting at 2.5 mo of age by looking for difficulty in breathing (a consequence of thymic hypertrophy). Tissues were fixed in 10% formalin and then paraffin-embedded for histology. Animal protocols were approved by the MIT, Harvard Medical School, and University of Groningen Medical Center Committees on Animal Care and by the U.K. Home Office.
MEF Isolation, Transduction, Flow Cytometry, and Time-Lapse Analysis.
MEFs were isolated as described previously (40) at E13.5 from Mps1f/f embryos and were cultured under low-oxygen (3%) conditions. Retroviral particles (pRetrox system; Clontech) were produced in Lipofectamine 2000-transfected (Life Technologies) 293T cells. MEFs were first transduced with pRetrox-rtTA virus and next with pRetrox-GFP-T2A-Cre virus, allowing us to assess Cre expression without linking Cre to GFP directly (41), or with pRetrox GFP-Mps1WT or GFP-Mps1DK virus (constructs are described in detail in SI Materials and Methods). For time-lapse imaging, cells subsequently were transduced with pRetrox-H2B-Cherry or pRetrox-H2B-GFP to visualize the DNA and were treated with 250 ng/mL nocodazole (Sigma) where indicated. For flow cytometry, transduced cells were treated with 1 μg/mL Dox (Sigma) for 48–72 h to induce the retroviral inserts, then were treated with 250 ng/mL nocodazole for 6 h, and were fixed in 70% ethanol. Cells then were stained with phycoerythrin-labeled pHistoneH3 antibody (Cell Signaling) and FxCycle Brilliant blue (Life Technologies) and analyzed on an LSRII analyzer (BD Biosciences). Data were analyzed using FlowJo software. For time-lapse analysis, cells were seeded on LabTek imaging chambers (Thermo Fisher), induced with 1 μg/mL Dox for 48–72 h, and imaged for up to 36 h on a DeltaVision Elite imaging station (Applied Precision, GE Healthcare) in a low-oxygen imaging chamber (Oko Laboratories). Movies were deconvolved and analyzed using SoftWorx suite (GE Healthcare).
DNA/RNA/Protein Isolation, Array, and Blotting Experiments.
Genomic DNA from tumor samples and control liver was isolated using genomic DNA tissue kits (Qiagen). Genotyping primers are listed in Dataset S5. Protein was isolated using a total protein extraction kit (Millipore). For protein detection, proteins were blotted using standard Western blotting protocols. The antibodies used were Mps1 (TTK-C19, Santa Cruz) and HRP-conjugated goat anti-rabbit (Cell Signaling). For aCGH experiments, labeled DNA (SI Materials and Methods) was hybridized to 244K mouse genome CGH arrays (Agilent) according to the manufacturer’s protocol and was analyzed as described in SI Materials and Methods. RNA was isolated from tumor samples and from the thymuses from 6-wk-old control mice using the RNeasy kit (Qiagen). Labeled RNA (SI Materials and Methods) was hybridized to Illumina v6.2 BeadChip arrays and was scanned and analyzed as described in SI Materials and Methods. qPCR primers are listed in Dataset S5. All array data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession no. GSE57334.
Supplementary Material
Acknowledgments
We thank members of the P.K.S. laboratory, S. W. M. Bruggeman, L. Albacker, and L. Kleiman for critically reading the manuscript and fruitful discussions; B. Bakker for assistance with cloning; Roderick Bronson for help with pathology; and A. Burds for sharing Lck-Cre and MMTV-Cre mice. This work was supported by Dutch Cancer Society Grant RUG 2012-5549; by grants from Stichting Kinder Oncologie Groningen and European Molecular Biology Organization (to F.F.); by National Institutes of Health (NIH) Grants CA084179 and CA139980 (to P.K.S.); by NIH Grant P30-CA14051 to the MIT mouse transgenic facility; and by the Wellcome Trust (F.F. and A.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE57334).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400892111/-/DCSupplemental.
References
- 1.Duijf PH, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer. 2013;132(10):2316–2326. doi: 10.1002/ijc.27924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: Mouse modelling of the human disease. Nat Rev Cancer. 2010;10(2):102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland AJ, Cleveland DW. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10(7):478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foijer F, Draviam VM, Sorger PK. Studying chromosome instability in the mouse. Biochim Biophys Acta. 2008;1786(1):73–82. doi: 10.1016/j.bbcan.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16(6):475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322(5902):703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones L, et al. Gain of MYC underlies recurrent trisomy of the MYC chromosome in acute promyelocytic leukemia. J Exp Med. 2010;207(12):2581–2594. doi: 10.1084/jem.20091071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 9.Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA. 2004;101(23):8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao R, Zielke CL, Zielke HR, Pevsner J. Global up-regulation of chromosome 21 gene expression in the developing Down syndrome brain. Genomics. 2003;81(5):457–467. doi: 10.1016/s0888-7543(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 11.Baker DJ, et al. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet. 2004;36(7):744–749. doi: 10.1038/ng1382. [DOI] [PubMed] [Google Scholar]
- 12.Baker DJ, et al. Early aging-associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol. 2006;172(4):529–540. doi: 10.1083/jcb.200507081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baker DJ, et al. Increased expression of BubR1 protects against aneuploidy and cancer and extends healthy lifespan. Nat Cell Biol. 2013;15(1):96–102. doi: 10.1038/ncb2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jallepalli PV, Lengauer C. Chromosome segregation and cancer: Cutting through the mystery. Nat Rev Cancer. 2001;1(2):109–117. doi: 10.1038/35101065. [DOI] [PubMed] [Google Scholar]
- 15.Taylor SS, Scott MI, Holland AJ. The spindle checkpoint: A quality control mechanism which ensures accurate chromosome segregation. Chromosome Res. 2004;12(6):599–616. doi: 10.1023/B:CHRO.0000036610.78380.51. [DOI] [PubMed] [Google Scholar]
- 16.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130(4):941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8(5):379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 18.Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11(1):25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nat Rev Cancer. 2002;2(4):251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 20.Janssen A, Kops GJ, Medema RH. Elevating the frequency of chromosome mis-segregation as a strategy to kill tumor cells. Proc Natl Acad Sci USA. 2009;106(45):19108–19113. doi: 10.1073/pnas.0904343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hached K, et al. Mps1 at kinetochores is essential for female mouse meiosis I. Development. 2011;138(11):2261–2271. doi: 10.1242/dev.061317. [DOI] [PubMed] [Google Scholar]
- 22.Nijenhuis W, et al. A TPR domain-containing N-terminal module of MPS1 is required for its kinetochore localization by Aurora B. J Cell Biol. 2013;201(2):217–231. doi: 10.1083/jcb.201210033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rudner AD, Murray AW. The spindle assembly checkpoint. Curr Opin Cell Biol. 1996;8(6):773–780. doi: 10.1016/s0955-0674(96)80077-9. [DOI] [PubMed] [Google Scholar]
- 24.Hennet T, Hagen FK, Tabak LA, Marth JD. T-cell-specific deletion of a polypeptide N-acetylgalactosaminyl-transferase gene by site-directed recombination. Proc Natl Acad Sci USA. 1995;92(26):12070–12074. doi: 10.1073/pnas.92.26.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 26.Burds AA, Lutum AS, Sorger PK. Generating chromosome instability through the simultaneous deletion of Mad2 and p53. Proc Natl Acad Sci USA. 2005;102(32):11296–11301. doi: 10.1073/pnas.0505053102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson SL, Compton DA. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol. 2010;188(3):369–381. doi: 10.1083/jcb.200905057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara T, et al. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437(7061):1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 29.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356(6366):215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 30.Purdie CA, et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene. 1994;9(2):603–609. [PubMed] [Google Scholar]
- 31.Jacks T, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4(1):1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 32.Wagner KU, et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10(6):545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 33.Zha S, et al. ATM-deficient thymic lymphoma is associated with aberrant tcrd rearrangement and gene amplification. J Exp Med. 2010;207(7):1369–1380. doi: 10.1084/jem.20100285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takabatake T, et al. Analysis of changes in DNA copy number in radiation-induced thymic lymphomas of susceptible C57BL/6, resistant C3H and hybrid F1 Mice. Radiat Res. 2008;169(4):426–436. doi: 10.1667/RR1180.1. [DOI] [PubMed] [Google Scholar]
- 35.Zhang B, Kirov S, Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33(Web Server issue):W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foijer F, et al. Spindle checkpoint deficiency is tolerated by murine epidermal cells but not hair follicle stem cells. Proc Natl Acad Sci USA. 2013;110(8):2928–2933. doi: 10.1073/pnas.1217388110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109(31):12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/s1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 39.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25(21):4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foijer F, Wolthuis RM, Doodeman V, Medema RH, te Riele H. Mitogen requirement for cell cycle progression in the absence of pocket protein activity. Cancer Cell. 2005;8(6):455–466. doi: 10.1016/j.ccr.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 41.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.