Significance
In many eukaryotes, two types of meiotic crossovers (COs) coexist: class I COs that show CO interference and class II COs that do not show interference. Little is known about the separate properties of these two CO pathways or their interaction in wild-type organisms because individual COs cannot be assigned by class using marker segregation. We present an improved cytogenetic approach to identify COs from each class simultaneously on tomato meiotic chromosomes to reveal that the two CO classes have different distributions, with class II COs enriched in short arms and heterochromatin. Although class II CO distributions are consistent with no interference, interference between the two pathways was detected, with suppression of close class I and class II COs.
Keywords: genetic interference, MLH1, recombination nodule, synaptonemal complex, MUS81
Abstract
Crossovers (COs) shuffle genetic information and allow balanced segregation of homologous chromosomes during the first division of meiosis. In several organisms, mutants demonstrate that two molecularly distinct pathways produce COs. One pathway produces class I COs that exhibit interference (lowered probability of nearby COs), and the other pathway produces class II COs with little or no interference. However, the relative contributions, genomic distributions, and interactions of these two pathways are essentially unknown in nonmutant organisms because marker segregation only indicates that a CO has occurred, not its class type. Here, we combine the efficiency of light microscopy for revealing cellular functions using fluorescent probes with the high resolution of electron microscopy to localize and characterize COs in the same sample of meiotic pachytene chromosomes from wild-type tomato. To our knowledge, for the first time, every CO along each chromosome can be identified by class to unveil specific characteristics of each pathway. We find that class I and II COs have different recombination profiles along chromosomes. In particular, class II COs, which represent about 18% of all COs, exhibit no interference and are disproportionately represented in pericentric heterochromatin, a feature potentially exploitable in plant breeding. Finally, our results demonstrate that the two pathways are not independent because there is interference between class I and II COs.
Eukaryotic sexual reproduction involves meiosis, a specialized cell division in which DNA duplication in a diploid cell is followed by two cell divisions to produce four haploid cells. The first division, Meiosis I, involves crossing over and chiasmata formation between each pair of homologous chromosomes, thereby ensuring separation of the homologs and formation of two haploid cells, each with one complete set of replicated chromosomes. The second division, Meiosis II, is a mitosis-like division in which the two sister chromatids separate to yield four haploid cells that directly or indirectly form gametes. Because these four products are genetically unique due to crossing over and independent segregation of homologous chromosomes during Meiosis I, meiosis plays an important role in creating genetic diversity in sexually reproducing organisms.
Crossing over during meiosis is tightly controlled so each pair of homologs has at least one “obligate” crossover (CO) that ensures balanced reductional segregation, but the presence of a CO reduces the likelihood of another CO in its vicinity, a phenomenon referred to as CO interference (1, 2). Significant progress has been made recently in illuminating the molecular events of meiotic recombination and the control of crossing over (3–8). The initiating event of meiotic recombination in most organisms is formation of numerous DNA double-strand breaks (DSBs). Homolog-dependent repair of a DSB may follow any one of at least three pathways: (i) non-CO that may result in a short gene conversion; (ii) CO with interference (class I COs, produced by pathway P1); or (iii) CO without interference (class II COs, produced by pathway P2) (6, 7, 9). The interfering CO pathway involves the resolution of double Holliday junctions, which requires many proteins including the ZMM group (ZIP1-4, MSH4-5, MER3) and the MutL homolog 1 (MLH1)/MLH3 complex (6, 10). The noninterfering CO pathway depends primarily on the Mus81/Mms4 endonuclease complex in budding yeast (MUS81/EME1 complex in plants and animals) (5–7, 11–14).
Meiotic COs occur in association with two cytological structures, synaptonemal complexes (SCs) that link each pair of homologous chromosomes throughout their lengths during pachytene and late recombination nodules (RNs) that are ellipsoidal structures on SCs (15). Every SC has at least one RN, each RN marks a CO site, and most RNs contain MLH1 protein (16–19). RNs are too small (50-100 nm) to be resolved using light microscopy (LM), but they can be readily visualized by transmission electron microscopy (EM), particularly in 2D spreads of SCs (18). Antibodies to MLH1 protein have been used as immunofluorescent probes to map class I COs on SCs (e.g., refs. 19 and 20). Pathway 2 (P2), which was revealed using mutants of the P1 pathway, produces class II COs, and these class II COs showed no interference in the marker intervals studied (21–23). The P1 pathway produces the majority of COs, and the P2 pathway accounts for ∼5–30% of COs (8, 11, 21). CO distributions have been effectively modeled by assuming that class II COs are independent from class I COs (24). However, class II COs have not been independently mapped on chromosomes (12), and little is known about the properties of each pathway or whether they interact in wild-type organisms.
Here, we describe an advanced approach that uses SC spreads from wild-type tomato (Solanum lycopersicum, 2n = 2x = 24) to directly identify the pathway of origin for each CO in individual meiotic nuclei. For this, we superimposed the immunofluorescent LM image of an SC spread showing MLH1 foci (class I COs) onto an EM image of the same SC spread showing RN locations (all COs). RNs that coincide with MLH1 foci (MLH1-positive RNs) mark class I COs, and RNs that do not coincide with MLH1 foci (MLH1-negative RNs) are considered to mark class II COs. Because EM is time-consuming, this approach takes advantage of both the relative speed of LM and the high resolution of EM, allowing us to analyze RNs on 1882 tomato SCs.
Results and Discussion
Characteristics of MLH1-Positive and MLH1-Negative RNs.
RNs were identified by EM based on their association with SCs, size, morphology, and staining intensity (18), and then RNs were classified as MLH1-positive or MLH1–negative using the corresponding LM image superimposed over the EM image (Fig. 1 and SI Appendix, Fig. S1). Using this same approach, we were unable to detect a distinct or reproducible MUS81 immunolabeling signal on tomato SC spreads, so we were not able to determine whether MLH1-negative RNs contain MUS81 protein (SI Appendix, Fig. S2). Perhaps our inability to cytologically detect MUS81 is related to the observation that Mus81–Mms4 acts as a single heterodimer (25), whereas Mlh1 appears to have a structural role also (26). Regardless, in the absence of an effective antibody to MUS81 protein, we classified and mapped RNs from nuclei in which each of the twelve chromosomes had at least one MLH1 focus to minimize the chance that an RN was not labeled with MLH1 antibodies for technical reasons (see SI Appendix, SI Materials and Methods for details and tests of selection criteria). This criterion is based on the observation that SCs without MLH1 foci are rare in tomato and other organisms, indicating that Class I COs are responsible for the obligate CO in each bivalent (3, 19, 27, 28). However, we cannot exclude the possibility that some of the unlabeled RNs may have contained MLH1 protein that was not detected in our assay. Using 162 nuclei that fit the criterion mentioned, we were able to map RN positions on 1882 individual chromosomes that were identified based on relative length and kinetochore position (arm ratio; SI Appendix, Tables S1 and S2 and Dataset S1). We observed a mean of 18.8 RNs per nucleus, with 15.4 (82%) MLH1-positive RNs and 3.4 (18%) MLH1-negative RNs (SI Appendix, Fig. S3). This frequency of MLH1 foci is similar to the 15.0 MLH1 foci per nucleus observed earlier for tomato (19). We found that a large fraction of SCs (1,419 of 1,882, 75%) had no MLH1-negative RN, and about 85% (392 of 463) of the remaining SCs had only one MLH1-negative RN (SI Appendix, Table S3).
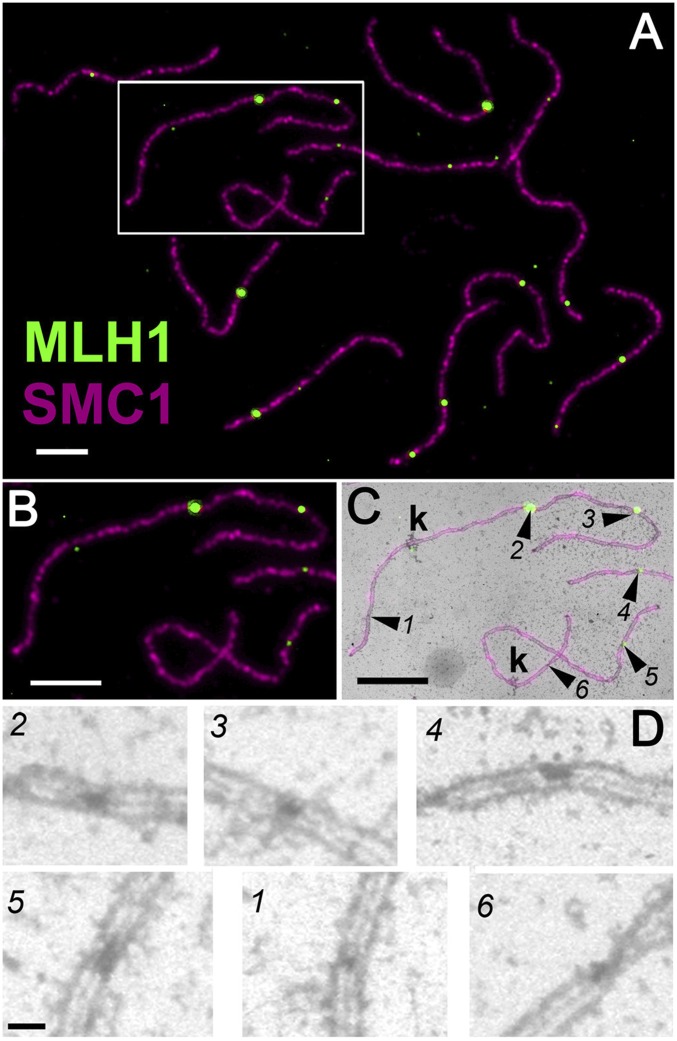
Fig. 1.
Identifying MLH1-positive and MLH1-negative RNs using consecutive light and electron microscopy of the same SC spread (SI Appendix, Fig. S1). (A) LM view of a complete set of 12 tomato SCs labeled with antibodies to MLH1 (green) and SMC1, an SC component (magenta). (B) Area boxed in A, enlarged. (C) Fluorescent LM image from B superimposed over the corresponding portion of the EM image. Two complete SCs, each with a kinetochore (k), are shown along with a part of another SC. Numbered arrowheads point to RNs that are visible by EM. Four RNs (numbers 2–5) correspond to sites of MLH1 foci (MLH1-positive RNs), whereas two RNs (numbers 1 and 6) do not correspond to MLH1 foci (MLH1-negative RNs). (D) Enlarged view of RNs without the LM overlay. MLH1-positive RNs (numbers 2–5) are larger than MLH1-negative RNs (numbers 1 and 6). (Scale bars: 4 µm for A–C and 250 nm for D.)
Several studies (13, 14) have shown that class II COs are capable of forming functional chiasmata, so we compared diakinesis chiasmata patterns with those predicted from RNs to evaluate whether MHL1-negative RNs in tomato mark crossover sites (Table 1 and SI Appendix, Fig. S4). We found that MLH1-positive RNs alone were not good predictors of chiasma configurations, whereas including both RN types was a good predictor of chiasma configurations. Thus, it is probable that both MLH1-negative and MLH1-positive RNs mark crossover sites, which is consistent with evidence from a number of organisms that (all) RNs give rise to COs (3). Presumably, any recombination intermediates that are resolved by MUS81 as non-COs (6, 29) are not cytologically distinguishable as RNs on mid- to late-pachytene SCs from tomato.
Table 1.
Chiasmata (Xta) configurations observed from diakinesis nuclei compared to chiasmata configurations predicted using all RNs on SCs or only MLH1-positive (MLH1+) RNs on SCs
| Average no. per nucleus (SD) | ||||
| Chiasmata | No. nuclei | Rods* | Rings† | Total Xta‡ |
| Observed | 100 | 7.9 (1.8) | 4.1 (1.7) | 16.0 (1.8) |
| Predicted | ||||
| All RNs | 132§ | 7.6 (2.0) | 4.4 (2.0) | 16.4 (2.0) |
| MLH1+ RNs only | 132§ | 9.3 (1.7) | 2.6 (1.7) | 14.6 (1.8) |
Rods = bivalents with at least one chiasma (one or more RNs) in only one arm. Chromosome 2 was always counted as a rod with one chiasma in the long arm.
Rings = bivalents with at least one chiasma in both chromosome arms.
The average total number of chiasmata observed per diakinesis nucleus is not significantly different from the value predicted using all RNs (P > 0.1, t test) but is significantly different from the value obtained using only MLH1-positive RNs (P < 0.001, t test).
The same 132 nuclei (each a complete set of 12 individually identified SCs) were evaluated for predicted chiasma configurations based on RN patterns.
Once the two RN types were distinguished by immunolabeling, we evaluated their morphology and staining characteristics and observed clear differences. MLH1-positive RNs stained more darkly than MLH1-negative RNs and were significantly larger in both length (parallel to the SC) and width (transverse to the SC; Fig. 1 and SI Appendix, Fig. S5 and Table S4). These differences in RN size and density are likely to indicate differences in protein components (other than MLH1) between the two RN types and are consistent with molecular and genetic studies showing that different proteins are involved in the two CO pathways (6, 7, 30). More speculatively, RN size differences might indicate DNA tract length differences in the two CO pathways.
Distribution of MLH1-Positive and MLH1-Negative RNs.
A common CO pattern observed in many organisms is a high level of COs in distal euchromatin and a low level of COs in pericentric heterochromatin, whether assayed using linkage maps, chiasmata, RNs, or MLH1 foci (e.g., refs. 16, 19, 20, and 31–33). Similarly, we found that 95% of RNs occurred in euchromatin and only 5% of all RNs were located in the roughly 30% of SC length in pericentric heterochromatin (Fig. 2 and SI Appendix, Table S5). We found, however, that the presence of pericentric heterochromatin does not affect both RN types equally because only 3% of all MLH1-positive RNs were located in pericentric heterochromatin compared with nearly 17% of all MLH1-negative RNs. Class II COs predominated over class I COs in heterochromatin in absolute numbers also (90–72, respectively). Possibly, the comparatively higher frequency of MLH1-negative RNs in heterochromatin is related to the high frequency of repetitive sequences (32). Such sequences may be more likely to form unusual and complex recombination intermediates that can be resolved as COs by the MUS81-dependent CO pathway (6, 29, 34, 35). At a more applied level, modifying local recombination frequencies in pericentric regions is an important challenge for plant breeding (e.g., for positional cloning), and knowing that heterochromatic regions are more prone to class II COs may lead to new strategies to promote COs in those regions, e.g., by mutating the FANCM gene that has been shown to specifically suppress class II COs in Arabidopsis (14).
Fig. 2.
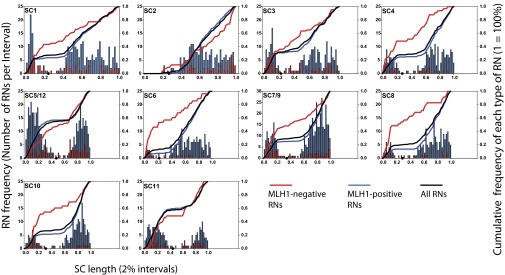
Distributions of MLH1-positive RNs (blue bars) and MLH1-negative RNs (red bars) for all tomato chromosomes and chromosome groups. Histogram bars are in 2% SC length intervals (X-axis) and show the number of RNs observed per interval (left y axis). The red histogram has been superimposed on the blue histogram for each chromosome. To highlight differences between the individual histograms, lines showing the cumulative frequency (right y axis) of all RNs (black), of MLH1-positive RNs (blue), and of MLH1-negative RNs (red) have been added. The length of each chromosome has been scaled to represent its appropriate length relative to the other chromosomes. The short arm of chromosome 2 is entirely heterochromatic, and the distal segment is usually asynapsed and/or broken off (and often lost) in SC spreads, explaining the lack of any RNs in that region. Only MLH1-negative RNs were observed on the retained short arm of chromosome 2. Except for the near-metacentric chromosomes 5/12 and 11, all chromosomes have a higher fraction of MLH1-negative RNs on the short arm and in the pericentric heterochromatin compared with MLH1-positive RNs (SI Appendix, Table S6). Black dots on the X-axis represent centromere positions.
Furthermore, the individual distributions of MLH1-positive RNs and MLH1-negative RNs were significantly different (Kolmogorov–Smirnov P value < 0.04) for all chromosomes except 11. In addition to the differences of RN distributions in heterochromatin, in most tomato chromosomes MLH1-negative RNs were disproportionately observed on the short arms, whereas MLH1-positive RNs were overrepresented on the long arms (Fig. 2 and SI Appendix, Table S6). This difference is particularly striking for chromosome 6 in which 54% of all MLH1-negative RNs were observed in the short arm (23% of chromosome length) compared with only 7% of all MLH1-positive RNs. We observed a significant positive correlation between RN frequency and SC length for MLH1-positive RNs (SI Appendix, Fig. S6; see also ref. 19). For MLH1-negative RNs, we were unable to detect a significant correlation (at the standard 5% level) between RN frequency and SC length. Nevertheless, the P value for the hypothesis of no correlation was low (0.07), suggesting that there might be a positive correlation that simply was not detectable given the low numbers of MLH1-negative RNs in our observations. However, when class I COs were blocked in Arabidopsis msh4 mutants (22), again no correlation between the numbers of residual chiasmata (from class II COs) and chromosome length was detected.
We also investigated whether synaptic initiation patterns correlate with the patterns of the two RN types. In tomato, synapsis usually begins in distal euchromatin, and pericentric heterochromatin synapses last (36). Using chromosome arm-specific DNA probes for markers in distal euchromatin of three chromosomes (9, 10, and 12), we observed that synapsis was initiated in long arms more often than short arms for all three chromosomes (SI Appendix, Table S7 and Fig. S7). Combining the observed distributions of RN types with these synaptic patterns indicates that MLH1-positive RNs are more likely to be in earlier synapsing parts of chromosomes, and MLH1-negative RNs are more likely to be in the later synapsing parts (short arms and pericentric heterochromatin). Assuming that recombination events are spatio-temporally coordinated with DNA replication in tomato as demonstrated for barley (37), one explanation for these RN patterns could be that distal euchromatic regions replicate and initiate recombination first, leading to early CO designation and a high frequency of class I COs. Interference from early class I COs would lead to a higher proportion of class II COs occurring away from the initiation regions (see below for evidence of interaction between the two pathways). Furthermore, any recombination intermediates that form among the repetitive sequences in pericentric heterochromatin may be more likely to have aberrant structures that require MUS81 for resolution (34), leading to a comparatively high frequency of MLH1-negative RNs in heterochromatin.
Interference Characteristics of the Two Pathways.
For all chromosomes, the numbers of MLH1-positive RNs per SC were not Poisson-distributed (P < 10−16), and MLH1-positive RNs showed significant interference (inferred Gamma model parameter ν ∼ 7; SI Appendix, Figs. S8 and S9 and Table S8). These results were expected because MLH1 marks class I COs that have been shown to interfere in animals and birds (27, 28, 38) and in tomato (19). In contrast, the numbers of MLH1-negative RNs per SC closely followed a Poisson distribution (P > 0.6), and we found no significant interference among MLH1-negative RNs for any chromosome (ν ∼ 1; SI Appendix, Fig. S8 and Table S8). These results are consistent with our hypothesis that MLH1-negative RNs mark class II COs because class II COs have been reported to be noninterfering and randomly distributed among chromosomes in mutants defective for the P1 pathway (8, 11, 21, 23). The novelty here is the demonstration that class II COs are not interfering with each other in a wild-type background in which both pathways are intact and this lack of interference extends over the whole genome.
The Two Crossover Pathways Interact.
We next asked whether there is any interaction between the two pathways. First, we tested the hypothesis that the distribution of the numbers of MLH1-negative RNs per SC is the same whether the SCs have one, two, or three MLH1-positive RNs. Pooling all chromosomes together, the P value was less than 10−9, indicating that the number of MLH1-negative RNs per SC is not independent from the number of MLH1-positive RNs on the same SC. This result was also significant for individual chromosomes 3, 5/12, 10, and 11 (Fisher’s exact test, all P values provided in SI Appendix, Table S9). Furthermore, the numbers of MLH1-negative and MLH1-positive RNs were negatively correlated (significant for six chromosomes individually; all P values provided in SI Appendix, Table S10).
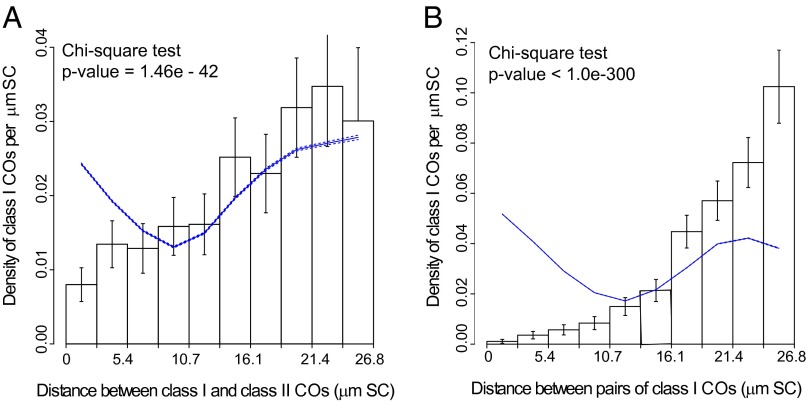
Second, we tested whether SC distances (measured in µm of SC length) between MLH1-positive and MLH1-negative RNs present on the same SC were compatible with no interference. This test takes into account the specific distributions and interference properties of each type of RN. When pooling all chromosomes together, we found a lower-than-expected frequency of MLH1-negative RNs close to MLH1-positive RNs (Fig. 3A), indicating significant interference between the two RN types (P value < 10−41). This result was confirmed for most individual chromosomes and also when genetic distances were used instead of SC distances (SI Appendix, SI Materials and Methods, Figs. S10 and S11, and Table S11).
Fig. 3.
Distribution of distances (in µm SC) between different types of RNs, pooled over all chromosomes. (A) Distribution of distances between MLH1-positive RNs (class I COs) and MLH1-negative RNs (class II COs). (B) Distribution of distances (in µm SC) between pairs of MLH1-positive RNs (class I COs). Histograms represent the experimental data. Vertical bars represent 95% confidence intervals for each bin of the histogram. The expected distribution in the absence of any interaction between class I and class II COs is plotted as a solid line with the associated 95% confidence intervals (dashed lines along the solid line). Interference between class I and class II COs (A) and between class I COs (B) is indicated by the lower than expected frequency of events at short distances. Similar figures for individual chromosomes are shown in SI Appendix, Figs. S10 and S12.
Comparison of Fig. 3A with Fig. 3B (a similar analysis as in Fig. 3A, but between MLH1-positive RNs) shows that interference extends further between pairs of MLH1-positive RNs (up to about 13 µm of SC, SI Appendix, SI Materials and Methods) than between mixed pairs of MLH1-positive RNs and MLH1-negative RNs (up to about 8 µm of SC). This pattern also holds true for all individual chromosomes and for genetic distances (SI Appendix, Figs. S12 and S13 and Table S12). Similar analysis between pairs of MLH1-negative RNs was not informative because of the low number (n = 70) of bivalents with two or more MLH1-negative RNs. The much smaller P values obtained when using SC distances compared with using genetic distances for both class I-class I COs and class I-class II COs are consistent with the hypothesis that interference between COs depends on physical distances rather than genetic distances, although the exact mechanism responsible for interference remains unsolved (2, 39, 40).
Previous work (19, 41) showed that early recombination nodules (ENs, that are believed to correspond to DSBs) have low levels of interference (ν ∼ 2). To investigate whether interference among ENs is sufficient to account for the level of interference between MLH1-positive and MLH1-negative RNs, we compared the physical SC distances over which interference extends in the two cases. For ENs, interference effects become negligible beyond a distance of 0.3 μm of zygotene SC axis (see figure 4 of ref. 41) compared with 8 µm distance between MLH1-positive and MLH1-negative RNs. Because the estimated physical lengths of zygotene and pachytene bivalents is similar (25 and 20 µm SC length, respectively; SI Appendix, Table S1; ref. 41), it is likely that interference between precursors would be negligible beyond 0.3 μm in pachytene chromosomes also, allowing us to conclude that precursor (DSB) interference is not sufficient to explain the interaction between interfering and noninterfering pathways.
The question of interference between class I and class II COs had not been amenable to investigation before our study because it was not possible to reliably specify individual class I and class II COs on the same chromosome. Our combined LM and EM method overcomes this hurdle to explicitly demonstrate interaction between the two CO pathways. Previously, it had been speculated that class I and class II COs might interfere because of a signal produced by class I COs to which class II COs would be sensitive (19, 42). Although we cannot confirm that such a signal exists, our results demonstrate that interpathway interference does occur, and that the physical extent of this interference is much greater than that observed between early RNs. Furthermore, we found that synapsis tends to occur first in the long arm of chromosomes. A plausible scenario linking these observations is that the first CO (presumably the obligatory one) is produced by pathway P1 and arises where synapsis initiates, predominantly on the long arm. Subsequently, interference resulting from the P1 CO would affect both class I and class II COs and lead to enrichment of class II COs on the short arms.
Given that interaction between the two CO pathways is inhibitory (the numbers of COs in each pathway are negatively correlated and there is interference between nearby COs), it seems likely that some kind of balance exists between the pathways that is probably related to the drive to maintain total CO levels per nucleus by CO homeostasis (43, 44). However, mutants in the P1 pathway do not appear to have any compensatory increase in class II COs and vice-versa (8). One exception to this generalization was reported for mus81−/− male mice in which elimination of class II COs resulted in a significant increase in the number of MLH1 foci (but not of chiasmata) per nucleus (12). Future experiments using our combined LM and EM method with mutants in P1 and/or P2 CO pathways should help to address this apparent contradiction and elucidate any balancing interactions between the two pathways.
Overall, simultaneous mapping of MLH1-positive RNs (class I COs) and MLH1-negative RNs (class II COs) has provided access, to our knowledge for the first time, to the characteristics of the separate CO formation pathways in a wild-type organism. This breakthrough reveals that (1) distribution patterns of class I and class II COs differ along chromosomes, (2) class II COs do not interfere with each other, and (3) there is interference between class I and class II COs.
Materials and Methods
Additional details of experimental and statistical procedures are provided in SI Appendix, SI Materials and Methods.
SC Spreading and Immunolabeling.
Primary microsporocytes in pachytene from the highly inbred cherry tomato line (LA4444) were used to prepare SC spreads on 0.6% Falcon plastic-coated slides as described in refs. 45 and 46. SC spreads were treated with DNase I, then immunolabeled with affinity-purified chicken anti-SlSMC1 diluted 1:50 and affinity-purified rabbit anti-SlMLH1 diluted 1:50 followed by goat anti-chicken Dylight 649 and goat anti-rabbit AlexaFluor 488, both from Jackson Labs and diluted 1:500 (19, 45–47). Fluorescence microscopy was performed using a 100× Plan-Apo objective with an adjustable iris and a Leica DM5000 microscope equipped for both phase contrast and fluorescence microscopy with narrow band-pass FITC and TRITC filter cubes and zero pixel shift.
Fluorescence LM and EM.
Red and green signals for each spread were captured individually using a cooled Hamamatsu monochrome 1,344 × 1,044 pixel camera and IP Lab software (ver 4). Because some MLH1 foci were quite small and dim, we used long exposure times to be sure that even dim foci would be imaged. Images for each spread were artificially colored using IPLab and merged using Photoshop CS2. After LM images were captured, the cover glass on each slide was removed carefully, and slides were stained with phosphotungstic acid (46). Plastic was lifted from each slide onto grids, and SC spreads previously imaged by fluorescence were photographed at a magnification of 3,000× (generally requiring three to six images each) using a JEOL 2000 EM.
LM–EM Image Analysis.
EM negatives were scanned at 800 dpi using an Epson Perfection V700 Photo scanner. A montage of each SC spread was assembled using Adobe Photoshop CS2, and RNs were identified (18). The corresponding fluorescent image was then layered over the EM montage, and each SC was analyzed individually by precisely aligning the red (SC) and green (MLH1) combined fluorescent image over the EM image of the same SC. Then, the MLH1 fluorescent signal at each previously identified RN position was assessed. Each “unlabeled” RN was then more carefully evaluated using only the green channel (instead of the red and green combined image), after additional temporary enhancement of the green signal. If no green signal was observed under these conditions, the RN was marked as an MLH1-negative RN. Only informative SC spreads in which each chromosome had at least one (obligate) MLH1 focus were selected for measurement using MicroMeasure 3.0 and subsequent analysis. In some cases, one or more SCs in a set had to be excluded due to a lack of distinct kinetochores or to the presence of stain precipitate at the EM level that partially obscured SCs. SCs from these groups were used for counting the number of RNs per SC set but were not used for mapping RN positions.
Genetic Coordinates and Local Recombination Rates Along Chromosomes.
SC coordinates of COs (MLH1 foci and/or RNs) and the kinetochore were constructed from the curvilinear distance. Given the SC positions of all COs, the genetic coordinate of an arbitrary point is defined as half the mean number of COs detected between the left end of the SC and the point of interest, and local recombination rates were measured in centiMorgan (cM) per µm along the SC by defining uniform intervals and taking 50 times the mean number of COs per µm in that interval.
Interference Strength Inferred Using the Gamma Model.
The Gamma model (48) describes CO formation and CO interference in a pathway. Its parameter ν quantifies the strength of interference: Absence of interference corresponds to the value ν = 1 and increasing interference corresponds to increasing ν > 1. For any realization of COs on an SC, it is possible to calculate its likelihood within the Gamma model assuming a value of ν (49); this allows us to use the maximum likelihood method to determine for each chromosome its “optimal” ν, i.e., that which best fits the experimental data (50). Note that even SCs with only zero or one RN contribute information to the Maximum Likelihood estimation of ν. Confidence intervals for the fitted ν's are computed based on the Fisher information matrix using CODA software (50).
Tests of No Cross-Talk Between the Two CO Formation Pathways.
Our first test is based on the numbers of MLH1-positive and MLH1-negative RNs on each SC and applying a Fisher’s exact test on the frequencies of these pairs of numbers. To perform this test on data pooled over all chromosomes, we wrote an implementation of Fisher’s exact test in which we compute the log-likelihood of the data by summing log-likelihoods over all chromosomes. We then obtain the P value of the test by comparing this log-likelihood with 109 log-likelihood values obtained after reshuffling the list of MLH1-negative RNs among all SCs to remove any correlation between MLH1-negative and MLH1-positive RNs while keeping the exact same distribution of numbers of MLH1-positive RNs and of MLH1-negative RNs. We checked this procedure by ensuring that for individual chromosomes, we obtained the same P values as when using the fisher.test() function in R.
Our second test is based on distances between MLH1-positive and MLH1-negative RNs. We test for independence of the two pathways by comparing the distributions of those distances when there is and when there is not shuffling. The χ2 [function chisq.test() in R] test applied to the histograms representing these distributions was used to produce P values.
Test of no P1-P1 Interference Based on Inter-CO Distances.
In this case, the observed distribution of distances between each class I CO and all other class I COs on the same SC was compared with the distribution expected in the absence of interactions, which was obtained again by a shuffling procedure. Specifically, while keeping the same number of MLH1-positive RNs for each SC, we shuffled their positions 1,000 times across the whole data set.
Supplementary Material
Acknowledgments
We thank Franck Gauthier for his help in data analyses and Christine Mézard, Raphaël Mercier, Stephen Stack, and Denise Zickler for helpful comments on the manuscript. This work was supported by National Science Foundation Grant MCB-1019708 (to L.K.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406846111/-/DCSupplemental.
References
- 1.Jones GH, Franklin FCH. Meiotic crossing-over: Obligation and interference. Cell. 2006;126(2):246–248. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Berchowitz LE, Copenhaver GP. Genetic interference: Don’t stand so close to me. Curr Genomics. 2010;11(2):91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zickler D, Kleckner N. Meiotic chromosomes: Integrating structure and function. Annu Rev Genet. 1999;33:603–754. doi: 10.1146/annurev.genet.33.1.603. [DOI] [PubMed] [Google Scholar]
- 4.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 5.Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: Generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18(2):117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149(2):334–347. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Muyt A, et al. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell. 2012;46(1):43–53. doi: 10.1016/j.molcel.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FC. Pathways to meiotic recombination in Arabidopsis thaliana. New Phytol. 2011;190(3):523–544. doi: 10.1111/j.1469-8137.2011.03665.x. [DOI] [PubMed] [Google Scholar]
- 9.Ehmsen KT, Heyer WD. Biochemistry of meiotic recombination: Formation, processing, and resolution of recombination intermediates. In: Egel R, Lankenau D-H, editors. Recombination and Meiosis. Berlin, Heidelberg: Springer-Verlag; 2008. pp. 91–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lynn A, Soucek R, Börner GV. ZMM proteins during meiosis: Crossover artists at work. Chromosome Res. 2007;15(5):591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 11.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of AtMUS81 in interference-insensitive crossovers in A. thaliana. PLoS Genet. 2007;3(8):e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4(9):e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgins JD, Buckling EF, Franklin FCH, Jones GH. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54(1):152–162. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- 14.Crismani W, et al. FANCM limits meiotic crossovers. Science. 2012;336(6088):1588–1590. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 15.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–558. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 16.Sherman JD, Stack SM. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High-resolution recombination nodule map for tomato (Lycopersicon esculentum) Genetics. 1995;141(2):683–708. doi: 10.1093/genetics/141.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165(4):2283–2287. doi: 10.1093/genetics/165.4.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson LK, Stack SM. Recombination nodules in plants. Cytogenet Genome Res. 2005;109(1-3):198–204. doi: 10.1159/000082400. [DOI] [PubMed] [Google Scholar]
- 19.Lhuissier FGP, Offenberg HH, Wittich PE, Vischer NO, Heyting C. The mismatch repair protein MLH1 marks a subset of strongly interfering crossovers in tomato. Plant Cell. 2007;19(3):862–876. doi: 10.1105/tpc.106.049106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froenicke L, Anderson LK, Wienberg J, Ashley T. Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet. 2002;71(6):1353–1368. doi: 10.1086/344714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de los Santos T, et al. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164(1):81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higgins JD, Armstrong SJ, Franklin FCH, Jones GH. The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: Evidence for two classes of recombination in Arabidopsis. Genes Dev. 2004;18(20):2557–2570. doi: 10.1101/gad.317504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chelysheva L, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8:e1002799. doi: 10.1371/journal.pgen.1002799. -doi:10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Copenhaver GP, Housworth EA, Stahl FW. Crossover interference in Arabidopsis. Genetics. 2002;160(4):1631–1639. doi: 10.1093/genetics/160.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz EK, et al. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol Cell Biol. 2012;32(15):3065–3080. doi: 10.1128/MCB.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argueso JL, et al. Systematic mutagenesis of the Saccharomyces cerevisiae MLH1 gene reveals distinct roles for Mlh1p in meiotic crossing over and in vegetative and meiotic mismatch repair. Mol Cell Biol. 2003;23(3):873–886. doi: 10.1128/MCB.23.3.873-886.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pigozzi MI. Distribution of MLH1 foci on the synaptonemal complexes of chicken oocytes. Cytogenet Cell Genet. 2001;95(3-4):129–133. doi: 10.1159/000059334. [DOI] [PubMed] [Google Scholar]
- 28.Basheva EA, Bidau CJ, Borodin PM. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res. 2008;16(5):709–719. doi: 10.1007/s10577-008-1221-y. [DOI] [PubMed] [Google Scholar]
- 29.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31(3):313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120(2):109–127. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giraut L, et al. Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet. 2011;7(11):e1002354. doi: 10.1371/journal.pgen.1002354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485(7400):635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer E, et al. Intraspecific variation of recombination rate in maize. Genome Biol. 2013;14(9):R103. doi: 10.1186/gb-2013-14-9-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ helicase, Sgs1, and XPF family endonuclease, Mus81-Mms4, resolve aberrant joint molecules during meiotic recombination. Mol Cell. 2008;31(3):324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20(4):563–573. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 36.Stack SM, Anderson LK. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. II. Synapsis in Lycopersicon esculentum (tomato) Am J Bot. 1986;73:264–281. [Google Scholar]
- 37.Higgins JD, et al. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell. 2012;24(10):4096–4109. doi: 10.1105/tpc.112.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falque M, Mercier R, Mézard C, de Vienne D, Martin OC. Patterns of recombination and MLH1 foci density along mouse chromosomes: Modeling effects of interference and obligate chiasma. Genetics. 2007;176(3):1453–1467. doi: 10.1534/genetics.106.070235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleckner N, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101(34):12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petkov PM, Broman KW, Szatkiewicz JP, Paigen K. Crossover interference underlies sex differences in recombination rates. Trends Genet. 2007;23(11):539–542. doi: 10.1016/j.tig.2007.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Anderson LK, Hooker KD, Stack SM. The distribution of early recombination nodules on zygotene bivalents from plants. Genetics. 2001;159(3):1259–1269. doi: 10.1093/genetics/159.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116(6):795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 43.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126(2):285–295. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cole F, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14(4):424–430. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stack SM, Anderson LK. Electron microscopic immunogold localization of recombination-related proteins in spreads of synaptonemal complexes from tomato microsporocytes. In: Keeney S, editor. Meiosis: Cytological Methods. Vol 2. Totowa, NJ: Humana; 2009. pp. 147–169. [DOI] [PubMed] [Google Scholar]
- 46.Anderson LK, Stack SM. Preparing SC spreads with RNs for EM analysis. In: Pawlowski WP, Grelon M, Armstrong S, editors. Plant Meiosis: Methods and Protocols. New York: Spring Science + Business Media; 2013. pp. 147–158. [Google Scholar]
- 47.Lohmiller LD, et al. Cytological analysis of MRE11 protein during early meiotic prophase I in Arabidopsis and tomato. Chromosoma. 2008;117(3):277–288. doi: 10.1007/s00412-007-0147-z. [DOI] [PubMed] [Google Scholar]
- 48.McPeek MS, Speed TP. Modeling interference in genetic recombination. Genetics. 1995;139(2):1031–1044. doi: 10.1093/genetics/139.2.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Broman KW, Weber JL. Characterization of human crossover interference. Am J Hum Genet. 2000;66(6):1911–1926. doi: 10.1086/302923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gauthier F, Martin OC, Falque M. CODA (crossover distribution analyzer): Quantitative characterization of crossover position patterns along chromosomes. BMC Bioinformatics. 2011;12:27. doi: 10.1186/1471-2105-12-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.